Abstract
After oral intake of drugs, drugs go through the first pass metabolism in the gut and the liver, which greatly affects the final outcome of the drugs’ efficacy and side effects. The first pass metabolism is a complex process involving the gut and the liver tissue, with transport and reaction occurring simultaneously at various locations, which makes it difficult to be reproduced in vitro with conventional cell culture systems. In an effort to tackle this challenge, here we have developed a microfluidic gut-liver chip that can reproduce the dynamics of the first pass metabolism. The microfluidic chip consists of two separate layers for gut epithelial cells (Caco-2) and the liver cells (HepG2), and is designed so that drugs go through a sequential absorption in the gut chamber and metabolic reaction in the liver chamber. We fabricated the chip and showed that the two different cell lines can be successfully co-cultured on chip. When the two cells are cultured on chip, changes in the physiological function of Caco-2 and HepG2 cells were noted. The cytochrome P450 metabolic activity of both cells were significantly enhanced, and the absorptive property of Caco-2 cells on chip also changed in response to the presence of flow. Finally, first pass metabolism of a flavonoid, apigenin, was evaluated as a model compound, and co-culture of gut and liver cells on chip resulted in a metabolic profile that is closer to the reported profile than a monoculture of gut cells. This microfluidic gut-liver chip can potentially be a useful platform to study the complex first pass metabolism of drugs in vitro.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Drug development process is a lengthy, costly process with an extremely low probability of success. It is thought to cost up to a billion dollar, and 10 to 12 years to develop one drug (Kaitin 2010). Despite this large amount of resources required, majority of drug candidates are opted out due to unforeseen side effects or lack of efficacy (Hughes 2009). In vitro cell-based models are widely used to predict the efficacy and toxicity of candidate molecules, especially in the early phases of drug development process (Strovel et al. 2004). The major drawback of in vitro model systems is that when cells are cultured in vitro, they often behave and response to drugs differently from when they were in the native tissue. It is often thought that this is because cells are not provided with various physiological cues that are present in the in vivo tissues, for example, 3D tissue structure (Abbott 2003). This makes it difficult to extrapolate the results from in vitro experiment to animals and humans. Animal models have their own limitations too, for example they often show different metabolic profiles from humans (Bhatia and Ingber 2014), let alone the ethical issues of sacrificing animals. Another limitation of in vitro model systems is that they are not able to reproduce organ-organ interactions, as only cells of a single tissue origin are cultured in a petri dish. One of the main examples of organ interactions in the body is the first pass metabolism, as drugs go through complex process of absorption and metabolism in the gut and the liver.
Organ-on-a-chip technology can be an alternative to the current cell-based models and animal models, as they can potentially overcome the aforementioned limitations. Organ-on-a-chip aims to reconstitute in vivo tissue environment for cells by mimicking the microscale tissue architecture and cell-cell, cell-matrix interactions (Chan et al. 2013). While most of organ-on-a-chip systems have focused on recapitulating the tissue functions of a specific organ, another approach focuses on the ability of organ-on-a-chip systems to reproduce the interactions between multiple organs. Often termed as human-on-a-chip, these systems aim to connect multiple tissue functions as observed in the human body (Sung et al. 2013).
One of the most important processes that determines the pharmacokinetics of drugs is the first pass metabolism, which consists of the absorption in the gut and the metabolism in the liver. Due to its importance in drug’s action in the body, a large amount of effort has been directed to improving current gut and liver models. The gut affects the fate of a drug by selectively absorbing drugs depending on their chemical and physical properties. While several in vitro gut absorption model is available, it has been thought that these model systems carry limitations in fully reproducing the human gut absorption (Artursson et al. 2001), and several attempts have been made to improve currently existing gut models. For example, co-culturing gut epithelial cells with other supporting cells (Hilgendorf et al. 2000), providing mechanical stimuli that are similar to what gut cells would experience in the native gut tissue (Kim et al. 2012), and mimicking the 3D topology of the intestine tissue (Wang et al. 2009) have been reported to modify the physiology of cultured cells compared to the conventional monolayer culture. Culturing gut cells in a fluidic environment has also been shown to improve the gut-related functions (Chi et al. 2015).
After drugs are absorbed in the gut, they are transported to the liver for subsequent metabolism. Although metabolism also occurs in the gut tissue to some extent by the epithelial cells and gut microbes, the liver is the main organ that is responsible for most of the metabolic conversion (Carlson and Fisher 2008). Diverse liver model systems exist, with varying degrees of complexity and physiological relevance. One of the simpler systems utilize metabolic enzyme fractions extracted from the liver tissue (Brandon et al. 2003). Although this extracted enzyme fractions are simple to use, and known to be rich in cytochrome P450 enzymes, which are the main metabolic enzyme family in the liver, distortion in the ratios of different metabolic enzymes can cause deviations from the human liver metabolism (Spaggiari et al. 2014). On the other end of the spectrum lies the liver tissue slices. Such tissue slices offer realistic estimate of human liver metabolism, but is difficult to use due to its short lifetime and scarcity of available tissues (Brandon et al. 2003). Attempts to improve liver model systems by manipulating the cellular environment have been reported, for example mimicking the hepatic blood flow, oxygen concentration gradient, or co-culturing hepatic cells with other cells (Allen et al. 2005; Prot et al. 2011).
While we have seen considerable achievement in improving the gut and the liver model systems, limitations still exist, because the first pass metabolism always occurs as a connected system of the gut and the liver. Not only drugs go through a sequence of absorption followed by metabolism, there also exists a cross-talk between the gut and the liver tissue via signaling molecules (van Midwoud et al. 2010). This raises the issue that any separate gut or liver model systems do not properly reflect the process that drugs go through in the body. A simple approach to this issue is to co-culture gut and liver cells in the same compartment (Choi et al. 2004). While this approach offers a simple solution, it does not fully capture the essence of the first pass metabolism, which is a time-dependent, dynamic crosstalk between two separate organs. The dynamic connection between different organs can play an important role and affect the final effect of drugs. For example, our previous study suggested that the flow-based connection of two organs results in a pharmacokinetic profile that is different from what would be expected from a traditional well-based system, resulting in different cell response to durgs (Lee et al. 2016). This suggests that a more advanced system is needed, in which gut and liver cells can be cultured separately while efficient cross-talk between the two organs is still possible, just as the gut and the liver in the human body. Several studies reported a microfluidic system to this end, where gut and liver compartment exist in separate spaces and connected via microfluidic channels (Imura et al. 2010). The most pioneering works for reproducing the sequential action of the gut and the liver were achieved by Shuler et al., who developed several different versions of ‘microscale cell culture analog’, where different cells are cultured in separate chambers on the same chip, which are connected via fluidic channels mimicking the blood circulation (Sung and Shuler 2009; Sung et al. 2010; Sung et al. 2013). This type of microfluidic chip enables interaction of different cells via signaling molecules traveling through the channels, and the desired type of interaction can be designed by arranging the chambers and channels. Often termed as ‘body-on-a-chip’, this device has been used as a gut-liver model to study the effect of nanoparticles on liver injury (Esch et al. 2014).
While these microfluidic gut-liver models are promising new tools for studying the first pass metabolism of drugs with improved physiological relevance than previous gut and liver systems, several limitations exist. First, reported cell incubation time in the chips was generally short. Gut cells were cultured for three days in the paper reported by Imura et al., and gut and liver slices were incubated for 1 day in the study by Groothuis et al. (Imura et al. 2010; van Midwoud et al. 2010). This can be a potential problem since often cells require a long incubation time of several weeks to properly differentiate (Artursson et al. 2001). Another potential limitation of previously reported systems is the complexity and difficulty of using the devices, which is partially an inherent problem of microfluidic systems requiring pumps and fluidic connections. This not only increases the risk of failure, for example fluid leaking or bacterial contamination, but also makes it difficult to perform high-throughput experiment. Thirdly, although a few proof-of-concept devices have been developed (Imura et al. 2010; van Midwoud et al. 2010; Imura et al. 2012; Esch et al. 2014), an attempt to compare the first pass metabolism of model drugs with that reported from in vivo experiment has been lacking.
Here, we report a novel, microfluidic gut-liver chip for reproducing the first pass metabolism. A two-layer design allows culturing gut and liver cells in separate compartments but in near vicinity, thus enabling direct and efficient crosstalk between the two. The chamber dimensions were further optimized to enable efficient transport by mathematical model simulation. We eliminated the need for a pump by utilizing a gravity-based flow, and the chips can be operated simply by placing them on a custom-made tilting stage. It was verified that gut (Caco-2) and liver (HepG2) cells can be cultured in the chip for several weeks while maintaining good viability and morphology, and the metabolic activity of both cells improved when cultured in the chip, compared to a monolayer culture in 96-well microplate. As a model molecule, flavonoid apigenin was administered to the gut-liver chip and the metabolites were analyzed to verify that the first pass metabolism can be successfully reproduced in the chip.
2 Materials and methods
2.1 Fabrication of gut-liver chip
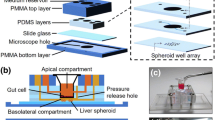
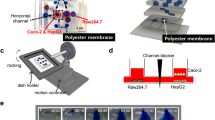
Soft-lithography was used to fabricate the gut-liver chip. SU-8 50 was spin-coated on a silicon wafer at a thickness of 120 μm to develop into microchannels that are 200 μm wide. The fabricated wafer was coated with Trichloro(1H,1H,2H,2H–perfluorooctyl)silane(448,931, Sigma Aldrich) to enable easier removal of Polydimethylsiloxane(PDMS) later. To fabricate the gut layer, PDMS perpolymer solution was poured onto a SU-8 master and cured for 2 h at 60 °C . For the liver layer, PDMS prepolymer solution was spin-coated on a master and cured to make a thin layer of PDMS. The same SU-8 master was used for the gut and the liver layer, as the shape of the fluidic channels were identical in both layers. When the two layers were bonded, channels were facing up in the gut layer, whereas channels were facing down (touching the slide glass bottom), and the two channels were aligned so that they cross in opposing directions (See Fig. 1 for more details).
Cell culture chambers for gut and liver cells and media reservoirs were made by punching holes with Biopsy punch (Miltex) with appropriate diameters. Four medium reservoirs were made, inlet and outlet reservoirs for upper (gut) and lower (liver) layers, respectively. Chips were assembled by bonding each layer on top of a slide glass, in the order of the liver layer, membrane, and the gut layer from the bottom. The porous membrane provides a support for Caco-2 cells in the top layer. The membrane is a polyester membrane with 0.4 μm pore size, which was manually cut out from 6-well plate insert (Corning CLS3450). Additional PDMS layer was bonded on top to provide sufficient volume for holding media in the reservoir. The gut chamber is sealed with a custom-made PDMS lid, which has a small opening for cell seeding. Assembled chips were sterilized by autoclave at 120 °C for 5 min before cell seeding. Fluidic channels were first wetted by injecting ethanol, and washed with Dulbecco’s Phosphate-Buffered Solution (DPBS) extensively to remove remaining ethanol. Finally, channels were filled with cell culture media.
2.2 Cell seeding and culture
Caco-2, human epithelial colorectal adenocarcinoma cell line, was used as gut cells, and HepG2, human hepatocellular carcinoma cell line was used as liver cells. For Caco-2 cells, 1.6 × 105 cells in 80 μL media were seeded into the gut chamber. For HepG2 cells, 5.5 × 104 cells were injected at 3.6 × 106 cell/mL density. After seeding cells, sterilized filter paper was wetted with sterile water and left in a dish containing the chips to minimize evaporation. Since Caco-2 cells require longer culture time to differentiate, they were seeded first. Cells were left in the chip for 3 days without flow to allow firm attachment, and then cultured under flow condition for 7 days. Before seeding HepG2 cells, cell culture chamber was coated with fibronectin at 5 μg/mL for 1 h to enhance cell attachment to the glass surface. HepG2 cells were seeded and incubated without flow for 24 h, and then cultured under flow condition for additional 2 days. The recommended media for each cell type was used when cultured alone (Low glucose DMEM (11885–084,Gibco) for Caco-2, High glucose DMEM (11995–065,Gibco) for HepG2). When co-cultured in the chip, High glucose DMEM was used for both cells. Gravity-driven flow was supplied by placing the chips on a tilting mechanical stage, which is controlled by a custom-built computer program (Fig. 1c). The default parameter was 0.1 degrees/s, 4 degrees, and 500 s.
2.3 Cell assays
Cells were washed with DPBS to eliminate remaining media. Calcein-AM (Life technologies, 1,235,336) was used to stain live cells and ethidium homodimer-1 was used to stain dead cells. Cells were stained for 1 h and images were taken with a confocal microscope. For immunostaining, chips were dissembled, washed with DPBS, and fixed with 4% (w/v) formaldehyde in DPBS for 1 h. After fixing, cells were washed with 0.1% BSA (Gibco) solution for 20 min. To facilitate diffusion through cell membrane, cells were incubated in 0.3% Triton-X 100 (Junsei), and washed with 0.1% BSA solution. For actin staining, cells were incubated in 1/200X Rhodamine phalloidin (Sigma Aldrich) for 1 h. After washing with DPBS for 10 min, nucleic acid was stained with 1/5000X Trihydrochloride trihydrate(DAPI) (Thermo scientific) solution for 30 min. After washing with DPBS for five minutes, cells were imaged under confocal microscope.
For P450 enzyme activity assay, BOMCC substrate (Life technologies) was diluted at 10 μM in Low glucose DMEM, which was inserted into the chip containing cells. Chips were operated under flow condition for 1 h for the reaction to take place. A 100 μL sample was taken from the reservoir and fluorescence intensity was measured using microplate reader with excitation wavelength of 460 nm and emission wavelength of 495 nm. Then chips were dissembled and the membrane with cells attached was immersed in Trypsin solution for 10 min. After cells detached, cell number was counted. Statistical significance was tested by student-t method, and the p-values are reported in the figure captions.
Permeability of intestinal Caco-2 cell layer was measured using model fluorescent molecule, fluorescein (Sigma Aldrich). Caco-2 cells were washed with HBSS buffer. After removing the lid that covered the gut chamber, the gut chamber and the media reservoir was filled with 5 μM fluorescein solution. The liver layer was filled with HBSS buffer. The lid was closed and the flow was initiated. The sample was taken from the liver layer at each time point, and the fluorescence intensity was measured with excitation wavelength of 490 nm and emission wavelength of 514 nm.
2.4 Metabolite analysis
To minimize the loss of apigenin, the inner surface of the PDMS chip was coated with BSA 1% (w/v), containing 50 μM apigenin in High glucose DMEM for 3 days. Then Caco-2 cells were seeded into the gut chamber and cultured for 7 days. The liver layer was coated with fibronectin solution (5 μg/mL) for 1 h and HepG2 cells were seeded. After HepG2 cells were seeded, media was switched to DMEM without phenol red with glucose (4.5 g/L, SH30284.01, Hyclone). When cells are ready, 50 μM apigenin in DMEM without phenol red was added to the gut layer and reservoirs. After 6 h and 24 h, 60 μL of media was taken from the gut and the liver layer, which was mixed immediately with 90 μL methanol to stop reaction.
2.5 Simulation of transport in the chip
A mathematical model to describe the transport inside the chip was developed using COMSOL Multiphysics, in a similar manner to our previous publication, using the same set of parameters for fluid properties (Kim et al. 2013).
3 Results and discussion
3.1 Design of the gut-liver chip
The main criteria for designing the gut-liver chip is that, 1) gut and liver cells can be easily seeded and co-cultured in the chip, 2) gut and liver cells can be cultured in separate compartments while allowing efficient transport between the two, 3) the dimensions of the cell culture chamber and reservoir should be sufficient that allow analysis of cells and metabolites in the media, 4) multiple chips should be operated easily and consistently. We designed our gut-liver chip with careful consideration to meet these criteria (Fig. 1). To allow easy cell seeding, we fabricated a PDMS lid that snug-fits into the open gut chamber. This enables precise and uniform seeding of the gut cells onto the membrane, which is important especially for gut cells since they need to form a homogeneous coverage of membrane to allow consistent transport across epithelial barrier. The presence of a hollow needle in the lid was necessary to allow small amount of media to fill up the needle and remove excess fluid pressure on the gut cells when closing the lid. Locating gut and liver cells in near vicinity is also important, since any molecules that passes gut cells should reach liver cells with efficiency. We decided that a two-layer design where gut cells are cultured on top of the liver cells, separated by a membrane would be the most suitable design to meet this criterion. To allow easy detection and analysis of the circulating metabolites, cell culture chambers were designed to be in a similar size to conventional 96-well microplate. An important factor is the volume of media circulating inside the chip. We wanted the volume to be at least 200 μL, to allow HPLC analysis of metabolites at multiple time points. Finally, chips should be operated in a relatively straightforward manner, and the need of a pump and connecting tubes would have added too much complexity to our system. The use of gravity-flow significantly enhanced consistency in our experiment.
A mathematical model of the transport inside the chip was developed to aid the design of the chip. The main concern was whether the chamber and channel dimensions would allow efficient transport inside the chip, and not suffer from any artificial transport limitation caused by poorly designed chip configurations. Simulation results suggested that the major factor that determines the transport efficiency between the gut and the liver chamber was the height of the gut chamber. We tested various gut chamber height ranging from 0.5 mm to 3 mm, and found that height that is greater than 1.5 mm caused significant limitation in transport from the gut layer channel to the membrane between the gut and the liver chamber (Fig. 2). Based on this simulation result, we designed the height of the gut chamber to be 1.5 mm.
The fluid velocity inside the chip was determined by simple calculation (Sung et al. 2010), which was verified by experimental measurement. Tilting at 10 degrees resulted in the flow rate of 240 μL/h, and 5 degrees resulted in 120 μL/h, which was roughly in consistency with calculations. Several different flow rates were tested. The main concern was to determine appropriate flow rate that would apply enough stimulus to the cells and allow efficient transport, while not applying excessive mechanical stimulus to cause damage to the cells. When the flow rate of 360 μL/h with the 15 tilting degrees was tested, we observed some detachment of HepG2 cells in some areas due to a high flow rate. We speculated that the high flow rate resulted in an excessively high shear rate in some areas, causing cells to detach. Also there is a report that a high flow rate can cause detrimental effect on the cell’s physiology (Vinci et al. 2011), and Kim et al. did not observe any detrimental effect on cells at the flow rate of 100 μL/h. Based on our observation and literatures, we determined the flow rate in the gut layer to be 96 μL/h. The liver layer experienced significantly slower flow rate, which is about one third of the flow rate in the gut layer, due to a difference in channel dimensions leading to the liver chamber. The main reason for this difference comes from the fact that the liver layer resides beneath the gut layer, making the liver channels longer than the gut channels. Longer channels resulted in higher fluidic resistance and hence slower flow rate in the liver layer.
The main parameter that determines the mechanical stimulus given to the cells by the fluid flow is the shear stress. The following equation was used for calculation, where w is the width, h is the height of the channel, and η is the viscosity, and Q is the volumetric flow rate (Young and Simmons 2010).
At given flow rate and channel dimension, the shear stress at the channel wall was calculated to be 7.89 × 10−5 (dyne/cm2). To compare this value with the actual shear stress in the human gut, we made a number of assumptions. Reported length of the small intestine is 7 m, and an average diameter of 2.75 cm was used (Helander and Fandriks 2014). The flow rate inside the small intestine was calculated using the average time it takes for ingested food to pass through the small intestine, which gave the shear stress inside the small intestine to be approximately 2.21 × 10−7 (dyne/cm2). This calculation suggests that the shear stress in our chip is significantly higher than that in the human gut, but local and temporary shear stress in the human intestine is probably higher, due to the peristaltic movement of the intestine. Also, this value is still closer to the human condition when compared with gut chip reported by other research groups. For example, Kim et al. developed a gut chip and reported a shear stress of 0.2 (dyne/cm2), which is significantly higher than ours, (7.89 × 10−5 (dyne/cm2)) (Kim et al. 2012).
One disadvantage of using gravity-flow in a microfluidic device is that it is difficult to accurately control the flow rate. To overcome this shortcoming, we also tested tube connections to a pump by inserting additional PDMS layer that can hold a needle. By inserting a needle into the additional layer and fixing it with a glue, the gut-liver chip could be connected to a pump for more precise fluid supply. Although in our study we mainly used gravity-flow due to its simplicity, we verified that in case where accurate flow control is necessary it is possible to switch the flow mode. Using gravity-flow mode, it was possible to operate the chip for up to four weeks without any noticeable problems. Chips could probably be operated even longer as long as cells remain viable.
3.2 Cell activity assays
To verify that cells were able to proliferate and differentiate in the gut-liver chip, we performed several assays. First, live-dead assay was done to visualize viable cells in the chip, using Calcein-AM and ethidium homodimer-1. This was done to verify that the flow condition in the chip was not causing significant damage to the cells in the chip. As shown in Fig. 3a, d, HepG2 and Caco-2 cells both maintained good viability over the culture period in the chip. As mentioned earlier, higher flow rate often resulted in partial cell detachment or losing viability (data not shown).
Secondly, we visualized actin filament and nucleus by staining cells with Phalloidin and DAPI, respectively. F-actin is a major protein in eukaryotic cells that forms cytoskeleton and abundant near cell membrane, and is frequently used to evaluate morphology of cultured cells (Ferruzza et al. 2012). DAPI binds to AT sequence of chromosomal DNA and frequently used to visualize the location of cell nucleus. For both HepG2 (Fig. 3b, c) and Caco-2 cell (Fig. 3e, f), actin and nucleus was clearly visualized. Formation of uniform monolayer was confirmed for both cell lines. Caco-2 cells showed relatively tighter attachment compared to HepG2 cells, probably due to a longer incubation time. Caco-2 cells are known to become polarized and form tight junctions when differentiated (Artursson et al. 2001).
The metabolic enzyme activity of both Caco-2 and HepG2 cells were evaluated by using a fluorogenic substrate. We chose P450 3A4 as a marker of metabolic activity, since P450 3A4 is known to be expressed both in the gut and the liver. P450 enzyme family is generally expressed in the liver tissue, and several sub-family members such as 1A1, 1A2, and 3A4 exist (Sivaraman et al. 2005). Among these members, 3A4 is known to be expressed in the intestine as well as the liver (Galetin and Houston 2006). A fluorogenic substrate, 7-benzyloxy-methyloxy-3-cyanocoumarin(Vivid BOMCC), reacts with P450 3A4 and generates fluorescence. By measuring the time-dependent intensity of fluorescence, one can quantify the activity of P450 3A4 enzyme (Moutinho et al. 2012).
We compared the P450 3A4 activity of Caco-2 cells cultured in static condition (microwell plate) with that of Caco-2 cells cultured in the gut-liver chip. For this experiment, Caco-2 cells were cultured alone without HepG2 cells. Figure 4a shows that the enzyme activity is enhanced more than two fold when Caco-2 cells are cultured in flow condition. In static condition, P450 activity of Caco-2 cells were measured at 3.14 μM/1010 cells/h, whereas in flow condition, it was measured at 6.48 μM/1010 cells/h. This enhancement by fluidic stimulus is even more dramatic in case of HepG2 cells, which showed non-detectable level of activity when cultured in static condition, but showed significant level of activity when cultured in the flow condition (Fig. 4b). In static condition, P450 activity of HepG2 cells was not detectable, whereas in flow condition it was measured at 0.99 μM/1010 cells/h. The most probable cause for such a dramatic increase in the enzyme activity is that the mechanical stimulus from the fluidic shear affects cell physiology in a positive manner. In fact, a similar observation has been reported by other research groups, who observed significant enhancement in the metabolic activity of liver cells when cultured in flow condition (Prot et al. 2011; Vinci et al. 2011). Interestingly, P450 3A4 activity was significantly higher in Caco-2 cells than HepG2 cells, which is rather contradictory to what one would expect. Generally, it is known that P450 activity is much higher in the liver than the intestine (Galetin and Houston 2006). We speculate that this is due to the cell origin, since HepG2 is well known for its weak expression of P450 activity, unless they are chemically induced to up-regulate expression of P450 enzymes (Westerink and Schoonen 2007). In addition, cases of higher metabolic activity in the intestine cells than hepatic cells have been reported (Martin et al. 2008). Another factor that needs to be considered is the fibronectin coating on the slide glass to facilitate attachment of HepG2 cells. It has been reported that the surface coating material can also affect the physiology of cells (Au et al. 2014).
3.3 Paracellular permeability of intestinal barrier
The absorption process in the gut can be roughly classified into transcellular transport and paracellular transport (Artursson et al. 2001). Transcellular transport occurs usually in case of lipophilic molecules, and is known to be generally faster than paracellular transport, since lipophilic materials can cross cell membrane more easily than hydrophilic materials. On the other hand, paracellular transport occurs through small openings between the cells. The small pores are made up of tight junction proteins, namely occludin and Claudin (Ulluwishewa et al. 2011). The tightness of these proteins affects pore sizes and how easily molecules can cross the epithelial barrier. Since drug absorption is one of the most important function of the intestine, we tested whether our gut-liver chip shows absorption properties that are comparable to the conventional Caco-2 model. Fluorescein is often used as a model marker to assess the paracellular permeability of gut epithelium (Aguirre et al. 2014). The permeability of a model molecule can be calculated from time-series measurement of concentration in the acceptor compartment, using the following equation, where C0 denotes initial concentration in the donor compartment, A denotes the surface area of absorption, Q is the mole flux of absorbed molecules (Masungi et al. 2004).
Figure 5 shows the permeability of fluorescein across the Caco-2 epithelial barrier, either cultured in static condition or flow condition in the chip. The liver cells were not cultured in the chip. In flow condition, the permeability of fluorescein was calculated to be 6.24 X 10−6 cm/s, whereas in static condition it was higher at 2.86 X 10−5 cm/s. Caco-2 cells in chip condition showed approximately 4.6 times lower permeability than the same cells in static well condition. The observed decrease in the absorption permeability is likely due to the formation of stronger tight junctions in the gut epithelium. It has been reported that fluidic shear stress can increase the expression of tight junction proteins such as ZO-1 in vascular endothelium (Booth and Kim 2012). A similar observation has also been made for Caco-2 cells, where the expression of Occludin increased when cultured in flow condition, resulting in tighter cell junctions and lower absorption permeability (Chi et al. 2015). Another plausible explanation is a change in tissue morphology resulting from the fluidic stimulus. It has been reported that combination of fluidic shear and strain resulted in villi-like structure of Caco-2 cells (Kim et al. 2012). Formation of such a 3D structure could increase the path that absorbed molecules has to travel, resulting in decreased permeability. Another factor that should be considered is a possible change in the thickness of unstirred water later (UWL) due to the presence of flow. It has been reported that the presence of fluidic shear stress can result in a decrease in the thickness of UWL, which can alter absorption kinetics of drugs (Naruhashi et al. 2003). The detailed mechanism is still unclear, and needs further investigation on these factors.
A consistent observation was also made in an in vivo experiment, where human intestine was challenged with varying flow rate (Fine et al. 1995). In this study, increasing the jejunal flow rate four-fold resulted in significantly decreased permeability of L-xylose/urea and average calculate pore size. Although exact mechanism of how permeability decreases under flow condition is uncertain, results obtained from our gut-liver chip is consistent with previously reported works. One possible reason suggested by Fine et al., is that the cells lining the villi and the cells at the villi tips have different permeability, and the flow alters the pattern of exposure to villi surface, changing the observed permeability. In the study by Chi et al., TEM images of Caco-2 cells exposed to flow condition showed smaller openings in the tight junctions between the cells, which implies that paracellular permeability would decrease in flow condition. These results suggest that our gut-liver chip provides a more physiologically relevant environment of gut cells than the conventional well condition.
3.4 First pass metabolism of apigenin
Apigenin is a natural flavonoid molecule found in many plants, and is known to have health promoting effect (Woodman and Chan 2004). Apigenin was selected as a model molecule to test if the gut-liver chip can reproduce the first pass metabolism of orally taken drugs. Apigenin is known to go through extensive metabolism in the intestine and the liver (Teng et al. 2012). It is also known to be unstable in aqueous solution and easily degraded (Gradolatto et al. 2004; Patel et al. 2007). It is also relatively hydrophobic and can potentially adsorb to PDMS surfaces (Berthier et al. 2012). Therefore, we tested running apigenin containing media in the chip for 24 h, and observed significant amount of apigenin disappearing from the media (data not shown). We have found that coating the PDMS surface with BSA can alleviate the adsorption issue (unpublished data), and we further tried to minimize absorption by pre-running media containing apigenin in the chip before cell seeding. Spontaneous degradation of apigenin was evaluated by measuring the concentration of apigenin in media after 6 and 24 h in the chip (Fig. 6a). We found that after 6 h, approximately 81% of initial apigenin remained, and after 24 h, 52% of initial apigenin remained. Comparing this result with apigenin containing media incubated in a tube, we found that similar amount of apigenin remained in the tube, suggesting that the PDMS absorption was reasonably prevented.
To examine the metabolic rate of apigenin, the gut-liver chip was run with two different conditions; one with gut cells only, and one with gut and the liver cells co-cultured in the chip. The purpose of this was to evaluate the respective contribution of the gut and liver cells to the first pass metabolism of apigenin in the chip. Analysis of remaining apigenin in the media in the upper (gut) and lower (liver) layer shows that in the chip with gut cells only, 33.5 μM of apigenin was detected in the gut layer and 1.7 μM of apigenin was detected in the liver layer after 6 h. After 24 h, 5.4 μM in the gut layer, and 1.4 μM in the liver layer was detected (Fig. 6b). Comparing this result with Fig. 6a, it can be deduced that significant amount of remaining apigenin was metabolized by the Caco-2 cells during 24-h period. In a chip with gut and liver cells co-cultured, after 6 h, 31.4 μM and 1.0 μM was detected in the gut and liver layer, respectively. After 24 h, 2.6 μM and 2.0 μM was detected in the gut and liver layer, respectively. Concentration of apigenin in the gut layer was slightly lower in case of co-culture, compared to the chip with gut cell monoculture. This suggests that majority of metabolic activity was exerted by Caco-2 cells in the gut layer, while HepG2 cells in the liver layer exerted additional metabolic activity. First pass metabolism of apigenin was studied by Teng et al., using both in vitro Caco-2 culture and in vivo rat. After oral administration of apigenin to a rat, it took about 24 h for complete removal of apigenin and its metabolites, similar to our result using the gut-liver chip. In their study, apigenin was also metabolized to a significant extent by Caco-2 cells, similar to our study. A similar observation was also made in a separate study, which reported that intestinal disposition may be more dominant than hepatic disposition (Chen et al. 2003).
Time-dependent concentrations of metabolites in the gut-liver chip was analyzed by HPLC to find the fractions of different metabolites produced. Chip experiment was done in two conditions; gut cell monoculture in the chip (Fig. 7a, b) and gut and liver cells co-cultured in the chip (Fig. 7c, d). In both conditions, apigenin was mainly metabolized to two types of metabolites, apigenin-7-glucuronide and apigenin-7-sulfate. In case of Caco-2 monoculture in the chip, majority of metabolites were sulfation products both in the gut and the liver layer (68.0% at 6 h and 73.7% at 24 h in the gut layer, and 27.8% at 6 h and 21.6% at 24 h in the liver layer). In case of gut and liver cell co-culture in the chip, glucuronide conjugated metabolite in both gut and liver layer increased significantly compared to the gut cell monoculture, although still majority of metabolites were sulfation conjugates (2.9% to 6.8% at 6 h in the gut layer, 1.3% to 8.3% at 6 h in the liver layer). Secondly, fraction of sulfation products increase mostly in the liver layer (27.8% to 40.2% at 6 h, 21.6% to 32.4% at 24 h). These results together suggest that Caco-2 cells metabolize apigenin mainly to sulfation products whereas HepG2 metabolize apigenin to both sulfation and glucuronide products.
In an In vivo analysis of apigenin metabolism in a rat, both sulfate and glucuronide conjugation was observed, while the fraction of glucuronide conjugation was higher than sulfation (Teng et al. 2012). Comparing this with our result, gut-liver chip seems to have produced sulfation conjugates more heavily than the rat, but co-culture with HepG2 cells seems to have moved the metabolism slightly to glucuronide conjugation, similar to the rat. This suggests that the gut-liver chip with gut and liver cell co-culture was able to show first pass metabolism that is closer to the rat than the chip with gut monoculture. This is still not in an accurate agreement with actual first pass metabolism of apigenin observed in vivo, and this can probably be improved by using more in vivo-like cells, such as primary cells and stem cells (Khetani et al. 2015). Another important consideration is to carefully adjust the relative contribution of gut and liver cells, that is, scale the two organ functions to accurately mimic in vivo situation (Moraes et al. 2013; Wikswo et al. 2013).
Another notable observation was that fraction of sulfation conjugates in the apical side (gut layer) increased significantly between 6 and 24 h (68.0% to 73.7% in Caco-2 monoculture condition, 44.7% to 60.8% in co-culture condition), while in the liver layer a decrease was observed (27.8% to 21.6% in monoculture condition, 40.2% to 32.4% in co-culture condition. This can be partly explained by previous reports that sulfation conjugates are less permeable than apigenin and its glucuronide conjugates (Hu et al. 2003), which could have led to accumulation of sulfation products in the apical side.
4 Conclusion
In this paper, we developed a microfluidic gut-liver with an aim of reproducing the first pass metabolism. A finite element model of transport inside the chip was used to set the design parameters. Caco-2 and HepG2 cells were successfully co-cultured in the chip, verified by the morphological evaluation of both cells. Changes in the metabolic and absorptive properties of both gut and liver cells were observed. Using apigenin as a model drug, we have shown that this device can be used as a tool to reproduce the first pass metabolism of drugs. Although several improvements are still needed for more accurate reproduction of a PK profile, the gut-liver chip showed the pharmacokinetic profile resembling that of in vivo models more closely in gut-liver co-culture condition than gut monoculture condition.
References
A. Abbott, Nature 424, 870 (2003)
T.A. Aguirre, M. Rosa, S.S. Guterres, A.R. Pohlmann, I. Coulter, D.J. Brayden, Eur. J. Pharm. Biopharm. 88, 856 (2014)
J.W. Allen, S.R. Khetani, S.N. Bhatia, Toxicol. Sci. 84, 110 (2005)
P. Artursson, K. Palm, K. Luthman, Adv. Drug Deliv. Rev. 46, 27 (2001)
S.H. Au, M.D. Chamberlain, S. Mahesh, M.V. Sefton, A.R. Wheeler, Lab Chip 14, 3290 (2014)
E. Berthier, E.W. Young, D. Beebe, Lab Chip 12, 1224 (2012)
S.N. Bhatia, D.E. Ingber, Nat. Biotechnol. 32, 760 (2014)
R. Booth, H. Kim, Lab Chip 12, 1784 (2012)
E.F. Brandon, C.D. Raap, I. Meijerman, J.H. Beijnen, J.H. Schellens, Toxicol. Appl. Pharmacol. 189, 233 (2003)
T.J. Carlson, M.B. Fisher, Comb. Chem. High Throughput Screen. 11, 258 (2008)
C.Y. Chan, P.H. Huang, F. Guo, X. Ding, V. Kapur, J.D. Mai, P.K. Yuen, T.J. Huang, Lab Chip 13, 4697 (2013)
J. Chen, H. Lin, M. Hu, J. Pharmacol. Exp. Ther. 304, 1228 (2003)
M. Chi, B. Yi, S. Oh, D.J. Park, J.H. Sung, S. Park, Biomed. Microdevices 17, 9966 (2015)
S. Choi, M. Nishikawa, A. Sakoda, Y. Sakai, Toxicol. In Vitro 18, 393 (2004)
M.B. Esch, G.J. Mahler, T. Stokol, M.L. Shuler, Lab Chip 14, 3081 (2014)
S. Ferruzza, C. Rossi, M.L. Scarino, Y. Sambuy, Toxicol. In Vitro 26, 1252 (2012)
K.D. Fine, C.A. Santa Ana, J.L. Porter, J.S. Fordtran, Gastroenterology 108, 983 (1995)
A. Galetin, J.B. Houston, J. Pharmacol. Exp. Ther. 318, 1220 (2006)
A. Gradolatto, M.C. Canivenc-Lavier, J.P. Basly, M.H. Siess, C. Teyssier, Drug Metab. Dispos. 32, 58 (2004)
H.F. Helander, L. Fandriks, Scand. J. Gastroenterol. 49, 681 (2014)
C. Hilgendorf, H. Spahn-Langguth, C.G. Regardh, E. Lipka, G.L. Amidon, P. Langguth, J. Pharm. Sci. 89, 63 (2000)
M. Hu, J. Chen, H. Lin, J. Pharmacol. Exp. Ther. 307, 314 (2003)
B. Hughes, Nat. Rev. Drug Discov. 8, 93 (2009)
Y. Imura, K. Sato, E. Yoshimura, Anal. Chem. 82, 9983 (2010)
Y. Imura, E. Yoshimura, K. Sato, Anal. Sci. 28, 197 (2012)
K.I. Kaitin, Clin. Pharmacol. Ther. 87, 356 (2010)
S.R. Khetani, D.R. Berger, K.R. Ballinger, M.D. Davidson, C. Lin, B.R. Ware, J Lab Autom. 20, 216 (2015)
H.J. Kim, D. Huh, G. Hamilton, D.E. Ingber, Lab Chip 12, 2165 (2012)
S.H. Kim, J.W. Lee, I. Choi, Y.C. Kim, J.B. Lee, J.H. Sung, J. Nanosci. Nanotechnol. 13, 7220 (2013)
H. Lee, D. S. Kim, S. K. Ha, I. Choi, J. M. Lee, J. H. Sung, Biotechnol. Bioeng. (2016)
P. Martin, R. Riley, D.J. Back, A. Owen, Br. J. Pharmacol. 153, 805 (2008)
C. Masungi, C. Borremans, B. Willems, J. Mensch, A. Van Dijck, P. Augustijns, M.E. Brewster, M. Noppe, J. Pharm. Sci. 93, 2507 (2004)
C. Moraes, J.M. Labuz, B.M. Leung, M. Inoue, T.H. Chun, S. Takayama, Integr Biol (Camb) 5, 1149 (2013)
D. Moutinho, C.C. Marohnic, S.P. Panda, J. Rueff, B.S. Masters, M. Kranendonk, Drug Metab. Dispos. 40, 754 (2012)
K. Naruhashi, I. Tamai, Q. Li, Y. Sai, A. Tsuji, J. Pharm. Sci. 92, 1502 (2003)
D. Patel, S. Shukla, S. Gupta, Int. J. Oncol. 30, 233 (2007)
J.M. Prot, C. Aninat, L. Griscom, F. Razan, C. Brochot, C.G. Guillouzo, C. Legallais, A. Corlu, E. Leclerc, Biotechnol. Bioeng. 108, 1704 (2011)
A. Sivaraman, J.K. Leach, S. Townsend, T. Iida, B.J. Hogan, D.B. Stolz, R. Fry, L.D. Samson, S.R. Tannenbaum, L.G. Griffith, Curr. Drug Metab. 6, 569 (2005)
D. Spaggiari, L. Geiser, Y. Daali, S. Rudaz, J. Pharm. Biomed. Anal. 101, 221 (2014)
J. Strovel, S. Sittampalam, N.P. Coussens, M. Hughes, J. Inglese, A. Kurtz, A. Andalibi, L. Patton, C. Austin, M. Baltezor, M. Beckloff, M. Weingarten, S. Weir, in Assay Guidance Manual, ed by G. S. Sittampalam, N. P. Coussens, H. Nelson, et al.. Early Drug Discovery and Development Guidelines: For Academic Researchers, Collaborators, and Start-up Companies (Eli Lilly & Company and the National Center for Advancing Translational Sciences, Bethesda, 2004)
J.H. Sung, M.L. Shuler, Lab Chip 9, 1385 (2009)
J.H. Sung, C. Kam, M.L. Shuler, Lab Chip 10, 446 (2010)
J.H. Sung, M.B. Esch, J.M. Prot, C.J. Long, A. Smith, J.J. Hickman, M.L. Shuler, Lab Chip 13, 1201 (2013)
Z. Teng, C. Yuan, F. Zhang, M. Huan, W. Cao, K. Li, J. Yang, D. Cao, S. Zhou, Q. Mei, PLoS One 7, e29647 (2012)
D. Ulluwishewa, R.C. Anderson, W.C. McNabb, P.J. Moughan, J.M. Wells, N.C. Roy, J. Nutr. 141, 769 (2011)
P.M. van Midwoud, M.T. Merema, E. Verpoorte, G.M. Groothuis, Lab Chip 10, 2778 (2010)
B. Vinci, C. Duret, S. Klieber, S. Gerbal-Chaloin, A. Sa-Cunha, S. Laporte, B. Suc, P. Maurel, A. Ahluwalia, M. Daujat-Chavanieu, Biotechnol. J. 6, 554 (2011)
L. Wang, S.K. Murthy, W.H. Fowle, G.A. Barabino, R.L. Carrier, Biomaterials 30, 6825 (2009)
W.M. Westerink, W.G. Schoonen, Toxicol. In Vitro 21, 1581 (2007)
J.P. Wikswo, E.L. Curtis, Z.E. Eagleton, B.C. Evans, A. Kole, L.H. Hofmeister, W.J. Matloff, Lab Chip 13, 3496 (2013)
O.L. Woodman, E. Chan, Clin. Exp. Pharmacol. Physiol. 31, 786 (2004)
E.W. Young, C.A. Simmons, Lab Chip 10, 143 (2010)
Acknowledgements
This work was supported by Ministry of Trade, Industry and Energy (MOTIE), Republic of Korea (10050154, Establishment of Infrastructure for industrialization of Korean Useful Microbes, R0004073), and KFRI (Korea Food Research Institute, grant no: E0121705), and Hongik University Research Fund. This work was also supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (No. NRF-2016R1A2B2008691) and by the KIST Institutional Program (No. 2 V04950).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Choe, A., Ha, S.K., Choi, I. et al. Microfluidic Gut-liver chip for reproducing the first pass metabolism. Biomed Microdevices 19, 4 (2017). https://doi.org/10.1007/s10544-016-0143-2
Published:
DOI: https://doi.org/10.1007/s10544-016-0143-2