Abstract
We have developed a microfluidic device capable of fully integrated sample preparation and gene analysis from crude biosamples such as whole blood. Our platform takes the advantage of the silica superparamagnetic particle based solid phase extraction to develop an all-in-one scheme that performs cell lysis, DNA binding, washing, elution and the PCR in the same reaction chamber. The device also employs a unique reagent loading scheme, allowing efficient preparation of multiple reactions via a single injection channel. In addition, PCR is performed in a droplet-in-oil manner, eliminating the need for chamber sealing. The combination of these design features greatly reduces the complexity in implementing fully integrated lab-on-a-chip systems for genetic detection, facilitating parallel analysis of multiple samples or genes on a single microchip. The capability of the device is demonstrated by performing DNA isolation from the human whole blood sample and analyzing the Rsf-1 gene using the TaqMan probe based gene specific PCR assays.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Downscaling biological assays and analytical devices has been a trend in bioanalytical sciences and instrumentation developments. Miniaturization of analytical systems has many advantages such as reduced sample and reagent consumption, shorter analysis time and less sample handling(Guzman 2003; Saito and Jinno 2003; Vilkner et al. 2004). Many miniaturized platforms have been shown to carry out biochemical synthesis and detection (Burns et al. 1998; Northrup et al. 1998; Erill et al. 2003; Shin et al. 2003; Cady et al. 2005; Chen et al. 2005; Dorfman et al. 2005; deMello 2006; Lehmann et al. 2006; Niu et al. 2006; Ohashi et al. 2007; Pipper et al. 2007; Pipper et al. 2008; Zhang et al. 2009), cell/particle microfluidic manipulation (El-Ali et al. 2006), biochemical sensing (Janasek et al. 2006), biosample preparation (Wen et al. 2008; Price et al. 2009), optofluidics (Psaltis et al. 2006), magnetic resonance imaging (Harel 2009) and mass spectroscopy (Grym et al. 2006). The combination of microfluidic device with confocal optical spectroscopy has created an ideal platform for single molecule study (Wang et al. 2005; Yeh et al. 2005; Rane et al. 2010).
Among all the aforementioned applications, the polymerase chain reaction (PCR) microchips have been extensively studied due to its important role in the clinical diagnostics and genetic analysis. Numerous studies have contributed to many aspects of the device, such as fabrication and packaging techniques, materials, microfluidic sample handling methods, surface passivation techniques and structural designs (Zhang et al. 2006). Many incorporate temperature control modules for thermal cycling and detection module for real time analysis(Woolley et al. 1996; Xiang et al. 2007). A limited number of platforms also integrate an upstream sample preparation module to process the biosamples and obtain the analytes of interest (Liu et al. 2004; Easley et al. 2006; Pipper et al. 2007; Pipper et al. 2008).
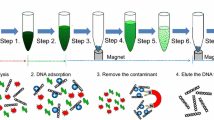
On-chip DNA sample preparation modules often employ the silica based solid phase extraction (SPE), either in the form of micropillars/microposts (Cady et al. 2003; West et al. 2007) or immobilized silica beads/particles(Breadmore et al. 2003; Gijs 2004; Pipper et al. 2007; Pipper et al. 2008). These methods involve the binding of the DNA to the silica surface in high-ionic-strength chaotropic salt, followed by a number of washing steps with organic solvent before eluting the DNA in buffers with low ionic strength. The pH and the temperature are also believed to play an important role in the process. The proposed mechanism of the DNA-silica interaction is thought as a combined effect of the electrostatic interaction, dehydration of the DNA molecule and the hydrogen bonding (Melzak et al. 1996). With the assistance of magnet based particle manipulation, the purification protocol avoids the use of centrifugation and vortexing, thus can be easily translated onto a microfluidic platform.
Since sample preparation involves multiple steps and various reagents, integrating it with PCR-based gene analysis requires establishing fairly complex microfluidic architecture to facilitate fluidic control and thermal cycling(Oleschuk et al. 2000; Yuen et al. 2001; Legendre et al. 2006).The added clinical demand to develop a total analysis system capable of processing multiple samples in parallel with high throughput would require multiple interconnected mixing and reaction chambers which further magnify the challenge. Conventional approach relies on separate bench-top instrumentations for sample preparation and multi-well PCR that consume large amount of samples and reagents. Although miniaturized sample analysis systems promise to enhance the cost-effectiveness, analysis speed and automation, their functions are often limited to either sample preparation or PCR, but not the combination. A few existing systems demonstrate the sample preparation and PCR on the same platform (Pipper et al. 2007; Pipper et al. 2008), but the lack of the capability to perform parallel analysis has limited their appeal to routine laboratory and clinical utilizations.
We report a crude sample analysis platform capable of sample-to-answer gene analysis by integrating silica superparamagnetic particle (SSP) based SPE and droplet-in-oil PCR (Zhang et al. 2009) to facilitate cell lysis, DNA extraction, purification, PCR amplification and detection in one single chamber. The all-in-one scheme greatly reduces the number of microfluidic components such as microvalves and microchambers needed for carrying out the entire sample preparation and gene detection process. This simple and yet fully integrated microchip platform therefore enables parallel gene analysis from crude biosamples with high throughput.
2 Methods and materials
2.1 Device fabrication
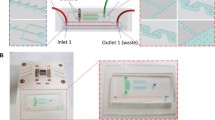
Two separate molds were fabricated for the fluidic layer and the valve layer respectively. Two different regions were present on the fluidic layer. The shallow channel region (Fig. 1) was fabricated by spin-coating a 15 μm positive photoresist (SPR 220-7, Dow Chemical Company) layer on a silicon substrate. After the UV exposure and the development, the wafer was hard baked at 110°C for 2 h. The shallow channel region, which facilitated easy valve closure, was located under the valve pad regions. The tall channels on the fluidic layer reduced the overall flow resistance, allowing for easy sample injection. To create the tall channel, another 50 μm layer of SU8-3025 (MicroChem Corp.) was spun on the same substrate. The tall channels were aligned to the positive photoresist layer. After the exposure and the development, the mold for the fluidic layer with both positive photoresist and negative photoresist were then hard baked at 110°C overnight. The mold for the valve layer was created by lithographically patterning a 50 μm SU8-3025 layer which was hard baked at 200°C overnight.
Before casting the PDMS (Corning), both molds were coated with Chlorotrimethylsilane (Sigma-Aldrich). The device was fabricated using the standard multiple layer soft lithography process. Buffer access ports and waste drain ports were created using a gauge 20 dispensing needle. The open reaction chambers were made by punching the PDMS with a 4 mm hollow puncher. The prepared PDMS was then bonded to the Grade 1 glass slide (Fisher Scientific) after oxygen plasma treatment. The bonded chip was baked at 75°C overnight. The magnet array was made by drilling through a piece of wooden board and inserting the permanent neodymium magnets into the holes. Each magnet was positioned right below one reaction chamber. The magnet array would not come into direct contact with the buffers, hence could be reused.
The pneumatic microvalves were actuated by the pressure source that is in turn controlled by electromagnetic valves (EMVs) with a MATLAB program (Supporting information Fig. S1). The EMVs were connected to the pressure source and the microvalves through the control lines. Once the EMVs were activated, the gas entered the control line, pressurized the PDMS membrane and closed the microvalves. Loaded buffer lines were plugged into the respective buffer access ports and connected to the pressure source controlled by EMVs through the same MATLAB program. The drain lines were plugged into the waste drain ports and connected to a Büchner flask that was hooked to the vacuum source.
2.1.1 Sample preparation
Human whole blood was collected from healthy volunteers after obtaining their informed consent. The SSP based SPE was carried out using Biosprint 15 blood kit (Qiagen Corp.). All buffers were prepared according to manufacturer’s instruction. For each preparation, 200 nL SSP and 5 μL whole blood were used. The other buffers were scaled down according to the sample volume (i.e. 5 μL binding buffer, 12.5 μL of washing buffer 1 and 12.5 μL of washing buffer 2). The DNA purification process was performed by first injecting buffer AL mixed with proteinase K. The mixture was incubated at room temperature for about 10 mins. The waste buffers were then discarded, leaving only the solid SSP, which was washed with washing buffer 1 and 2. After the final washing step, the SSP was air dried. Instead of eluting in the elution buffer, the purified DNA was directly eluted with 10 μL of PCR buffer. No SSP removal is required since the PCR could be performed in the presence of the SSP.
2.2 PCR recipe
PCR was performed using the Taq PCR core kit (Qiagen Corp.). The PCR mixture contained 1X Qiagen PCR buffer, 4 dNTPs each at 400 μM, total of 3.5 mM Mg2+, 0.05 unit/μL Taq polymerase, forward primer and reverse primer each at 400 nM, 100 nM TaqMan probe and 0.1% (w/v) bovine serum albumin (BSA). A touchdown PCR protocol was used to enhance the specificity (supporting information Tab. S1).
2.3 Gel electrophoresis
The purified human genomic DNA (gDNA) was collected and run on a 0.8% agarose gel at 8 V/cm for 90 mins. The PCR product was run on a 2% agarose gel at 8 V/cm for 90 mins. The gels were stained with the GelStar® nucleic acid gel stain (Lonza Group). The gel images were acquired using the Typhoon™ 9400 variable imager (GE healthcare) with the following settings: 488 nm laser excitation, 530SP emission filter, 500 V PMT gain and normal sensitivity.
2.4 Direct chip scan and image analysis
The TaqMan probe was dually labeled with the Quasar 670 and the Black Hole Quencher 2 (BHQ-2) (Table 1). The end point fluorescence image was acquired using the Typhoon™ 9400 variable imager with the following settings: 633 nm laser excitation, 670 nm BP30 emission filter, 500 V PMT gain and high sensitivity. The image analysis was carried out using ImageJ (NIH).
3 Results and discussion
Our device (Fig. 2(a)) has eight open reaction chambers arranged in a snowflake array (Zhang et al. 2009). Buffers injected from the buffer access ports enter the central line and split equally into each reaction chamber. The waste buffers in the reaction chambers are removed by applying vacuum from the waste drain ports located at the distal end of the reaction chambers. The valve layer and the fluidic layer are fabricated by standard polydimethylsiloxane (PDMS) based multilayer soft lithography. Two PDMS layers are bonded to a piece of glass slide that sits on top of a magnet array comprising 8 permanent neodymium magnets that are positioned right below the reaction chambers (Fig. 2(b)). The fluidic layer includes tall channel regions to minimize the flow resistance and shallow channel regions for easy valve closure. Tubings loaded with buffers are plugged into the designated buffer access ports and connected to a pressure source controlled by EMVs through a MATLAB program which also controls the pneumatic microvalves. By default, all the pneumatic microvalves are at CLOSE position. To inject the buffers, the buffer pressure source is turned on and the microvalves are switched to OPEN position. Waste buffers are later removed by the vacuum from the waste drain port while the air is injected from the buffer access port to clean the central line.
The all-in-one sample preparation scheme is illustrated in the cross section view along AA′ (Fig. 3(a)). The pneumatic microvalves are all by default set to CLOSE position (Fig. 3(b)). Sample mixed with SSP is dispensed into the reaction chambers. To inject the buffer, the pneumatic microvalve is first switched to OPEN position and the buffer pressure source is switched on in order to infuse the buffer into the central line. Meanwhile, the SSP is held in position by the permanent magnet located below the reaction chamber (Fig. 3(c)). The waste buffer is later removed from the waste drain port by the vacuum while the channel is flushed with the air. Same as the previous case, the SSP is held in position by the permanent magnet (Fig. 3(d)). Finally, the PCR reagent is injected followed by the immiscible companion oil (Fig. 3(e)). The companion oil fills the reaction chamber and encapsulates the PCR reagent together with the SSP into a droplet (Fig. 3(f)). The companion oil prevents the droplet from direct evaporation and isolates the reaction from the environment, avoiding possible contamination. Hitherto, the device is primed and ready for thermal cycling.
All-in-one DNA extraction and droplet-in-oil PCR preparation scheme. (a) Cross section along channel AA′. (b) The pneumatic microvalves are by default at CLOSE position. Clinical samples mixed with SSP a dispensed into the reaction chamber. (c) Valve is switched to OPEN position. Buffer is injected while the SSP is hold in position by a permanent magnet. (d) Waste buffer is removed from the waste drain port while the channel is cleaned by the air and the SSP is hold in position by the permanent magnet. (e) Elution/PCR buffer and immiscible companion oil is injected in sequence. (f) The PCR buffer with SSP embedded is encapsulated into droplets by the companion oil. The droplet-in-oil is ready for PCR
To aid visualization, we demonstrated the sample preparation scheme using food dye. The chip was first aligned to the magnet array (Fig. 4(a)). Food dye of three different colors (red, green and blue) and water were loaded into the tubings and plugged into respective buffer access ports. All the pneumatic microvalves were switched to the CLOSE position. Using the operation scheme described in the paragraph above, the red dye was first injected into the chip. Figure 4(b) demonstrated that the red dye entered the central line and split into the reaction chambers. The magnet arrays held the SSP in position, preventing the SSP from being washed away (Fig. 4(b)). The red dye was then removed from the waste drain port and the microfluidic network was cleaned before the injection of subsequent buffers (Fig. 4(c)). In a similar fashion, the green and blue dyes were infused into and drained from the chip (Fig. 4(d)-(f)). Last, the water was infused followed by the companion oil which encapsulated the water and the SSP into a droplet that sat on the bottom of the reaction chamber (Fig. 4(g)).
To demonstrate the capability of the proposed device, a fluorescence based gene analysis was performed using crude biosamples. Human whole blood samples mixed with SSP were dispensed into every other reaction chambers labeled with ‘S’ (Fig. 5(a)). The rest chambers did not contain any samples and served as the negative controls (labeled as ‘N’ in Fig. 5(a)). Buffer lines were loaded and plugged into the designated buffer access ports (Fig. 1). The lysis/binding buffer containing the proteinase K was first injected to lyse the cell and promote the binding of DNA to the SSP surface. The SSP was held in position by the magnet array below. DNA that adsorbed to the SSP surface was then washed with washing buffer 1 and washing buffer 2 sequentially. Next, the SSP was washed with ethanol and air dried before the PCR reagent was injected. It is noteworthy that the PCR reagent also served as the elution buffer. The PCR reagent had low ionic strength and neutral to high pH, hence favored the DNA desorption from the SSP. The PCR could be performed in the presence of SSP.
To facilitate PCR reactions in open chambers, the mixture of PCR reagent and DNA was encapsulated by mineral oil using the sample loading scheme described previously(Zhang et al. 2009). Thereafter, the chip was placed onto a flatbed thermal cycler for temperature cycling. Primers that targeted an 86 bp region of the Rsf-1 gene were used to amplify the genomic DNA together with a target specific TaqMan probe (Table 1). The Rsf-1 gene was a chromatin remodeling gene that was found to play an important role in the ovarian cancer(Shih et al. 2005). Patients with Rsf-1 amplification/overexpression had shorter overall survival than those without. The droplet-in-oil PCR was performed according to the recipe shown in supporting information (Tab. S1). The end point fluorescence image (Fig. 5(a)) confirmed the successful gene detection. The dark spots in the ‘S’ wells indicated successful amplification. The image analysis revealed significantly increase in fluorescence signal compared to the negative control for all 4 samples (Fig. 5(b)). The PCR product was further verified on a 2% agarose gel and the product band of the right size was observed (Supporting information Fig. S2). To validate the process of DNA extraction, we conducted a separate experiment to analyze the eluted DNA solution with gel electrophoresis. The purified DNA appeared as a band >23 kbp on the gel, which is the typical size using the Biosprint 15 kit, hence demonstrated successful genomic DNA extraction form the blood sample (Fig. 6).
As described above, gene analysis is a complicated process requiring multiple steps of biochemical reactions and mass transfers. To develop a fully integrated microsystem capable of providing sample-to-answer analysis would typically require multiple microfluidic components such as microchambers and microvalves to carry out the reaction, buffer exchange and material transfer. As such, most designs to date toward a total analysis microsystem has mainly been limited to processing a single sample. In our proposed all-in-one scheme, the entire process from genomic DNA extraction from crude blood sample to final PCR detection can be carried out in the same reaction chamber and thereby reduces the use of complex microfluidic components and greatly simplifies system integration. As a result, multiple functional units, each for analyzing a different sample, can be easily added onto the same platform for processing in parallel without complicating the device operation. The platform exploits the advantages associated with its snowflake and open chamber design as well as its sample loading scheme (Zhang et al. 2009). All controlling modules are connected to the computer. Once the device is primed with samples and buffers, the entire analysis process is controlled through a MATLAB program with minimum intervention by the operator. The semi-automated operation significantly minimizes human error, rendering the analysis more reliable. In addition, the proposed platform can also be applied to genetic based pathogen identification, mutation detection and epigenetic analysis.
4 Conclusion
In conclusion, we have developed an integrated microfluidic platform that is able to process multiple biosamples in parallel in a semi-automated fashion. The platform performs multiple tasks, such as cell lysis, DNA purification, and gene analysis through droplet-in-oil PCR, all in one single reaction chamber, which greatly reduces the complexity of the system and allows relatively high degree of parallelization. We have successfully demonstrated the capability of the platform by analyzing the Rsf-1 gene from the crude human total blood samples using a TaqMan based PCR assay.
References
M.C. Breadmore, K.A. Wolfe et al., Microchip-based purification of DNA from biological samples. Anal. Chem. 75(8), 1880–1886 (2003)
M.A. Burns, B.N. Johnson et al., An integrated nanoliter DNA analysis device. Science 282(5388), 484–487 (1998)
N.C. Cady, S. Stelick et al., Nucleic acid purification using microfabricated silicon structures. Biosens. Bioelectron. 19(1), 59–66 (2003)
N.C. Cady, S. Stelick et al., Real-time PCR detection of Listeria monocytogenes using an integrated microfluidics platform. Sens. Actuators, B 107(1), 332–341 (2005)
J.F. Chen, M. Wabuyele et al., Electrokinetically synchronized polymerase chain reaction microchip fabricated in polycarbonate. Anal. Chem. 77(2), 658–666 (2005)
A.J. deMello, Control and detection of chemical reactions in microfluidic systems. Nature 442(7101), 394–402 (2006)
K.D. Dorfman, M. Chabert et al., Contamination free continuous flow microfluidic polymerase chain reaction for quantitative and clinical applications. Anal. Chem. 77(11), 3700–3704 (2005)
C.J. Easley, J.M. Karlinsey et al., A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc. Natl. Acad. Sci. 103(51), 19272–19277 (2006)
J. El-Ali, P.K. Sorger et al., Cells on chips. Nature 442(7101), 403–411 (2006)
I. Erill, S. Campoy et al., Biochemical analysis and optimization of inhibition and adsorption phenomena in glass-silicon PCR-chips. Sens. Actuators, B 96(3), 685–692 (2003)
M.A.M. Gijs, Magnetic bead handling on-chip: new opportunities for analytical applications. Microfluid. Nanofluid. 1(1), 22–40 (2004)
J. Grym, M. Otevrel et al., Aerodynamic mass spectrometry interfacing of microdevices without electrospray tips. Lab Chip 6(10), 1306–1314 (2006)
N.A. Guzman, Improved solid-phase microextraction device for use in on-line immunoaffinity capillary electrophoresis. Electrophoresis 24(21), 3718–3727 (2003)
E. Harel, Magnetic resonance detection: spectroscopy and imaging of lab-on-a-chip. Lab Chip 9(1), 17–23 (2009)
D. Janasek, J. Franzke et al., Scaling and the design of miniaturized chemical-analysis systems. Nature 442(7101), 374–380 (2006)
L.A. Legendre, J.M. Bienvenue et al., A simple, valveless microfluidic sample preparation device for extraction and amplification of DNA from nanoliter-volume samples. Anal. Chem. 78(5), 1444–1451 (2006)
U. Lehmann, C. Vandevyver et al., Droplet-based DNA purification in a magnetic lab-on-a-chip. Angew. Chem. Int. Ed. 45(19), 3062–3067 (2006)
R.H. Liu, J. Yang et al., Self-contained, fully integrated biochip for sample preparation, polymerase chain reaction amplification, and DNA microarray detection. Anal. Chem. 76(7), 1824–1831 (2004)
K.A. Melzak, C.S. Sherwood et al., Driving forces for DNA adsorption to silica in perchlorate solutions. J. Colloid Interface Sci. 181(2), 635–644 (1996)
Z.Q. Niu, W.Y. Chen et al., DNA amplification on a PDMS-glass hybrid microchip. J. Micromech. Microeng. 16(2), 425–433 (2006)
M.A. Northrup, B. Benett et al., A miniature analytical instrument for nucleic acids based on micromachined silicon reaction chambers. Anal. Chem. 70(5), 918–922 (1998)
T. Ohashi, H. Kuyama et al., A simple device using magnetic transportation for droplet-based PCR. Biomed. Microdevices 9(5), 695–702 (2007)
R.D. Oleschuk, L.L. Shultz-Lockyear et al., Trapping of bead-based reagents within microfluidic systems: On-chip solid-phase extraction and electrochromatography. Anal. Chem. 72(3), 585–590 (2000)
J. Pipper, M. Inoue et al., Catching bird flu in a droplet. Nat. Med. 13, 1259–1263 (2007)
J. Pipper, Y. Zhang et al., Clockwork PCR including sample preparation. Angew. Chem. Int. Ed. 47(21), 3900–3904 (2008)
C.W. Price, D.C. Leslie et al, Nucleic acid extraction techniques and application to the microchip. Lab Chip 9(17), (2009)
D. Psaltis, S.R. Quake et al., Developing optofluidic technology through the fusion of microfluidics and optics. Nature 442(7101), 381–386 (2006)
T.D. Rane, C.M. Puleo et al., Counting single molecules in sub-nanolitre droplets. Lab Chip 10(2), 161–164 (2010)
Y. Saito, K. Jinno, Miniaturized sample preparation combined with liquid phase separations. J. Chromatogr. A 1000(1–2), 53–67 (2003)
L.M. Shih, J.J.C. Sheu et al., Amplification of a chromatin remodeling gene, Rsf-1/HBXAP, in ovarian carcinoma. Clin. Cancer Res. 11(24), 9161S–9161S (2005)
Y.S. Shin, K. Cho et al., PDMS-based micro PCR chip with parylene coating. J. Micromech. Microeng. 13(5), 768–774 (2003)
T. Vilkner, D. Janasek et al., Micro total analysis systems. Recent developments. Anal. Chem. 76(12), 3373–3385 (2004)
T.H. Wang, Y.H. Peng et al., Single-molecule tracing on a fluidic microchip for quantitative detection of low-abundance nucleic acids. J. Am. Chem. Soc. 127(15), 5354–5359 (2005)
J. Wen, L.A. Legendre et al., Purification of nucleic acids in microfluidic devices. Anal. Chem. 80(17), 6472–6479 (2008)
J. West, M. Boerlin et al., Silicon microstructure arrays for DNA extraction by solid phase sample contacting at high flow rates. Sens. Actuators, B 126, 664–671 (2007)
A.T. Woolley, D. Hadley et al., Functional integration of PCR amplification and capillary electrophoresis in a microfabricated DNA analysis device. Anal. Chem. 68(23), 4081–4086 (1996)
Q. Xiang, B. Xu et al., Miniature real time PCR on chip with multi-channel fiber optical fluorescence detection module. Biomed. Microdevices 9(4), 443–449 (2007)
H.C. Yeh, S.Y. Chao et al., Single-molecule detection and probe strategies for rapid and ultrasensitive genomic detection. Curr. Pharm. Biotechnol. 6(6), 453–461 (2005)
P.K. Yuen, L.J. Kricka et al., Microchip module for blood sample preparation and nucleic acid amplification reactions. Genome Res. 11(3), 405–412 (2001)
C.S. Zhang, J.L. Xu et al., PCR microfluidic devices for DNA amplification. Biotechnol. Adv. 24(3), 243–284 (2006)
Y. Zhang, V. Bailey et al., DNA methylation analysis on a droplet-in-oil PCR array. Lab Chip 9(8), 1059–1064 (2009)
Acknowledgement
The authors would like to thank the funding from NIH (R21-CA120742, U54-AI057168-07), NSF (0546012, 0725528, 0730503), and DARPA Micro/Nano Fluidics Fundamentals Focus (MF3) Center.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 720 kb)
Rights and permissions
About this article
Cite this article
Zhang, Y., Park, S., Yang, S. et al. An all-in-one microfluidic device for parallel DNA extraction and gene analysis. Biomed Microdevices 12, 1043–1049 (2010). https://doi.org/10.1007/s10544-010-9458-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10544-010-9458-6