Abstract
Effective conservation of rare species requires reasonable knowledge of population locations. However, surveys for rare species can be time-intensive and therefore expensive. We test a methodology using stacked species distribution models (S-SDMs) to efficiently discover the greatest number of new rare species’ occurrences possible. We used S-SDMs for 22 rare plant species in southern Ontario, Canada to predict the best survey locations among individual 1-ha cells. For each cell, we weighted distribution model outputs by accuracy and species rarity to create an efficiency value. We used these efficiency values as an index to determine the locations of our field surveys. We conducted field surveys in multi-species cells, “MSC” (areas with high predicted efficiency for multiple species) and single species cells, “SSC” (areas with high probability for only one species) to determine the relative efficiency of a multi-species survey approach. MSC were more than twice as likely as SSC to have at least one rare plant species discovered. Efficiency ranks were also useful in directing surveyors toward incidental discoveries of other rare species that were not modeled. Our technique of using S-SDMs can help direct surveys to more efficiently find rare species occurrences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
To effectively monitor and protect rare species, we must know the geographic locations of their populations, but many rare species lack precise distribution information. Field surveys are needed to fill these gaps (Peterson et al. 2011), but they are time-consuming and expensive (Lindenmayer et al. 2013). Thus, reliable protocols are needed to help efficiently direct survey efforts.
Species distribution models have recently proliferated as conservation tools (Guisan et al. 2013). By using species occurrence data along with environmental or spatial predictors, the habitat suitability or probability of occurrence for a species can be predicted across an area of interest (Guisan and Zimmerman 2000; Elith et al. 2006). SDMs have also been used to predict current species richness (e.g. Guisan and Theurillat 2000; Newbold et al. 2009; Parvianen et al. 2009), predict future species richness with changing climate (Thuiller et al. 2005), predict shifts in species’ distributions (e.g. Thuiller 2004; Elith et al. 2010; McKenney et al. 2015), assess the efficacy of current protected areas and propose locations for new ones (Loiselle et al. 2003; Koch et al. 2017; Amaral et al. 2017), prescribe management for invasive species (Bennett 2014), and find new occurrences of rare species (e.g. Williams et al. 2009; Rebelo and Jones 2010; Peterson et al. 2011; McCune 2016).
Outputs of SDMs for individual species can be stacked together to create one composite map (Ferrier and Guisan 2006); this technique is often referred to as S-SDM (where the “S” stands for stacking; Dubuis et al. 2011). S-SDMs offer an effective method for highlighting areas of special conservation importance based on predictions of species richness of a taxonomic group of interest (Newbold et al. 2009; Yu et al. 2017), and have been used to predict threatened species richness (Parvianen et al. 2009; Koch et al. 2017). Knowledge of which areas have the highest potential concentration of rare species can point to promising survey sites, thus leading to the discovery of previously unknown rare species locations (Williams et al. 2009). This in turn increases the accuracy of the estimate of the number of rare species populations, which allows for a more accurate assessment of a species’ conservation status and better-informed management.
Two aspects that have begun to receive attention in the S-SDM literature are: (1) accuracy of SDMs, and (2) species’ conservation status. The accuracy of distribution models for individual species (not stacked) has been discussed extensively (e.g. Segurado and Araaujo 2004; Elith and Leathwick 2009). A model that overpredicts suitable habitat and makes commission errors will waste survey resources by sending surveyors to unsuitable areas that were predicted to be suitable, while a model that underpredicts suitable habitat and makes omission errors will cause surveyors to overlook areas where the species is present (Loiselle 2003). Stacking individuals models together in a S-SDM can lead to inaccurate predictions in the form of species richness (usually, but not always, as an overestimation, Guisan and Rahbek 2011), for which correction methods have been proposed (Calabrese et al. 2014; Del Toro et al. 2019; Pouteau et al. 2015). Fewer studies have accounted for individual model accuracy, within the resulting S-SDMs (but see Dunn et al. 2016). When working within an S-SDM framework, there will be variation in the accuracy of the individual models being stacked due to the differing characteristics of the species being modeled (Guisan et al. 2007; Syphard and Franklin 2010), the quality of the data for each species (e.g. Graham et al. 2004; Moudrý and Šímová 2012), and the relative prevalence of the species (Hernandez et al. 2006; Le Lay et al. 2010). Fernandes et al. 2018 examined the effects on the predictive accuracy of S-SDMs for virtual species by varying individual model components known to affect accuracy at the single model level and found that sampling size and modeling technique had the largest impacts. However, we have yet to see individual SDM accuracy for real species quantitatively estimated based on data independent of the records used to build the SDM, rigorously evaluated, and weighted within the S-SDM framework itself. In fact, assessing the accuracy of SDMs using independently collected presence and absence data is rarely done in any context. It is important to take individual model accuracy into account in the stacking process if the goal is to focus on survey locations with the highest probabilities of containing at least one of the species of interest, as would be the case in attempts at rare species discovery.
Secondly, many agencies prioritize management of species based on rarity (e.g. ESA 1973; SARA 2002), so knowing where less prevalent species are may be more important than knowing where more prevalent species are. Some modelers have noted this fact and have weighted individual models in S-SDMs by species’ relative rarity (Albuquerque and Beier 2016; Tukainen et al. 2017; Yu et al. 2017), or have used the habitat specificity of the species (Miličić et al. 2017) to highlight potential areas important for threatened species conservation. However, to our knowledge, no study has tested whether weighting individual model outputs by both rarity and accuracy within an S-SDM framework leads to an increased detection rate of priority threatened and rare species.
To do so, we built SDMs for 22 rare vascular plant species in southern Ontario, Canada and stacked them, weighting each individual model by accuracy, species conservation status, both, or neither. This weighting procedure allowed us to create an index based on the resulting efficiency values, from which we conducted field surveys in areas predicted to be suitable for one or more species to test the effectiveness of these S-SDM maps in leading to the discovery of new rare plant occurrences. We aimed to test (i) How much more likely are surveys to lead to the discovery of at least one rare species in cells with high predicted probability of occurrence for multiple rare species, compared to cells with high predicted probability of occurrence for only one species (i.e. how much do we gain in survey efficiency by stacking SDMs), and (ii) What effect does weighting individual species’ models by model accuracy and/or species rarity have on survey efficiency?
Methods
Study region and species
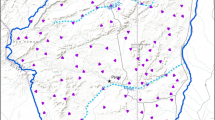
We focused our study on the forests of Southern Ontario (Fig. 1), predominantly in the Carolinian forest zone of the extreme southwest (Crins et al. 2009), which is characterized by deciduous canopy cover. We also located some study sites in the forests north and east of the Carolinian zone, which are characterized by mixed deciduous-evergreen cover (Crins et al. 2009). Southern Ontario is important for vascular plants at both the provincial and national level, as approximately 72% of Ontario’s and more than 40% of Canada’s plant species occur here (Oldham 2017), despite the dominance of urban development and agricultural land use (Crins et al. 2009). The total study area encompasses approximately 7 million hectares.
We initially selected 27 vascular plant species native to Ontario and growing in woodland habitats, based on their conservation importance, and their relative ease of identification in the field. We defined rare species as those ranked S1, S2, or S3 at the provincial level, corresponding to critically imperiled, imperiled, and vulnerable species (Faber-Langendoen et al. 2012, Table 1). Most of the species included here are at the northern edge of their range within southern Ontario, with large portions of their ranges extending south into the United States. The species represent a wide taxonomic range and include trees, shrubs, herbs and ferns. Nomenclature follows the Ontario Natural Heritage Information Centre (NHIC, https://www.ontario.ca/page/get-natural-heritage-information).
Building and evaluating individual models
We obtained presence-only occurrence records for each species from the NHIC. These records include herbarium records, field surveys by Ministry of Natural Resources (MNRF) biologists, and other confirmed sightings. We used only records that had < 100 m accuracy to build the SDMs, to correspond with the resolution of the environmental variables (see below). The total number of occurrence records we used to build the models ranged from 4 to 1594 with a mean of 83.7 records per species and a median of 17.5 (Table 1). Species with occurrence records on the lower end of this range are potentially subject to overfitting (Merow et al. 2013); however, SDMs can perform well even with sample sizes as low as 5 (Hernandez et al. 2006; Pearson et al. 2007; van Proosdij et al. 2016) and we are specifically studying rare species with limited available records. Additionally, species that have lower prevalence (i.e. percent area occupied within the study region) require fewer records than higher prevalence species to achieve similar model accuracy (van Proosdij et al. 2016). In the case of most of the species we modeled here, they can reasonably be assumed to have low prevalence within our study region given the infrequency of sightings (see results for further validation of this assumption). Records date from 1897 to 2012. We considered high spatial accuracy to be the most important criterion for including occurrence records. However, we explored the effect of record age on the performance of SDMs and found minimal effect on model accuracy when using records with < 100 m spatial accuracy (unpublished data). While we only used the partial range for each species in their respective SDMs (the study region as opposed to their entire extent of occurrence), our study region represents the edge of the range for most species included here, which has been found to produce more accurate model results than including the complete range (Luoto et al. 2005). Because we evaluated each SDM prior to including it in the S-SDM based on independently collected presence and absence records, we are able to exclude SDMs that had low accuracy as a result of few occurrence records, sampling bias in the records, or any of the many other factors that can influence SDM accuracy (see Merow et al. 2013).
We collected data on climatic, topographic, soil, and surficial geology environmental variables to use as predictors in the models (Online Resource 1). Previous research (McCune 2016) suggests that forest type and the amount of forest on the landscape surrounding a site can influence whether a threatened plant species is present. Therefore, we also collected data on forest contiguity (number of 1-hectare cells out of the 9 × 9 cell area immediately surrounding the focal cell that are forested) and land cover type (deciduous forest, mixed forest, swamp, etc.) across the study region. We tested for multicollinearity between environmental predictors and used only those that were not highly correlated (r < 0.7), following Gogol-Prokurat (2011). Where 2 variables were correlated at r > 0.7, the more generalized variable was used (e.g., mean annual temperature and mean temperature of the warmest quarter were correlated so the former was kept). We resampled each variable to a 100 m × 100 m resolution using the Resample function in ArcGIS and the “Majority” resampling technique (see Online Resource 1 for original spatial resolutions for each variable).
Because the occurrence data were presence-only records and were limited in number, we chose to use Maxent (Phillips et al. 2006) to build SDMs. Maxent has been shown to perform very well using a range of performance measures (Elith and Graham 2009), even when few species occurrence records are available (Hernandez et al. 2006; Pearson et al. 2007; van Proosdij et al. 2016). We built 8 different SDMs for each species (Fig. 2). As a beginning foundation, all models for each species contained the climatic, topographic, soil, and surficial geology predictor variables (14 predictors). Additionally, we built models for each species that included either the forest contiguity or land cover variables or both to determine if these less commonly used variables would help improve the accuracy of models for any species. We ran each of these model types twice: once with a regularization multiplier set to 1 and once with regularization set to 0.5. The regularization parameter can correct for model overfitting, with higher values allowing for a more generous inclusion of predicted suitable habitat and lower values providing a more conservative estimate of predicted suitable habitat. Given the desire to narrow down the area for potential survey sites as much as possible, we chose to test a lower regularization parameter value in addition to the default value of 1.Testing different regularization parameters is recommended, in order to arrive at the best-performing model (Merow et al. 2013). We set aside a random subsample of 25% of available records to test each model and fit the model 10 times with different model fitting and test subsamples, making the final result an average of the outputs from the subsamples. We used Maxent’s cumulative output in which each grid cell receives a score from 0 to 100, which can be interpreted as the percentage of cells that have a value equal to or less than that cell’s value (Merow et al. 2013). We set features in Maxent to auto (which includes linear, quadratic, product, threshold, and hinge functions), jackknife to measure variable important was selected, and set the background as the complete study area. We set the random test percentage to twenty-five percent with replicated run type of subsample.
We evaluated the models with independent presence and absence data originating from two sources. First, we obtained independent presences from the NHIC’s central holdings database, which consists of records that have not yet been added to the main database of occurrences. Second, we obtained independent presences and absences from 2014 and 2015 field data in which botanists surveyed 156 100 m × 100 m cells throughout Southern Ontario predicted to be suitable for one or more rare plant species (see McCune 2016; McCune et al. 2017). We excluded any independent presence located within the same grid cell as a record used to build the SDM, using the raster package in R (R Core Team 2016; Hijmans and van Etten 2017). We also excluded absences if the grid cell was surveyed outside the time of year during which the plant is present and identifiable.
We chose one SDM for each species based on three criteria: (1) first, we chose the model with the highest sensitivity (the percent of actual presences correctly classified as presences by the model) based on calculations with the independent data (i.e. the model that predicted the highest number of independent presence records as suitable) using a threshold that achieved 10% omission of training presences for species with at least 15 records, and 0% omission (i.e. threshold = minimum training presence suitability) for species with fewer than 15 records.; (2) if more than one model was tied for the highest sensitivity, we chose the model with the highest AUC (a threshold independent measure of model performance, Fielding and Bell 1997); (3) if there was still a tie between models, we chose the model with the lowest area predicted suitable (Engler et al. 2004). We considered sensitivity to be the most important measure of predictive performance for this study because we wanted to minimize the number of false negatives produced by each model. If an area is falsely characterized as unsuitable, then it will not be surveyed and rare occurrences will be missed as a result of omission error (Liu et al. 2016). We used a 0% omission rate for species with very few occurrences because with fewer known occurrences, it is less likely that the lowest 10% suitability values represent areas unsuitable for the species; this approach is recommended by Pearson et al. (2007). We chose model AUC as a selection factor because of its independence from a threshold, its ubiquitous use by modelers, and its generally effective measure of model discrimination ability (e.g. Pearce and Ferrier 2000). Smallest suitable area was also used as a selection factor because narrowing down the area considered suitable for a species narrows down the potential survey area, an important consideration when time is a limiting factor.
It is important to recognize that the MaxEnt predicted habitat suitability does not necessarily correlate linearly with species probability of occurrence (Gogol-Prokurat 2011; Vaughan and Ormerod 2005). Therefore, we converted MaxEnt suitability outputs from the best SDM for each species (which range from values of 0 to 100) to estimated probabilities of occurrence (0 to 1 value range) based on the independent presence and absence data described above, using generalized linear models (GLMs) with a binomial link function. Gogol-Prokurat (2011) tested for a linear relationship between SDM output and probability of occurrence using a modified Hosmer–Lemeshow deciles of risk test. However, we recognize that these relationships may be significant but not linear, so we used binomial GLMs. We extracted Maxent suitability values for each presence and absence location, using the suitability values as the explanatory factor in the logistic model. We used the resulting GLMs to create new rasters for each species in which each cell represents the probability of occurrence of that species in that cell. This allowed us to estimate the likelihood of species presence in a cell based on independent data not used to build the SDM. To test the usefulness of the GLM for predicting species probability of occurrence, we compared it to an intercept-only model and deemed habitat suitability a significant predictor of probability of occurrence when ΔAIC was less than 2 (Burnham and Anderson, 2002).
S-SDM efficiency maps
We used only the species that had acceptable AUC values and GLM probability of occurrence models that were significantly better than an intercept-only model (ΔAIC > 2; Burnham and Anderson 2002) in the stacking process, to avoid using models that were not useful for predicting species distributions. We defined acceptable AUC values as those ≥ 0.6 since an AUC value of 0.5 is no better than random. Although a common practice is to consider AUC values ≥ 0.7 as adequate (Swets 1988), we adopted 0.6 as the cutoff because the species modeled here are rare, and we assumed that any information that is better than random is potentially important for creating useful models that could be used in management. Additionally, evaluating the models with independently collected data from field surveys (as opposed to data separated from the original dataset) further helped to ensure all models included in the stack produced reasonable results.
To create the efficiency maps, we stacked the estimated probability of occurrence model outputs for these 22 species in four ways: (1) individual estimated probability of occurrence maps added together with no weighting; (2) probabilities of occurrence added together and weighted by accuracy of the SDM (as measured by AUC) and by the S-rank (threat level) of the species, using the following equation:
where Ei is the resulting Efficiency Index of cell i, Pij is the estimated probability of occurrence of species j occurring in cell i, Rj is the rarity weight (i.e., provincial S-rank where S1 = 3, S2 = 2, S3 = 1) of species j, and Aj is the accuracy of the SDM for species j as measured by AUC; (3) models weighted by accuracy but not by rarity; and (4) models weighted by rarity but not by accuracy. We used a rarity weighting because rarer species are of greater priority and urgency for locating new populations. We used an accuracy weighting because when field survey time is limited, it is best to visit areas with greater confidence in the model predicting the species’ probability of occurrence.
Field surveys
We selected candidate cells for surveys that were either highly suitable for multiple species or suitable for just one species. We term the former multi-species cells (MSC) and the latter single-species cells (SSC). There has been some debate about the appropriateness of applying a suitability threshold and obtaining binary predicted presence/absence maps for individual species (bS-SDM) compared to using raw estimated probability of occurrence values (pS-SDM) (Guisan and Rahbek 2011; Calabrese et al. 2014; D’Amen et al. 2015); we used both methods in identifying MSC and SSC. Specifically, we defined MSC as being suitable for 2 or more species using a threshold allowing for a 10% omission rate for species with greater than 15 records used to build their SDM, and a 0% omission rate for species with fewer than 15 records (bS-SDM), while also having an efficiency index within the top 5% of all grid cells according to the s-SDM weighted by both accuracy and rarity (pS-SDM) (Parviainen et al. 2009). These two constraints taken together ensured that cells were not chosen for surveys that had low to moderate probability of species presence for many species. We defined SSC as being suitable for only one species (using the same omission rate threshold rules as defined above) independent of the cell’s efficiency value. We attempted to survey the same number of MSC and SSC within each tertiary level watershed (subdivisions of secondary watersheds which are mostly made up of large river systems) to reduce spatial segregation of MSC and SSC. On each survey day, we randomly chose several cells among the possible MSC/SSC within a given watershed and surveyed the first site for which we could obtain landowner permission. Because of the limited amount of time available to conduct these searches, both in our study and commonly in practice by field botanists, we did not survey sites with low habitat suitability for all species. These sites are unlikely to harbor any of our modeled species, and the goal of our study was to test the efficiency of S-SDMs versus single SDMs to direct surveys for rare species. Thus, these would not have been useful to survey.
We surveyed 70 cells (including 33 MSC and 37 SSC) on privately owned sites as well as protected areas (e.g. Nature Conservancy of Canada, Nature Trust, or Provincial Park land), between May and August 2017. We obtained written or verbal permission from the landowners for all privately owned sites, and research permits for protected areas. At each site, we navigated to the center of the 100 m by 100 m grid cell using a handheld GPS unit and used flagging tape and a compass to delineate four quadrants. The field team included one to four people with at least one trained botanist present. We walked the entire square grid cell systematically, recording all vascular plant species observed. Each survey lasted 2.5–5 h.
Comparison of S-SDM methods
We built logistic regression models to test the significance of the relationship between the efficiency values (four versions for each surveyed cell) and the probability of finding any rare plant species at that location based on our field surveys. We also tested for differences between MSC and SSC in the total, native, and exotic species richness recorded, using t-tests assuming unequal variances. We performed all data analysis in R 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria 2016).
Results
The independent AUC value for the best MaxEnt models for each species ranged from 0.442 to 0.999 with a mean of 0.83 (Online Resource 2). Five species were excluded from further analysis due to low model accuracy: three based on low independent AUC values and two based on ΔAIC < 2 (Burnham and Anderson 2002) when comparing their GLMs to an intercept-only model. This left 22 species included in the S-SDM efficiency maps.
Of the 70 cells surveyed, 22 had at least one rare plant species (ranked S1, S2, or S3). Fifteen had one species, five plots had 2, and two plots had 3. We found a total of 30 occurrences of 17 rare plant species. Only 4 out of these 17 species were those we modeled and incorporated into the efficiency maps (Castanea dentata, Celtis tenuifolia, Cornus florida, and Lithospermum latifolium), the rest being incidental discoveries of species not modeled by the SDMs (Online Resource 3).
The probability of finding at least one rare plant species was approximately double in MSC compared to SSC. Including incidental rare plant discoveries, MSC had a 42.4% success rate (presence of at least one rare species) while the SSC had a 21.6% success rate. Not including the incidental species, the MSC had an 18.2% success rate while the SSC had an 8.1% success rate (Fig. 3). Results were qualitatively the same when excluding SSC sites where we had no paired MSC. None of the measures of species richness (total, native, or exotic) were significantly different between the two site types.
a The percent of multi-species cell (MSC) and single species cell (SSC) plots that had at least one rare plant species discovered, either including or excluding incidental species discoveries (species which were not modeled) and b Visualization of logistic regression model showing estimated probability of presence of at least rare plant species across efficiency index values of the S-SDM weighted by both rarity and accuracy with 95% confidence intervals
GLMs relating the estimated probability of occurrence of at least one rare plant species to the efficiency index showed little difference among weighting procedures used to create the S-SDMs. All ΔAIC values were < 5 and percent deviance explained by the full models was similar (Table 2). There was a significant positive relationship in the logistic regression between the efficiency index values of all the S-SDMs and the probability of at least one rare species (target and incidental species included) being present (p = 0.01). Results were similar when only target species were included. There was no significant relationship between the calculated efficiency index values and the field-measured total species richness (p = 0.21, R2 = 0.01) or total species richness and estimated probability of occurrence of a rare species (p = 0.23).
Discussion
The efficiency maps we created by stacking the probability of occurrence model outputs for 22 rare plant species allowed for the discovery of new occurrences of 17 rare species. These discoveries were approximately twice as likely to occur in multi-species (MSC) sites, where multiple species were predicted to have suitable habitat, than in single-species (SSC) sites, where only one species was predicted to have suitable habitat. While we found new occurrences of only 4 out of the 22 species modeled, because we were working with rare species this discovery rate is not unusual (MacDougall and Loo 2002; Williams et al. 2009; McCune 2016). Our results point to the usefulness of pairing modeling with field surveys for uncovering previously unknown rare species occurrences, thereby increasing our knowledge of rare species distributions and habitat occupancy.
We found 11 rare plant species which were not modeled and not incorporated into the efficiency maps. This preponderance of incidental finds suggests that the efficiency index not only helps to find species explicitly modeled, but also those which are not modeled but also rare. Thus, the efficiency index is capturing something shared in the ecological niches of the species modeled as well as in some rare species that were not modeled. We noticed that the incidental rare species that we discovered tend to prefer mesic, moderately shaded, floodplain conditions in older woodlands. Therefore, our efficiency index may be especially helpful at highlighting these areas for surveys.
The most important variables included in the individual models according to percent contribution were surficial geology, land cover, and soil texture. The most important variables according to permutation importance (Online Resource 4) were annual mean temperature, precipitation of the warmest quarter, forest contiguity, and mean temperature of the growing season. It seems that these environmental variables largely dictate the presence or absence of rare species in our study area. The importance of the land cover variable to the performance of the majority of the models is of particular note given the unresolved question of its inclusion as an appropriate and useful predictor in SDMs; of especial concern can be its categorical (rather than continuous) nature and its resolution size in relation to species’ scale of interaction with their environments (Bucklin et al. 2015; Cord et al. 2014; Eskildsen et al. 2013; Wilson et al. 2013). Our results agree with those of Pearson et al. (2004), who found that adding land cover to their distribution models for plant species improved predictive performance Additionally, the results of Luoto et al. (2007) showed increased model accuracy including land cover as a predictor when spatial resolution was fine. These results also agree with those of this study given the high resolution of our variables and resulting cells within the models. It may be possible to even further improve model accuracy with the addition of continuous remote sensing predictors (Cord et al. 2014), however the good to excellent performance of most of our models indicates that a categorical land cover variable may be sufficient, especially for rare species that prefer distinct land cover type(s). We are not surprised that land cover was important for many of our modeled species, as it likely allowed MaxEnt to focus on areas of the proper forest type (coniferous, mixed, or deciduous) and remove areas of agricultural or urban land use.
The efficiency index performed better at predicting rare species presence than a simple measure of total species richness: there was no significant relationship between total species richness and probability of rare species presence in the logistic regression, while there was a significant positive relationship between the efficiency index and rare species probability of presence (Fig. 3b). This is important because, at least for rare plants in our study region, creating SDMs for a subset of rare species may be sufficient if the main goal is to locate new rare species sharing a given habitat type., rather than creating models to predict total species richness, which may require more information.
It must be noted that this efficiency index will not be useful for finding new rare plant species occurrences in every situation. Firstly, the efficiency index did not result in the discovery of any species ranked S1, those which are the most imperiled in the province. This is likely because these species are the least prevalent and thus any new population locations (if they exist) will be the most difficult to find. Le Lay et al. (2010) had similar results when attempting to discover new occurrences of two extremely rare plant species based on ensembles of SDMs.
Secondly, the efficiency index will not aid in the discovery of rare species that have very distinct habitat requirements and thus are not likely to occur in the MSC of our efficiency maps. The MSC represent areas that have especially suitable habitat based on the ecological requirements of many of the species modeled. Species that do not share these ecological requirements will not overlap in distribution. Consequently, new locations of this type of species will be more likely to occur in the SSC areas of the efficiency map. For example, Asplenium scolopendrium, a fern species that can only grow on limestone substrate (Oldham and Brinker 2009), was not found at any MSC locations we searched in 2017. There are contrasting results in the literature concerning whether or not species with distinct habitat requirements are more easily modeled with SDMs than generalists (Elith and Burgman 2002; Hernandez et al. 2006; Le Lay et al. 2010; Grenouillet et al. 2011; McCune 2016; Soultan and Safi 2017; Rhoden et al. 2017). If species with distinct habitats are in fact easily modeled, then their lack of discovery in our efficiency maps is not a major point of concern because any specialized species of particular interest can be separately modeled. The methodology presented here focuses on efficiently finding the most occurrences of rare plant species and not necessarily on finding individual species. If the latter is the desired conservation outcome, an individual SDM should be used.
Of course, our models, like all SDMs, were imperfect. For example, the field surveys that informed the models likely did not sample the full niche space, and the AUC of our models may not perfectly reflect model accuracy, in part due to this measure’s effective lowering of species prevalence (Raes and ter Steege 2007). However, our results clearly indicate that even such imperfect models can be very good at efficiently indicating potential survey sites for rare species.
Like Williams et al. (2009), we found that many surveyed sites with high probability of occurrence lacked our rare species, which is likely a result of factors other than habitat suitability leading to species absence. Dispersal limitation could act as one of these additional distribution determinants. Suitable habitat may exist outside of where the species of interest is currently found, but the species’ physical inability to spread to these areas through seeds or spores restricts its distribution to a smaller area. Previous research has found differences in the amount of predicted suitable habitat when dispersal is or is not accounted for in plant distribution models, with less habitat available when dispersal is restricted (Krause et al. 2015). Given that the ability of plants to disperse is limited and that dispersal can be especially problematic for rare plant species (Primack and Miao 1992), dispersal limitations should be kept in mind when creating models for rare plant species. Another limit to plant species real distributions beyond the abiotic environment could be biotic interactions, including pollinators (Giannini et al. 2013), pathogens (Bueno de Mesquita et al. 2016), and competition from other plant species (Meier et al. 2010; Pellissier et al. 2010). Modelers may also choose to include the distributions of one or more interacting species to create more realistic models.
Our results did not show a clear difference among the weighting systems (species S-rank, model accuracy, both, neither) in predicting the presence of at least one rare plant species using our ΔAIC criterion. However, there was a strong relationship between the efficiency index and the estimated probability of occurrence of rare species. Thus, although the stacking of individual model outputs was useful for discovering new rare plant occurrences, the weighting of model outputs by threat level and model accuracy was unnecessary in this study for the goal of field site survey prioritization. Dunn et al. (2016) also found that weighting SDMs did not change the areas highlighted for conservation very much compared to unweighted SDM stacking. This does not mean that model weighting by either threat level or accuracy should be completely discounted for future S-SDM analysis. It is possible that a similar weighting system used in another study area, with a different combination of species, or with more surveyed cells would have different results.
References
Albuquerque F, Beier P (2016) Predicted rarity-weighted richness, a new tool to prioritize sites for species representation. Ecol Evol 6:8107–8114
Amaral AG, Munhoz CB, Walter BM, Aguirre-Gutiérrez J, Raes N (2017) Richness pattern and phytogeography of the Cerrado's herb-shrub flora and implications for conservation. J Veg Sci 28:848–858
Bennett JR (2014) Comparison of native and exotic distribution and richness models across scales reveals essential conservation lessons. Ecography 37:120–129
Bucklin DN, Basille M, Benscoter AM, Brandt LA, Mazzotti FJ, Romanach SS, Speroterra C, Watling JI (2015) Comparing species distribution models constructed with different subsets of environmental predictors. Divers Distrib 21:23–35
Bueno de Mesquita CP, King AJ, Schmidt SK, Farrer EC, Suding KN (2016) Incorporating biotic factors in species distribution modeling: are interactions with soil microbes important? Ecography 39(10):970–980
Burnham KP, Anderson DR (2002) Model selection and multi-model inference: a practical information-theoretic approach, 2nd edn. Springer-Verlag, New York, USA
Calabrese JM, Certain G, Kraan C, Dormann CF (2014) Stacking species distribution models and adjusting bias by linking them to macroecological models. Glob Ecol Biogeogr 23:99–112
Cord AF, Klein D, Mora F, Dech S (2014) Comparing the suitability of classified land cover data and remote sensing variables for modeling distribution patterns of plants. Ecol Model 272:129–140
Crins WJ, Gray PA, Uhlig PWC, Wester MC (2009) The ecosystems of Ontario, part I: ecozones and ecoregions. Ontario Ministry of Natural Resources, Peterborough, ON
D’Amen M, Dubuis A, Fernandes RF, Pottier J, Pellissier L, Guisan A (2015) Using species richness and functional traits predictions to constrain assemblage predictions from stacked species distribution models. J Biogeogr 42:1255–1266
Del Toro I, Ribbons RR, Hayward J, Andersen AN (2019) Are stacked species distribution models accurate at predicting multiple levels of diversity along a rainfall gradient? Austral Ecol 44(1):105–113
Dunn JC, Buchanan GM, Stein RW, Whittingham MJ, McGowan PJ (2016) Optimising different types of biodiversity coverage of protected areas with a case study using Himalayan Galliformes. Biol Conserv 196:22–30
Dubuis A, Pottier J, Rion V, Pellissier L, Theurillat JP, Guisan A (2011) Predicting spatial patterns of plant species richness: a comparison of direct macroecological and species stacking modelling approaches. Divers Distrib 17:1122–1131
Elith J, Burgman MA (2002) Predictions and their validation: rare plants in the Central Highlands, Victoria, Australia. In: Scott JM, Heglund PJ, Morrison ML, Haufler JB, Raphael MG, Wall WA, Samson FB (eds) Predicting species occurrences: issues of accuracy and scale. Island Press, Washington, DC, pp 303–313
Elith J, Graham CH (2009) Do they? How do they? WHY do they differ? On finding reasons for differing performances of species distribution models. Ecography 32:66–77
Elith J et al (2006) Novel methods improve prediction of species' distributions from occurrence data. Ecography 29:129–151
Elith J, Kearney M, Phillips S (2010) The art of modelling range-shifting species. Methods Ecol Evol 1:330–342
Elith J, Leathwick JR (2009) Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst 40:677–697
Engler R, Guisan A, Rechsteiner L (2004) An improved approach for predicting the distribution of rare and endangered species from occurrence and pseudo-absence data. J Appl Ecol 41:263–274
ESA (Endangered Species Act) (1973) Endangered Species Act of 1973, enacted through the 93rd United States Congress. U.S. Department of the Interior
Eskildsen A, le Roux PC, Heikkinen RK, Høye TT, Kissling WD, Pöyry J, Wisz MS, Luoto M (2013) Testing species distribution models across space and time: high latitude butterflies and recent warming. Glob Ecol Biogeogr 22:1293–1303
Faber-Langendoen D, Nichols J, Master L, Snow K, Tomaino A, Bittman R, Hammerson G, Heidel B, Ramsay L, Teucher A, Young B (2012) NatureServe Conservation Status Assessments: methodology for assigning ranks. NatureServe, Arlington, VA
Fernandes RF, Scherrer D, Guisan A (2018) How much should one sample to accurately predict the distribution of species assemblages? A virtual community approach. Ecol Inform 48:125–134
Ferrier S, Guisan A (2006) Spatial modelling of biodiversity at the community level. J Appl Ecol 43:393–404
Fielding AH, Bell JF (1997) A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ Conserv 24:38–49
Giannini TC, Chapman DS, Saraiva AM, Alves-dos-Santos I, Biesmeijer JC (2013) Improving species distribution models using biotic interactions: a case study of parasites, pollinators and plants. Ecography 36(6):649–656
Gogol-Prokurat M (2011) Predicting habitat suitability for rare plants at local spatial scales using a species distribution model. Ecol Appl 21:33–47
Graham CH, Ferrier S, Huettman F, Moritz C, Peterson AT (2004) New developments in museum-based informatics and applications in biodiversity analysis. Trends Ecol Evol 19:497–503
Grenouillet G, Buisson L, Casajus N, Lek S (2011) Ensemble modelling of species distribution: the effects of geographical and environmental ranges. Ecography 34:9–17
Guisan A, Rahbek C (2011) SESAM—a new framework integrating macroecological and species distribution models for predicting spatio-temporal patterns of species assemblages. J Biogeogr 38:1433–1444
Guisan A et al (2013) Predicting species distributions for conservation decisions. Ecol Lett 16:1424–1435
Guisan A, Theurillat JP (2000) Equilibrium modeling of alpine plant distribution: how far can we go? Phytocoenologia 30:353–384
Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecol Model 135:147–186
Guisan A, Zimmermann N, Elith J, Graham C, Phillips S, Peterson A (2007) What matters for predicting spatial distributions of tree occurrences: techniques, data, or species' characteristics. Ecol Monogr 77:615–630
Hernandez PA, Graham CH, Master LL, Albert DL (2006) The effect of sample size and species characteristics on performance of different species distribution modelling methods. Ecography 29:773–785
Hijmans RJ, van Etten J (2017). raster: geographic analysis and modeling with raster data. https://CRAN.R-project.org/package=raster
Krause CM, Cobb NS, Pennington DD (2015) Range shifts under future scenarios of climate change: dispersal ability matters for Colorado Plateau endemic plants. Nat Areas J 35(3):428–438
Koch R, Almeida-Cortez JS, Kleinschmit B (2017) Revealing areas of high nature conservation importance in a seasonally dry tropical forest in Brazil: combination of modelled plant diversity hot spots and threat patterns. J Nat Conserv 35:24–39
Le Lay G, Engler R, Franc E, Guisan A (2010) Prospective sampling based on model ensembles improves the detection of rare species. Ecography 33:1015–1027
Lindenmayer DB, Piggott MP, Wintle BA (2013) Counting the books while the library burns: why conservation monitoring programs need a plan for action. Front Ecol Environ 11:549–555
Liu C, Newell G, White M (2016) On the selection of thresholds for predicting species occurrence with presence-only data. Ecol Evol 6:337–348
Loiselle BA, Howell CA, Graham CH, Goerck JM, Brooks T, Smith KG, Williams PH (2003) Avoiding pitfalls of using species distribution models in conservation planning. Conserv Biol 17:1591–1600
Luoto M, Pöyry J, Heikkinen RK, Saarinen K (2005) Uncertainty of bioclimate envelope models based on the geographical distribution of species. Glob Ecol Biogeogr 14:575–584
Luoto M, Virkkala R, Heikkinen RK (2007) The role of land cover in bioclimatic models depends on spatial resolution. Glob Ecol Biogeogr 16:34–42
MacDougall A, Loo J (2002) Land use history, plant rarity, and protected area adequacy in an intensively managed forest landscape. J Nat Conserv 10:171–183
McCune JL (2016) Species distribution models predict rare species occurrences despite significant effects of landscape context. J Appl Ecol 53:1871–1879
McCune JL, Van Natto A, MacDougall AS (2017) The efficacy of protected areas and private land for plant conservation in a fragmented landscape. Landsc Ecol 32:871–882
McKenney DW, Pedlar JH, Lawrence K, Papadopol P, Campbell K (2015) Hardiness zones and bioclimatic modelling of plant species distributions in North America. Acta Hortic 1085:139–148
Meier ES, Kienast F, Pearman PB, Svenning JC, Thuiller W, Araújo MB, Guisan A, Zimmermann NE (2010) Biotic and abiotic variables show little redundancy in explaining tree species distributions. Ecography 33(6):1038–1048
Merow C, Smith MJ, Silander JA (2013) A practical guide to MaxEnt for modeling species’ distributions: what it does, and why inputs and settings matter. Ecography 36:1058–1069
Miličić M, Vujić A, Jurca T, Cardoso P (2017) Designating conservation priorities for Southeast European hoverflies (Diptera: Syrphidae) based on species distribution models and species vulnerability. Insect Conserv Divers 10:354–366
Moudrý V, Šímová P (2012) Influence of positional accuracy, sample size and scale on modelling species distributions: a review. Int J Geogr Inf Sci 26:2083–2095
Newbold T, Gilbert F, Zalat S, El-Gabbas A, Reader T (2009) Climate-based models of spatial patterns of species richness in Egypt’s butterfly and mammal fauna. J Biogeogr 36:2085–2095
Oldham MJ, Brinker SR (2009) Rare vascular plants of Ontario, 4th edn. Natural Heritage Information Centre, Ontario Ministry of Natural Resources, Peterborough, ON
Oldham MJ (2017) List of the Vascular Plants of Ontario’s Carolinian Zone (Ecoregion 7E). Carolinian Canada and Ontario Ministry of Natural Resources and Forestry, Peterborough, ON
Parviainen M, Marmion M, Luoto M, Thuiller W, Heikkinen RK (2009) Using summed individual species models and state-of-the-art modelling techniques to identify threatened plant species hotspots. Biol Conserv 142:2501–2509
Pearce J, Ferrier S (2000) Evaluating the predictive performanceof habitat models developed using logistic regression. Ecol Model 133:225–245
Pearson RG, Raxworthy CJ, Nakamura M, Peterson AT (2007) Predicting species distributions from small numbers of occurrence records: a test case using cryptic geckos in Madagascar. J Biogeogr 34:102–117
Pellissier L, Anne Bråthen K, Pottier J, Randin CF, Vittoz P, Dubuis A, Yoccoz NG, Alm T, Zimmermann NE, Guisan A (2010) Species distribution models reveal apparent competitive and facilitative effects of a dominant species on the distribution of tundra plants. Ecography 33(6):1004–1014
Peterson AT, Soberon J, Pearson RG, Anderson RP, Martínez-Meyer E, Nakamura M, Araujo MB (2011) Ecological niches and geographic distributions. Princeton University Press, Princeton, NJ
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pouteau R, Bayle É, Blanchard É, Birnbaum P, Cassan JJ, Hequet V, Ibanez T, Vandrot H (2015) Accounting for the indirect area effect in stacked species distribution models to map species richness in a montane biodiversity hotspot. Divers Distrib 21:1329–1338
Primack RB, Miao SL (1992) Dispersal can limit local plant distribution. Conserv Biol 6(4):513–519
R Core Team (2016) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Raes N, ter Steege H (2007) A null-model for significance testing of presence-only species distribution models. Ecography 30:727–736
Rebelo H, Jones G (2010) Ground validation of presence-only modelling with rare species: a case study on barbastelles Barbastella barbastellus (Chiroptera: Vespertilionidae). J Appl Ecol 47:410–420
Rhoden CM, Peterman WE, Taylor CA (2017) Maxent-directed field surveys identify new populations of narrowly endemic habitat specialists. PeerJ 5:e3632. https://doi.org/10.7717/peerj.3632
SARA (Species at Risk Act) (2002) Bill C-5, an Act Respecting the Protection of Wildlife Species at Risk in Canada. Government of Canada, Ottawa, Ontario
Segurado P, Araujo MB (2004) An evaluation of methods for modelling species distributions. J Biogeogr 31:1555–1568
Soultan A, Safi K (2017) The interplay of various sources of noise on reliability of species distribution models hinges on ecological specialisation. PLoS ONE 12:e0187906. https://doi.org/10.1371/journal.pone.0187906
Swets JA (1988) Measuring the accuracy of diagnostic systems. Science 240:1285–1293
Syphard AD, Franklin J (2010) Species traits affect the performance of species distribution models for plants in southern California. J Veg Sci 21:177–189
Thuiller W (2004) Patterns and uncertainties of species' range shifts under climate change. Glob Change Biol 10:2020–2027
Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC (2005) Climate change threats to plant diversity in Europe. Proc Natl Acad Sci USA 102:8245–8250
Tukiainen H, Bailey JJ, Field R, Kangas K, Hjort J (2017) Combining geodiversity with climate and topography to account for threatened species richness. Conserv Biol 31:364–375
van Proosdij AS, Sosef MS, Wieringa JJ, Raes N (2016) Minimum required number of specimen records to develop accurate species distribution models. Ecography 39:542–552
Vaughan IP, Ormerod SJ (2005) The continuing challenges of testing species distribution models. J Appl Ecol 42:720–730
Williams JN, Seo C, Thorne J, Nelson JK, Erwin S, O’Brien JM, Schwartz MW (2009) Using species distribution models to predict new occurrences for rare plants. Divers Distrib 15:565–576
Wilson JW, Sexton JO, Jobe RT, Haddad NM (2013) The relative contribution of terrain, land cover, and vegetation structure indices to species distribution models. Biol Conserv 164:170–176
Yu F, Skidmore AK, Wang T, Huang J, Ma K, Groen TA (2017) Rhododendron diversity patterns and priority conservation areas in China. Divers Distrib 23:1143–1156
Acknowledgements
We thank J. Lloren, J. Pon, and C. Raymond for field work assistance. M. Oldham, T. Smith, and E. Snyder provided plant identification advice. The Natural Heritage Information Centre of Ontario provided the occurrence records for all species. We also thank the private landowners who allowed us access to their woodlots, as well as to The Nature Conservancy of Canada, Ontario Nature, the Province of Ontario, and the University of Waterloo, for granting permits to access protected areas. This research was funded by the Ontario Ministry of Natural Resources and Forestry’s Species at Risk Stewardship Fund, the Natural Science and Engineering Research Council of Canada (NSERC) through a Postdoctoral Fellowship to JLM and a Discovery Grant to JRB, and a Liber Ero fellowship to JLM.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Daniel Sanchez Mata.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rosner-Katz, H., McCune, J.L. & Bennett, J.R. Using stacked SDMs with accuracy and rarity weighting to optimize surveys for rare plant species. Biodivers Conserv 29, 3209–3225 (2020). https://doi.org/10.1007/s10531-020-02018-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10531-020-02018-1