Abstract
Phenol-degrading pseudomonads possessing different phenol hydroxylases (PH) were evaluated by the values of apparent half-saturation constant for phenol-oxygenating activity (K S ), maximum specific growth rate (μ max), lag-time length (λ), inhibition constant (K I ) and growth yield factor (Y X/S ). Strains of the same PH type showed similar kinetic parameters: single-component PH (sPH) harbouring strains had higher values of K S and lower values of μ max than the strains having multicomponent PH (mPH). However, the values of K I and the dependencies of the lag-time length on initial phenol concentration were strain-specific. The elevated ratio between specific activities of catechol 1,2-dioxygenase (C12O) and muconate cycloisomerase in sPH-strains caused irreversible accumulation of a high amount of exogenous cis,cis-muconate (CCM) which resulted in decreased Y X/S values. Co-presence of sPH and mPH genes did not give the strains PC16 and P69 any extra advantage and according to determined kinetic parameters only one PH was active during phenol degradation. At the same time simultaneous functioning of catechol ortho and meta cleavage pathways (strain PC20) resulted in higher μ max and Y X/S values. Evaluation of strains showed that the type of PH determined the efficiency of phenol degradation, whereas the tolerance to elevated phenol concentrations was strain-specific.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenol and its derivatives are common constituents in wastewaters produced in many industrial processes. It has been reported that phenol-degrading bacteria can adapt to biological wastewater treatment processes and that Pseudomonas strains are dominant in degradation of these aromatic compounds (Watanabe et al. 1998; Whiteley et al. 2001; Watanabe et al. 2002; El Fantroussi and Agathos 2005).
The first step in phenol degradation pathway is conversion of phenol into catechol by phenol hydroxylase (PH), which may determine the kinetic constants for cellular phenol-oxygenating activity (Hino et al. 1998). This flavoprotein monooxygenase uses NAD(P)H and O2 as co-substrates and is composed of either one (single-component phenol hydroxylase (sPH)) or several (multicomponent phenol hydroxylase (mPH)) components (Pessione et al. 1999). The ring cleavage of catechol can occur either via an ortho pathway catalysed by catechol 1,2-dioxygenase (C12O) or meta pathway catalyzed by catechol 2,3-dioxygenase (C23O). The cleavage products of these pathways are cis,cis-muconate (CCM) and 2-hydroxymuconic semialdehyde, respectively (Ornston and Stanier 1966; Dagley et al. 1960). Both catechol ortho- and meta-cleavage types can be combined with either sPH or mPH. The mPHs are classified into two groups according to genetic organisation of the operon—the dmp type (from Pseudomonas sp. CF600) and the mop type (from Acinetobacter calcoaceticus NCIB 8250), in which the mPH genes are linked to the genes for either meta (Shingler et al. 1989) or ortho pathway (Ehrt et al. 1995), respectively. In addition to the plasmid-borne dmpN gene cluster (Shingler et al. 1989) almost identical chromosome-encoded mPHs (encoded by phhN and pheA genes) have been found in P. putida strains P35X and BH, respectively (Ng et al. 1994; Takeo et al. 1995). We have previously characterised different types of mPHs in P. putida, P. mendocina and P. fluorescens PC strains and have shown that the strains phylogenetically close to those with the mop type of mPH, contain genes for ortho cleavage (Merimaa et al. 2006). Among the same set of strains, the catechol ortho-cleavage type was also characteristic to the sPH-possessing strains, in which the structure of the corresponding operon was similar to the pheBA operon of Pseudomonas sp. EST1001 (Nurk et al. 1991; Kivisaar et al. 1991). However, in P. pickettii PKO1 the sPH gene tbuD was shown to be co-transcribed with the C23O (tbuE) gene (Kukor and Olsen 1990, 1991). We also demonstrated the co-presence of genes for mPH and sPH combined with the cleavage of aromatic ring via ortho pathway in some Pseudomonas strains (Merimaa et al. 2006). The presence of two different monooxygenases, which are involved in catalysis of identical or similar reactions in the same microorganism was also shown by Cafaro et al. (2004). These authors assumed that the presence of two different enzymes confers a particular selective advantage to strains concerning the ability of a microorganism to optimise the use of aromatic compounds (Cafaro et al. 2005).
The multiplicity of phenol degradation pathways and organisation of the corresponding genes as well as the presence of both mPH and sPH enzymes in the same strain rises question about the efficiency of these bacteria to degrade phenolic compounds in polluted wastewaters. It was found that the mPH-containing phenol-degrading bacteria are predominant in natural environment (Peters et al. 1997). The catalytic domain of multicomponent phenol hydroxylase resides within the largest subunit of mPH (LmPH) (Hino et al. 1998). Watanabe et al. (1998, 2002) showed that phylogenetic grouping of bacteria on the basis of the DNA fragments encoding LmPH was in good correlation with physiological grouping based on kinetics of phenol-oxygenating activity of a whole cell. It was shown that in Pseudomonas strains the affinity and inhibition constants for phenol (K S and K SI , respectively) were similar and correlated with phylogenetic diversity of mPHs (Watanabe et al. 2002). However, the same group recently reported that the Variovorax strains possessing low-K S -type LmPH had a very low affinity for phenol (high K S values) (Futamata et al. 2005).
The phenol-degrading strains possessing different types of PHs and catechol ortho- or meta-cleavage pathways hold different potential for practical use in bioaugmentation. In carbon limited chemostat cultures it has been shown that strains possessing catechol meta pathway revealed higher growth rate values than strains having catechol ortho pathway (Kiesel and Müller 2002). According to the analysis of oxygen utilisation kinetics for phenol, the strain possessing only C23O activity expressed almost similar K S values as the strain with C12O activity, whereas specific growth rate was higher in the former case (Jiang et al. 2006). However, the respective kinetic parameters of genetically different phenol-degrading strains have not been investigated systematically yet. In the present study we performed quantitative and qualitative analysis of phenol degradation data obtained in batch experiments and we evaluated different phenol-hydroxylase-possessing phenol-degrading pseudomonads according to kinetic parameters.
Materials and methods
Bacterial strains and culture conditions
The phenol-degrading strains used in this study were isolated previously from phenol-polluted river water (Heinaru et al. 2000) and are listed in Table 1. P. putida EST1412 and Pseudomonas sp. CF600 were used as reference strains (Kivisaar et al. 1990; Shingler et al. 1989). The strains represent three PH groups: sPH- (PC24, PC31, PC20, EST1412), mPH- (PC1, PC18, CF600, PC30, PC17) and sPH + mPH-possessing strains (PC16, P69). In sPH-strains catechol is cleaved through ortho pathway and these strains contain the pheBA operon. The strains of the present study represent all groups revealed by phylogenetic grouping of relevant catabolic genotypes by sequence analysis of LmPH (Table 1) and C23O genes (Merimaa et al. 2006). Catechol cleavage is catalysed by C23O or C12O in mPH-strains PC1, PC18, CF600 and PC17, PC30, respectively.
The strains were grown in mineral salts medium supplemented with 2.5 mM phenol, as described previously (Heinaru et al. 2000).
Phenol-oxygenating activity
Oxygen-consumption rate by the bacterial cells pregrown on 2.5 mM phenol was determined by using Clark-type oxygen electrode. Endogenous oxygen consumption by bacteria was measured in a cuvette (1.5 ml) of the electrode pre-stabilized at 30°C in air-saturated M9 medium without phenol. After the addition of phenol (0.4–170 μM) the phenol-oxygenation-dependent oxygen-consumption rate was determined. Phenol-oxygenating activity was calculated by subtraction of the endogenous oxygen consumption rate from the substrate-dependent one. A unit of the activity was defined as μmol of oxygen consumed per minute. The specific activity (V) was defined as acivity per gram of dry cell weight (DW) of the culture that was determined gravimetrically by filtration through a 0.22 μm pore-size membrane filter. Apparent kinetic constants, half saturation constant (K S ) and maximum specific activity (V max), in Michaelis–Mentens equation (1) were determined by a non-linear regression method (SigmaPlot 2001 Enzyme Kinetic Module, SYSTAT).
where S is concentration of phenol.
Growth kinetics and lag-time
The growth of the strains was measured spectrophotometrically. Into each well of sterile flat-bottomed 96-wells microtiter plates (NuncTM) 140 μl of minimal medium supplemented with phenol (0.2–10.6 mM) and 10 μl of inoculum (∼107 CFU/ml) was added. The plates were sealed with parafilm and incubated on a Titramax 1000 (Heidolph) thermostated shaker table (600 rpm) at 30°C. To minimize border effects, the water baths were also placed on shaker table. Absorbance was measured by Labsystem Multiskan® MCC/340 microplate autoreader at 492 nm after every 30 min. All experiments were run in triplicate. The specific growth rate (μ) of cultures grown on different initial phenol concentrations was calculated from exponential growth phase of semilogarithmic absorbance growth curves using linear regression. The Aiba–Edwards equation was used to describe the inhibition of specific growth rate (μ) by phenol:
where μ max is maximum specific growth rate, S is the initial concentration of phenol, K SG is half-saturation constant and K I is inhibition constant.
The lag-time (λ) of the growth curve was calculated using the modified Gompertz model (Zwietering et al. 1990):
where μ G is specific growth rate, A is asymptotic phase, ABS t and ABS 0 are absorbance values at time t and 0, respectively.
Yield factor (Y X/S ), enzyme assays and determination of phenol and cis,cis-muconate
The yield factors (g DW g−1 PHE), Y X/S , of the strains grown in mineral salts medium (total volume 50 ml) supplemented with 2.7 mM phenol (except strain P69 that was grown on 1.4 mM phenol) were obtained using the following equation:
where X 0 and X M are initial and maximum dry cell concentrations (DW) and C 0 and C S are initial phenol concentration and phenol concentration at maximum cell density, respectively. For every strain correlation between the absorption at 492 nm and the dry weight of the cells was determined.
The samples for enzyme assays were taken during the middle and late exponential growth phase of batch cultures on 2.7 mM phenol minimal medium. Crude extracts were prepared and enzyme activity assays were carried out as described previously (Heinaru et al. 2000). The activities of C12O and C23O were measured spectrophotometrically (Feist and Hegeman 1969; Hegeman 1966). The activity of cis,cis-muconate cycloisomerase (MCI) was measured by determining the decrease of absorption at 260 nm in solution containing 33 mM Tris–HCl (pH 8.0), 1 mM MnCl2, and 0.1 mM cis,cis-muconate (CCM) as a substrate (Vollmer et al. 1998). Protein concentration was measured by the method of Bradford with bovine serum albumin as a standard (Bradford 1976). The values of apparent kinetic constants (half saturation constant K M ) of C12O for catechol were determined from initial velocities of enzymatic reactions at substrate concentrations 0.6…12.0 μM using the Michaelis–Menten equation by a non-linear regression method (SigmaPlot 2001 Enzyme Kinetic Module, SYSTAT). Induction experiments were performed as described previously (Heinaru et al. 2000).
The samples for substrate assay were filtered through a 0.22-μm pore size Millipore filter and stored frozen until analysis. For quantification of substrates and intermediates, Agilent 1100 series LC system with DAD (270 nm, bandwidth 4 nm, reference 380 nm, bandwidth 40 nm; with full spectrum registered between 190 and 900 nm) was used. Chromatographic separation was performed on an Agilent Zorbax Eclipse XDB-C18 analytical column (250 mm in length, ID 4.6 mm and 5 μm particle size) with an Agilent precolumn Eclipse XDB-C18 (12.5 mm in length, ID 4.6 mm and 5 μm particle size). Elution was carried out at a flow rate 0.8 ml min−1 with the mobile phase acetonitrile—buffer solution (1 mM ammonium acetate with 0.1% formic acid pH ≈ 2.8) (25:75). Phenol and its catabolic intermediates (catechol, CCM) were quantified using calibration graphs. Combined standard uncertainty of the results takes into account all significant uncertainty sources and is expressed as a standard deviation, i.e. at approximately 68% confidence level (Ellison et al. 2000). The uncertainties include contributions from sample preparation, instrumental measurement (both samples and calibration standards) and peak integration.
Results and discussion
Kinetic analysis of the phenol-oxygenating activity
Kinetic parameters of phenol-oxygenating activity of nine strains of different Pseudomonas species and biotypes alongside with those of two reference strains are presented in Table 2. The values of half-saturation constant for phenol-oxygenating activity, K S , were by almost one order of magnitude lower in the strains harbouring mPH (1.3…4.9 μM for the strains PC1, PC18, PC17, PC30 and the reference strain CF600) compared to the strains having sPH (12.8…21.4 μM for the strains PC24, PC31, PC20 and the reference strain EST1412) (Table 2). Similar grouping of strains was observed on the basis of V max/K S ratio, referred to as specific affinity and proposed by Healey (1980) as a more suited parameter that should reflect both the affinity and catalytic activity. In our study, the V max/K S values of the mPH-strains (33.2…200 U g−1 DW μM−1) were higher than those of the sPH-strains (3.9…24.5 U g−1 DW μM−1). The type of catechol cleavage pathway did not affect the kinetics of phenol oxidation—the K S and V max/K S values were similar for mPH-harbouring strains regardless to whether catechol was subjected to ortho (PC17, PC30) or meta (PC1, PC18, CF600) cleavage pathway (Table 2).
The strains PC16 and P69 possessing both sPH (encoded by pheA) and mPH genes (Merimaa et al. 2006) revealed different kinetic constants. The K S value of PC16 for phenol (2.4 μM) was about ten times lower compared to P69 (27.1 μM). These values as well as the corresponding V max/K S values (31.3 and 3.9 U g−1 DW μM−1) are similar to those of the strains possessing only mPH or sPH, respectively (Table 2). These results allow us to speculate that only mPH is active in the strain PC16 while in the strain P69 only sPH is functional. This assumption is in good concordance with our previous finding that pheA expression (sPH) in PC16 was by one order of magnitude lower than in P69 (Merimaa et al. 2006).
According to our data we conclude that in pseudomonads with different catabolic genotypes, the apparent half-saturation constants for phenol-oxygenating activity depends on the type of PH (sPH or mPH) and not on the type of subsequent catechol cleavage pathway.
Maximum specific growth rate (μ max) on phenol
Among kinetic parameters of growth, the maximum specific growth rate (μ max) significantly affects the rate of contaminant degradation and is therefore an important parameter to characterise microbes in biological treatment systems. As both μ max and half-saturation constant for growth affect the biodegradation efficiency of a pollutant, they both must be considered to address biodegradation rates (Goudar and Strevett 1998). Kovárová- Kovar and Egli (1998) found that the only parameter which rigidly fixes the growth behaviour of strains in a batch culture, is μ max. Our experiments showed that there was up to four-time difference between the μ max values (from 0.231 to 0.931 h−1) of different strains (Table 2). The mPH-strains (including PC16) had significantly higher μ max values (0.535…0.914 h−1) than sPH-strains PC24, PC31 and P69 (0.231…0.381 h−1). The sPH-strains PC20 and EST1412 were exceptional as they had high μ max values (0.699 and 0.931 h−1, respectively) characteristic to mPH-strains. It can be caused by a particular genetic background and genetic regulation in these strains. Namely, the strain PC20 has two cleavage pathways for catechol, i.e. besides C12O activity, it reveals constitutive expression of C23O (Heinaru et al. 2005). In comparing the expression of C23O during the growth of strain PC20 on 0.2% Casamino acids and on the same medium with inducer phenol (2.7 mM), the values of specific activity were 498 ± 7 and 624 ± 17 nmol min−1 mg−1, respectively, whereas the value of specific activity for C12O in uninduced conditions was <10 and with phenol 378 ± 11 nmol min−1 mg−1. We suppose that constitutive expression of C23O enables PC20 to grow faster on phenol as compared with other sPH-strains. The latter, laboratory-constructed strain EST1412 has two sets of genes for C12O (specific activity with inducer phenol 1,165 nmol min−1 mg−1), i.e. the chromosomally encoded catA gene and the plasmid-borne pheB gene of the pheBA operon (Kivisaar et al. 1990). According to Chen et al. (2004) the Ralstonia taiwanensis strain TJ86 which degrades phenol by mPH and catechol through the meta cleavage pathway, has μ max value of 0.52 h−1 (batch cultivations). In the Wautersia eutropha (formerly Alcanigenes eutrophus and Ralstonia eutropha) strain JMP134 possessing both catechol cleavage pathways, two μ max values were determined in carbon limited chemostat cultivations, whereas a higher value (0.41 h−1) was obtained in case of expression of the meta pathway compared to the μ max of 0.27 h−1 in case of expression of the ortho pathway (Müller and Babel 1996). The μ max value determined for an ortho pathway possessing Pseudomonas sp. strain in batch experiments was 0.27 h−1 (Polymenakou and Stephanou 2005). Herein it is important to underline that we did not observe differences between the μ max values of the mPH-strains possessing either ortho (PC17 and PC30) or meta (PC1, PC18 and CF600) cleavage type, whereas the strains having sPH showed a lower rate of phenol degradation.
Thus, both half-saturation constant for phenol-oxygenating activity (K S ) and maximum specific growth rate (μ max) values permit grouping of phenol-degrading strains according to the type of PH. Our data also indicated that strains with low K S values for phenol had higher μ max values (Table 2).
Inhibition constant (K I ) and the length of the lag-time (λ)
The inhibiting effect of phenol on growth of bacteria was evaluated by inhibition constant K I and by duration of the lag-time. The K I values revealed strain-specific tolerance to phenol but did not allow differentiation of the strains according to mPH and sPH types (Table 2). The highest K I values (7.44…13.15 mM) were determined for the strains CF600, PC1, EST1412 and PC24. The lowest K I values (1.99…2.52 mM) were obtained for the strains P69 and PC18. The length of the lag-time (λ) increased exponentially with phenol concentration (Fig. 1) and the effect was much stronger in strains with low K I values. Indeed, the lowest tolerance against phenol was revealed for the strains P69, PC20, PC17, PC30 and PC18, while the strains PC1, PC16, C600 and EST1412 were highly tolerant (Fig. 1). The mPH + C12O and mPH + C23O strains (PC17, PC30 and PC1, CF600, respectively), which are the representatives of two different LmPH groups, had similar phenol tolerance. The strain PC18 being a representative of a unique phylogenetic group of LmPH (Merimaa et al. 2006) was more sensitive to phenol than other mPH + C23O-strains. Strains with two PHs, PC16 and P69, had very different tolerance to phenol.
It was shown that a high growth rate and a short lag-time are needed to outcompete other strains in mixed cultures (Franck-Mokroβ and Schmidt 1998). In a separate study we have shown that in a leachate microcosm inoculated with the mixture of strains PC1, PC18, PC20 and PC24, the strain PC1 became dominant among the microbial population (Heinaru et al. 2005). The strain PC1 has a high value of both μ max and K I , and the length of the lag-time showed only minor dependence on initial phenol concentration in the medium.
Yield factor (Y X/S )
The lowest yield factor (Y X/S ) values were determined for the sPH-possessing strains EST1412 and PC24, in which catechol is degraded via ortho pathway (Table 2). This indicates that the sPH-strains with functional pheBA operon have lower Y X/S values than the mPH-strains. However, the strain PC31 from the same group had much higher Y X/S value. Among the mPH-strains the Y X/S , as well as μ max, did not depend on the type of catechol cleavage pathway of the strain. These data, determined from batch cultivations, contradict those by Kiesel and Müller (2002) obtained in chemostat systems under carbon limitation, according to which the catechol meta pathway results in a high growth rate and a low growth yield.
Accumulation of cis,cis-muconate in the growth medium and specific activities of enzymes
The flux of phenol into ortho and meta pathways is governed mainly by the affinities of the cleavage enzymes towards the substrate (Pieper et al. 1995). Therefore we determined the affinities of C12O to catechol, expressed as apparent half saturation constants (K M ), for sPH- and mPH-possessing strains. The obtained K M values were similar (from 2.8 to 3.6 μM), which means that studied strains cannot be differentiated by this value.
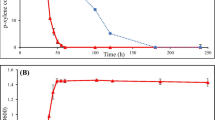
It is known that a cleavage product of catechol, cis,cis-muconate (CCM), is a critical intermediate of ortho decay in Pseudomonas strains (Feist and Hegeman 1969; Ornston 1971). In previous experiments we detected the irreversible accumulation of CCM in the growth medium reaching 35…55 μM concentration during growth of the strain PC24 on phenol-containing mixtures, whereas transient accumulation of catechol was observed only during growth of this strain on the mixture of phenol and benzoate (Heinaru et al. 2001). A low growth yield of the strain PC24 can be explained by non-productive accumulation of CCM, as exogenous CCM can be used for growth only by permeable mutants (Williams and Shaw 1997). The overflow of CCM hints to the pathway bottleneck localised at the level of C12O and muconate cycloisomerase, MCI, also known as cis,cis-muconate lactonising enzyme, converts CCM to muconolactone (Ornston and Stanier 1966). This is why we studied the ratio between the activities of C12O and MCI in middle and late log-phase during the growth of the strains expressing the catechol ortho pathway on 2.7 mM phenol (Fig. 2). The experiments revealed that at the end of active growth phase, the concentration of CCM was 55 μM and 70 μM in case of sPH-strains PC24 and PC31, respectively (Fig. 2). The mPH-strains with catechol ortho cleavage pathway (PC17 and PC30), which had a high growth yield, accumulated less CCM than the sPH-strains (Fig. 2). The ratio between specific activities of C12O and MCI showed that in sPH-strains the activity of MCI was always lower than that of C12O. The imbalance between the levels of the enzymes determining sequential catabolic reactions may cause intracellular accumulation of CCM as well as its exogenous accumulation in case of free diffusion. The conversion of phenol to CCM and accumulation of the latter in the growth medium due to the increased expression of C12O may be an important advantage for a strain enabling survival at high substrate concentrations. However, a high growth rate (Table 2) and short lag-phase (Fig. 2) of the sPH-strains PC31 and EST1412 improved the growth yield.
Accumulation of cis,cis-muconate (◊) in the medium and ratio of specific activities of catechol 1,2-dioxygenase and muconate cycloisomerase (columns) in strains PC17, PC30, PC16, PC24, PC31 and EST1412. Insets show biomass growth (●) and phenol consumption (▴) as a function of time. Values are the averages of triplicate determinations. Error bars indicate the combined standard uncertainties (Ellison et al. 2000) of phenol and cis,cis-muconate concentrations or standard deviations of C12O/MCI data
The strain PC20 is exceptional and very specific because it possesses two catabolic plasmids, NAH and PHE, and consequently, two catechol cleavage pathways (Heinaru et al. 2000, 2005; Merimaa et al. 2006). Phenol degradation via sPH and C12O is encoded by the pheBA operon induced by phenol, whereas the synthesis of C23O (from nah operon) is constitutive (Heinaru et al. 2005). So, catechol as the first intermediate of phenol catabolism can be further cleaved by both pathways in this strain. We measured consumption of phenol, accumulation of CCM and the activities of C12O, C23O and MCI in exponentially growing cells of PC20 (Fig. 3). Expectedly, the accumulation of CCM in the growth medium was accompanied by a decrease in specific activity of MCI. The inverse correlation was established between the activities of C12O and C23O (Fig. 3), and the activation of the meta pathway resulted in reversible accumulation of a yellow coloured product, 2-hydroxymuconic semialdehyde (detected visually) in the growth medium. The ortho cleavage pathway is the main route of phenol degradation in PC20 and after disappearance of phenol the synthesis of C23O will be derepressed. The co-presence of two cleavage pathways in PC20 led to higher μ max and Y X/S values compared to other sPH-strains.
Biomass growth (●), phenol consumption (▴), accumulation of cis,cis-muconate (◊) (a) and specific activities of catechol 1,2-dioxygenase (■), catechol 2,3-dioxygenase (Δ), and muconate cycloisomerase (▼) as a function of time in strain PC20 (b). Values are the averages of triplicate determinations. Error bars indicate the combined standard uncertainties (Ellison et al. 2000) of phenol and cis,cis-muconate concentrations or standard deviations of C12O, C23O and MCI data
Taken together, these results suggest that increased intracellular accumulation of CCM may be caused by altered activities of functional enzymes and may result in change of the growth yield.
Conclusions
Different phenol hydroxylase-possessing pseudomonads isolated from river water continuously polluted with phenolic compounds were evaluated by kinetic parameters for phenol degradation. It was shown that the values of apparent half-saturation constant for phenol-oxygenating activity (K S ) in the strains harbouring mPH were almost by one order of magnitude lower than in the strains having sPH. Additionally, the mPH-possessing strains exhibiting high affinity towards phenol had also higher values of maximum specific growth rate (μ max) than sPH-possessing strains.
The mPH-strains representing the same phylogenetic group of LmPH had similar kinetic parameters. For example, the phenol degradation parameters determined for the strain PC18, belonging to an unique phylogenetic group, differed from those of other mPH + C23O strains (PC1 and CF600). Our data showed that the type of catechol cleavage does not affect kinetic constants of phenol degradation.
We consider that the low growth yield (Y X/S ) of sPH-strains is caused by imbalance between the production and degradation of the intermediate CCM as well as by the increased length of the lag-phase. Due to the co-presence of ortho (from pheBA operon) and meta (from nah operon) cleavage pathways for the catechol in the strain PC20, higher μ max and Y X/S values were detected despite the accumulation of CCM.
The strains PC16 and P69 possessing two different phenol hydroxylases, sPH (encoded by pheA) and mPH (LmPH similar to MopN), had highly dissimilar kinetic parameters. Kinetic parameters of these strains revealed that only one of two PHs was functional in degradation of phenol. Thus, the strain PC16, which had a low level of expression of the pheA, degraded phenol similarly to the other strains possessing Mop type of LmPH (PC17 and PC30), whereas the strain P69 with high pheA activity had kinetic parameters similar to sPH-strains.
References
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem 72(1–2):248–254
Cafaro V, Izzo V, Scognamiglio R, Notomista E, Capasso P, Casbarra A, Pucci P, Di Donato A (2004) Phenol hydroxylase and toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1: Interplay between two enzymes. Appl Environ Microbiol 70(4):2211–2219
Cafaro V, Notomista E, Capasso P, Di Donato A (2005) Regiospecificity of two multicomponent monooxygenases from Pseudomonas stutzeri OX1: Molecular basis for catabolic adaptation of this microorganism to methylated aromatic compounds. Appl Environ Microbiol 71(8):4736–4743
Chen W-M, Chang J-S, Wu C-H, Chang S-C (2004) Characterization of phenol and trichloroethene degradation by the rhizobium Ralstonia taiwanensis. Res Microbiol 155(8):672–680
Dagley S, Evans WC, Ribbons DW (1960) New pathways in the oxidative metabolism of aromatic compounds by micro-organsms. Nature 188:560–566
Ehrt S, Schirmer F, Hillen W (1995) Genetic organization, nucleotide sequence and regulation of expression of genes encoding phenol hydroxylase and catechol 1,2-dioxygenase in Acinetobacter calcoaceticus NCIB8250. Mol Microbiol 18(1):13–20
El Fantroussi S, Agathos SN (2005) Is bioaugmentation a feasible strategy for pollutant removal and site remediation? Curr Opin Microbiol 8:1–8
Ellison SLR, Rösslein M, Williams A (2000) Eurachem/CITAC guide, quantifying uncertainty in analytical measurement, 2nd edn. Eurachem, Berlin
Feist CF, Hegeman GD (1969) Phenol and benzoate metabolism by Pseudomonas putida: regulation of tangential pathways. J Bacteriol 100(2):869–877
Franck-Mokroβ AC, Schmidt E (1998) Simultaneous degradation of chloro- and methylsubstituted aromatic compounds: competition between Pseudomonas strains using the ortho and meta pathway or the ortho pathway exclusively. Appl Microbiol Biotechnol 50:233–240
Futamata H, Nagano Y, Watanabe K, Hiraishi A (2005) Unique kinetic properties of phenol-degrading Variovorax strains resposible for efficient trichloroethylene degradation in a chemostat enrichment culture. Appl Environ Microbiol 71(2):904–911
Goudar CT, Strevett KA (1998) Comparison of relative rates of BTEX biodegradation using respirometry. J Ind Microbiol Biotechnol 21:11–18
Healey FP (1980) Slope of the Monod equation as an indicator of advantage in nutrient competition. Microb Ecol 5(4):281–286
Hegeman GD (1966) Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol 91(3):1140–1154
Heinaru E, Truu J, Stottmeister U, Heinaru A (2000) Three types of phenol and p-cresol-degrading bacteria isolated from river water continuously polluted with phenolic compounds. FEMS Microbiol Ecol 31:195–205
Heinaru E, Viggor S, Vedler E, Truu J, Merimaa M, Heinaru A (2001) Reversible accumulation of p-hydroxybenzoate and catechol determines the sequential decomposition of phenolic compounds in mixed substrate cultivations in pseudomonads. FEMS Microbiol Ecol 37:79–89
Heinaru E, Merimaa M, Viggor S, Lehiste M, Leito I, Truu J, Heinaru A (2005) Biodegradation efficiency of functionally important population selected for bioaugmentation in phenol- and oil-polluted area. FEMS Microbiol Ecol 51:363–373
Hino S, Watanabe K, Takahashi N (1998) Phenol hydroxylase cloned from Ralstonia eutropha strain E2 exhibits novel kinetic properties. Microbiology 144:1765–1772
Jiang H-L, Tay ST-L, Maszenan AM, Tay J-H (2006) Physiological traits of bacterial strains isolated from phenol-degrading aerobic granules. FEMS Microbiol Ecol 57:182–191
Kivisaar M, Horak R, Kasak L, Heinaru A, Habicht J (1990) Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid 24(1):25–36
Kivisaar M, Kasak L, Nurk A (1991) Sequence of the plasmid-encoded catechol 1,2-dioxygenase-expressing gene, pheB, of phenol-degrading Pseudomonas sp. strain EST1001. Gene 98(1):15–20
Kiesel B, Müller RH (2002) The meta pathway as a potential energy-generating secuence and its effects on the growth rate during the utilisation of aromatics. Acta Biotechol 22(3–4):221–134
Kovárová- Kovar K, Egli T (1998) Growth kinetics of suspended microbial cells: from single-substrate-controlled growth to mixed-substrate kinetics. Microbiol Mol Biol Rev 62(3):646–666
Kukor JJ, Olsen RH (1990) Molecular cloning, characterization, and regulation of a Pseudomonas pickettii PKO1 gene encoding phenol hydroxylase and expression of the gene in Pseudomonas aeruginosa PAO1c. J Bacteriol 172:4624–4630
Kukor JJ, Olsen RH (1991) Genetic organization and regulation of a meta cleavage pathway for catechols produced from catabolism of toluene, benzene, phenol and cresols by Pseudomonas pickettii PKO1. J Bacteriol 173(15):4587–4594
Merimaa M, Heinaru E, Liivak M, Vedler E, Heinaru A (2006) Grouping of phenol hydroxylase and catechol 2,3-dioxygenase genes among phenol- and p-cresol-degrading Pseudomonas species and biotypes. Arch Microbiol 186:287–296
Müller RH, Babel W (1996) Growth rate-dependent expression of phenol-assimilation pathways in Alcaligenes eutrophus JMP 134—the influence of formate as an auxiliary energy source on phenol conversion characteristics. Appl Microbiol Biotechnol 46:156–162
Ng LC, Shingler V, Sze CC, Poh CL (1994) Cloning and sequences of the first eight genes of the chromosomally encoded (methyl) phenol degradation pathway from Pseudomonas putida P35X. Gene 151(1–2):29–36
Nurk A, Kasak L, Kivisaar M (1991) Sequence of the gene (pheA) encoding phenol monooxygenase from Pseudomonas sp. EST1001: expression in Escherichia coli and Pseudomonas putida. Gene 102(1):13–18
Ornston LN (1971) Regulation of catabolic pathways in Pseudomonas. Bacteriol Rev 35(2):87–116
Ornston LN, Stanier RY (1966) The conversion of catechol and protocatechuate to beta-ketoadipate by Pseudomonas putida. I. Biochemistry. J Biol Chem 241(16):3776–3786
Peters M, Heinaru E, Talpsep E, Wand H, Stottmeister U, Heinaru A, Nurk A (1997) Acquisition of a deliberately introduced phenol degradation operon, pheBA, by different indigenous Pseudomonas species. Appl Environ Microbiol 63(12):4899–4906
Pessione E, Divari S, Griva E, Cavaletto M, Rossi GL, Gilardi G, Gilunta C (1999) Phenol hydroxylase from Acinetobacter radioresistens is a multicomponent enzyme. Purification and characterization of the reductase moiety. Eur J Biochem 265(2):549–555
Pieper DH, Stadler-Fritzsche K, Knackmuss H-J, Timmis KN (1995) Formation of dimethylmuconolactones from dimethylphenols by Alcaligenes eutrophus JMP 134. Appl Environ Microbiol 61(6):2159–2165
Polymenakou PN, Stephanou EG (2005) Effect of temperature and additional carbon sources on phenol degradation by an indigenous soil Pseudomonad. Biodegradation 16:403–413
Shingler V, Franklin CH, Tsuda M, Holroyd D, Bagdasarian M (1989) Molecular analysis of a plasmid-encoded phenol hydroxylase from Pseudomonas CF600. J Gen Microbiol 135:1083–1092
Takeo M, Maeda Y, Okada H, Miyama K, Mori K, Ike M, Fujita M (1995) Molecular cloning and sequencing of the phenol hydroxylase gene from Pseudomonas putida BH. J Ferment Bioeng 79(5):485–488
Vollmer MD, Hoier H, Hecht H-J, Schell U, Gröning J, Goldman A, Schlömann M (1998) Substrate specificity of and product formation by muconate cycloisomerases: an analysis of wild-type enzymes and engineered variants. Appl Environ Microbiol 64(9):3290–3299
Watanabe K, Teramoto M, Futamata H, Harayama S (1998) Molecular detection, isolation, and physiological characterization of functionally dominant phenol-degrading bacteria in activated sludge. Appl Environ Microbiol 64(11):4396–4402
Watanabe K, Futamata H, Harayama S (2002) Understanding the diversity in catabolic potential of microorganisms for the development of bioremediation strategies. Antonie Van Leeuwenhoek 81:655–663
Whiteley AS, Wiles S, Lilley AK, Philp J, Bailey MJ (2001) Ecological and physiological analyses of Pseudomonad species within a phenol remediation system. J Microbiol Methods 44:79–88
Williams PA, Shaw LE (1997) mucK, a gene in Acinetobacter calcoaceticus ADP1 (BD413), encodes the ability to grow on exogenous cis,cis-muconate as the sole carbon source. J Bacteriol 179(18):5935–5942
Zwietering MH, Jongenburger I, Rombouts FM, van´t Riet K (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56(6):1875–1881
Acknowledgments
The Estonian Science Foundation, Grant 5682, and the Institute of Molecular and Cell Biology, University of Tartu, supported this research. We gratefully acknowledge Prof. I. Leito, Dr. T. Alamäe and Dr. K. Orupõld for helpful discussions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Viggor, S., Heinaru, E., Künnapas, A. et al. Evaluation of different phenol hydroxylase-possessing phenol-degrading pseudomonads by kinetic parameters. Biodegradation 19, 759–769 (2008). https://doi.org/10.1007/s10532-008-9180-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10532-008-9180-8