Abstract
Do host invaders and their associated symbiont co-invaders have different genetic responses to the same invasion process? To answer this question, we compared genetic patterns of native and exotic populations of an invasive symbiont-host association. This is an approach applied by very few studies, of which most are based on parasites with complex life cycles. We used the mitochondrial genetic marker cytochrome oxidase subunit I (COI) to investigate a non-parasitic freshwater ectosymbiont with direct life-cycle, low host specificity and well-documented invasion history. The study system was the crayfish Procambarus clarkii and its commensal ostracod Ankylocythere sinuosa, sampled in native (N American) and exotic (European) ranges. Results of analyses indicated: (1) higher genetic diversity in the symbiont than its host; (2) genetic diversity loss in the exotic range for both species, but less pronounced in the symbiont; (3) native populations genetically structured in space, with stronger patterns in the symbiont and (4) loss of spatial genetic structure in the exotic range in both species. The combination of historical, demographic and genetic data supports a higher genetic diversity of source populations and a higher propagule size that allowed the symbiont to overcome founder effects better than its host co-invader. Thus, the symbiont might be endowed with a higher adaptive potential to new hosts or off-host environmental pressures expected in the invasive range. We highlight the usefulness of this relatively unexplored kind of symbiont-host systems in the invasion context to test important ecological and evolutionary questions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Biological invasions serve as unplanned experiments for understanding fundamental ecological and evolutionary processes (Sax et al. 2007). The potential harmful effects of small population sizes, an important concern in conservation genetics, have been tested in the context of invasions, leading to a genetic paradox (Allendorf and Lundquist 2003). Invasive species are expected to suffer a bottleneck during the invasion process due to founder effects. Conservation genetics predicts that the loss of genetic diversity from a bottleneck will produce two adverse effects: inbreeding depression will decrease fitness, and the reduced genetic diversity will limit the ability of invasive species to evolve in the new environment. Contradicting the theory, invasive species often overcome these problems and, in some cases, are favoured by rapid evolution (Huey et al. 2000).
Empirical data from both plants (Novak and Mack 2005) and animals (Wares et al. 2005) show that, in practice, invasive species often do not appear to be affected by founder effects. The most important mechanism invoked to explain this phenomenon is propagule pressure (Lockwood et al. 2005), which has two components: propagule size (i.e. number of individuals in any one release event) and propagule number (number of release events). Native populations are often genetically structured, so that in the invasive range, multiple propagule events from different source populations promote admixture, which increases within-population and reduces among-population genetic variance (Kolbe et al. 2008). In the particular case of a unique invasion event or a low number of events, propagule size (together with genetic diversity of source populations and exotic population growth rates) is expected to play a major role in determining the initial effective population size of the invasive species and, therefore, the size of the bottleneck effects the invader will suffer (Cox 2004).
Co-invasions by host organisms and their symbionts may provide novel insights into these issues. Symbiont-host associations are subjected to macro and micro-evolutionary processes that make them models of special interest for evolutionary ecologists (Poulin 2007). The co-evolution of symbiont-host systems has produced a great diversity of interaction types (a continuous gradient between parasitism and mutualism), symbiont life cycles (from very simple to highly complex, including various intermediate hosts), reproductive modes (including hermaphroditism, gonochorism and parthenogenesis), dispersal mechanisms (depending on the kinds of hosts exploited and off-host dispersal) and levels of host specificity (Poulin and Morand 2005). All of these factors may influence genetic diversity and population structure, and consequently the evolutionary responses of the symbiont-host system. For example, during the co-evolutionary process of mutualist interactions, both associates can evolve at the same speed, whereas parasites may have the need to evolve faster than their hosts (Combes 2004). Symbionts with recurrent generations on a single host, like many phytophagous insects, are expected to be largely homozygous and to have low genetic diversity. In contrast, symbionts that release offspring into the external environment, where they are mixed and then recruited back into new hosts, have similar or higher levels of genetic diversity than their hosts, as occurs in most animal macroparasites (Criscione et al. 2005). Outcrossing species tend to have higher levels of diversity than selfing species (Jarne 1995; Charlesworth 2003). Whereas effective population sizes (related to adaptive potential) of symbionts with direct life cycles are expected to be directly correlated to the number of adult symbionts in the host population, the existence of intermediate hosts in symbionts with complex life cycles produces more complex demographic patterns (Criscione et al. 2005). Host dispersal is a key determinant of gene flow and genetic structure among symbiont populations (Blouin et al. 1995; Criscione and Blouin 2004). Additionally, low host specificity can promote sympatric patterns of genetic structure as a result of host-related selective pressures (McCoy et al. 2001). Thus, there is a wide variety of life history traits exhibited by symbionts that are likely to affect their population genetics and, therefore, their genetic responses to bottlenecks (including invasion bottlenecks).
Symbionts accompanying invasive hosts are known to be affected by strong filters during the invasion process, acting at the interspecific level to reduce symbiont richness of the host species (Torchin et al. 2003). However, do filters act at the intraspecific level, making symbiont invaders more sensitive to founder effects than their host co-invaders? In the absence of strong intraspecific filters, differences in effective population size and genetic diversity between the symbiont and its host in the source populations, and their differences in evolutionary rates, are expected to be key factors determining whether the symbiont and host respond differently to an invasion bottleneck (Cox 2004). Strength of founder effects may challenge the adaptive potential of symbiont invaders to new environmental challenges emerging in the invasion context. Given that symbionts often influence the invasion success of their hosts and produce dramatic effects on native biota (Roy and Handley 2012), understanding genetic responses of symbiont-host co-invaders that share the same invasion process is not only of general evolutionary interest, but also a crucial topic in invasion biology. Surprisingly, very few studies have compared intraspecific genetic diversity patterns of native and exotic populations of a symbiont-host association, all of them based on parasites of marine or intertidal environments (i.e. Miura et al. 2006; Blakeslee et al. 2008; Stefani et al. 2012; Gaither et al. 2013). Three of these studies were based on parasitic symbionts with complex life cycles (trematodes and nematodes) and intermediate (Miura et al. 2006; Blakeslee et al. 2008) or definitive (Gaither et al. 2013) hosts, whereas the other is based on a monogenean parasite, with direct life cycle (Stefani et al. 2012). Results generally indicated congruent genetic diversity patterns in the exotic symbiont and its host, with either a similar decrease in diversity (Miura et al. 2006; Blakeslee et al. 2008; Gaither et al. 2013) or an absence of significant bottleneck effects (Stefani et al. 2012) in both co-invaders. The only exception was an exotic trematode that had similar genetic diversity to native populations, even though its intermediate host, a marine mud snail, showed a severe diversity drop in the invasive range (Miura et al. 2006). This incongruous pattern was explained by an alternative introduction pathway for the symbiont, different from the snail, via migrant birds (definitive hosts).
The crustaceans Ankylocythere sinuosa (Rioja, 1942) and Procambarus clarkii (Girard, 1852) form a symbiont-host association that co-invaded Europe after two consecutive human-mediated introductions for aquaculture purposes in 1973 and 1974 respectively (Gutiérrez-Yurrita et al. 1999; Mestre et al. 2013). P. clarkii is native to NE Mexico and S Central USA, extending westward to Texas, eastwards to Alabama, and northwards to Tennessee and Illinois (Hobbs 1972). Introduction of P. clarkii into Europe was thoroughly documented by various authors (see Gutiérrez-Yurrita et al. 1999). In June 1973, a batch of P. clarkii (100 kg) was transported from New Orleans (Louisiana, USA), to a crayfish farm in Badajoz (SW Iberian Peninsula), and a year later a second batch (100 kg) of P. clarkii was released into a pond located in the lower Guadalquivir (Puebla del Río, Seville, S Iberian Peninsula). Subsequent human translocations, stimulated by elevated prices for the crayfish, accelerated its expansion into the entire Iberian Peninsula, then to the Azores, the Balearic and the Canary islands. Human-assisted dispersal within the invasive range continued, giving rise to a wide current distribution that includes several European countries (Kouba et al. 2014).
The ostracod A. sinuosa is a commensal ectosymbiont that belongs to the family Entocytheridae, and inhabits American crayfish (Hart and Hart 1974). It has a native distribution similar to that of P. clarkii, a common host of this symbiont species. Exotic populations of A. sinuosa are widely distributed around the Iberian Peninsula, associated with P. clarkii (Mestre et al. 2013). Entocytherids, inhabiting crustacean hosts, mainly crayfish, have a direct life cycle and sexual reproduction. They are characterised by low degrees of host specificity (Mestre et al. 2014a) and are horizontally transmitted, without the need of contact between hosts (Young 1971). However, they have limited off-host dispersal, as they do not swim but move by crawling through the sediment (Mestre et al. 2015). Exotic populations of A. sinuosa have very high levels of prevalence (Mestre et al. 2014b), and there is evidence of the same pattern in native populations (Young 1971). Despite A. sinuosa populations benefiting from a certain independence from the external environment offered by the commensal life-style, they remain sensitive to variation in off-host environmental conditions such as specific conductance, NH4 + concentration, and alkalinity (Mestre et al. 2014b).
In this study, we investigated patterns of genetic diversity and structure of an invasive symbiont-host association for the first time based on a non-parasitic freshwater ectosymbiont with a direct life cycle. The invasion history for this species is relatively well-known, and our analyses included wide-ranging samples from the native region. Mitochondrial genes are expected to be particularly sensitive to bottleneck events, because mtDNA is haploid with uniparental inheritance and thus has only one quarter the effective population size of nuclear genes (Avise 1994), although it can be affected by selective sweeps (Bazin et al. 2006). A single mtDNA marker was used here taking into account that genomic resources for ostracods are very limited, COI is highly polymorphic in both studied species and no sex-biased migration is expected for both species. We show clear change in genetic diversity associated with invasion, discuss the potential evolutionary implications of these patterns, and highlight the usefulness of this kind of model to test important ecological and evolutionary questions.
Materials and methods
Field sampling
The target species of this study were the entocytherid A. sinuosa and its main host, the crayfish P. clarkii. During 2010 and 2011, crayfish were sampled from two regions: 14 localities from Louisiana plus one locality from Memphis, Tennessee (USA; Fig. 1a, c), representing the native region, and 29 localities widely distributed across the Iberian Peninsula, in the European exotic range (Fig. 1b, d). The sampling range of native populations was focused on Louisiana because it represents the core of P. clarkii’s native distribution and is documented as the source area for initial introductions that originated the exotic populations in the Iberian Peninsula (Gutiérrez-Yurrita et al. 1999). A variety of water body types were sampled in both native and exotic ranges (See Table S1 in ESM), representing different habitat characteristics. Up to 20 crayfish from each locality were individually subjected, in situ, to a protocol of ectosymbiont removal by submersion in commercial carbonated water (Mestre et al. 2011), with crayfish subsequently preserved in ethanol (96–100 %). The carbonated water was filtered through 63-μm mesh to isolate symbionts from each crayfish, and the residue was preserved in 50-mL containers filled with ethanol (96–100 %). Entocytherid samples were checked in the lab for species identification based on adult males, following Hart and Hart (1974). Some entocytherid samples from the native range were obtained from other crayfish species (i.e. Procambarus zonangulus, P. vioscai and Orconectes palmeri; see Table S1 in ESM).
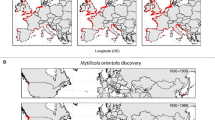
Geographical distribution of COI haplotype frequencies of the crayfish host Procambarus clarkii and the ectosymbiotic ostracod Ankylocythere sinuosa in their native range in Louisiana, USA (a, c), and exotic range in the Iberian Peninsula, Europe (b, d). An additional native crayfish sample (free of entocytherids) from out of Louisiana (i.e. Memphis) was also obtained, represented in an additional map of wider extent (inset, top-left of a), where the location of the main native sampling region (i.e. more detailed map) is also highlighted in a square frame. Sampling site names are coded as in Table S1 in ESM. Haplotypes restricted to one sampling location (i.e. private haplotypes) are represented in white while haplotypes shared between locations are color-coded. The size of pie charts is proportional to sample size. The map has a Mollweide equal-area projection
DNA extraction, mtDNA amplification and sequencing
The DNA of P. clarkii and A. sinuosa was extracted by the ammonium-acetate precipitation method. Specific primers were designed for amplification of mitochondrial cytochrome c oxidase subunit I (COI) in each species with Primer 3 v. 0.4.0 (Untergrasser et al. 2012), based on general primers for invertebrates and crayfish. Primers for P. clarkii were ClarkF (forward; 5′-GAGGAGTTGGAACAGGATGG-3′) and ClarkR (reverse; 5′-TCAGCAGGAGGATAAGAATGC-3′), which amplify a COI fragment of 1147 bp. Primers for A. sinuosa were EntoF (forward; 5′-TCGTGTAGAATTGGGTCATCC-3′) and EntoR (reverse; 5′-AGGATCTCCACCACCATTAGG-3′), which amplify a 605-bp fragment. The PCR amplifications were performed with an MJ Research PTC-225 Tetrad Thermal Cycler. The protocol used for crayfish was a starting denaturation at 94 °C for 3 min, followed by 35 cycles of 45 s at 94 °C, 60 °C for 45 s annealing time, 90 s at 72 °C for extension and a final extension stage of 10 min at 72 °C. The 10-μl volume reactions for P. clarkii contained 1 μl of DNA (~10 ng), 0.25 U of BIOTAQ™ (Bioline) DNA polymerase, 1 × NH4 Reaction Buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 μM of each primer and ddH2O. The PCR conditions for A. sinuosa were an initial denaturation step at 95 °C for 15 min followed by 34 cycles of 30 s denaturation at 94 °C, 90 s annealing at 59 °C, and 90 s extension at 72 °C, with a final extension at 72 °C for 10 min. In this case, the 11-μl reaction contained 6 μl Qiagen Multiplex PCR Mastermix (with HotStar Taq), 4 μl primer master mix (21 μl Low TE plus 2 μl of each 10 μM primer) and 1 μl of ddH2O.
The PCR products were cleaned with ExoSAP-IT® (USB Corporation, Cleveland, OH), and 2 μl of purified PCR product were used in each 10-μl sequencing reaction (in both directions) with the Big Dye® Terminator v. 3.1 cycle sequencing kit (Applied Biosystems) and the corresponding PCR primer (0.33 μM). Sequencing conditions were 1 min at 96 °C followed by 39 cycles of 10 s at 94 °C, 5 s at 50 °C and 4 min at 60 °C. Finally, products were further purified with an ethanol/EDTA precipitation method (ABI cycle sequencing kit manual) to be run on an ABI 3730xl Genetic Analyser.
Data analyses
Resulting chromatograms were edited in CodonCode Aligner v. 4.0.4 and aligned through the ClustalW algorithm in MEGA v. 6.0.6 (Tamura et al. 2013). Sequences were checked for identity with BLASTN v. 2.2.31 (Zhang et al. 2000). As expected, putative crayfish COI fragments scored highly with other COI sequences of P. clarkii in Genbank (e.g. 99 % identity). So far, there have been no submissions of COI from species of the family Entocytheridae to Genbank. However, search results for A. sinuosa sequences showed high similarity with other Ostracoda COI sequences (e.g. 80 % identity with Macroscapha opaca; 79 % with Eucypris virens). We used the invertebrate mtDNA code to translate COI fragments into protein sequences with MEGA, and check for presence of stop codons within the reading frame, which would indicate undesired amplifications of nuclear copies of mitochondrial sequences. Two crayfish sequences with stop codons were removed from the study. Preliminary visualization of pairwise genetic distances between sequences through classical multidimensional scaling (Fig. S1 in ESM) allowed us to detect species misidentifications or cryptic species evidenced by outlier sequences, which were also removed from the study. Some sequenced entocytherids were obtained from the same host individual (Table S1 in ESM). The genetic sequences from this study were deposited into Genbank: Accession numbers KX450622–KX450774 (crayfish) and KX450478–KX450621 (entocytherids).
Genetic diversity and structure
Genetic diversity for crayfish and entocytherids in both native and exotic regions was estimated by standard haplotype (h) and nucleotide (л) diversity indices, with DnaSP v. 5.10.01 (Librado and Rozas 2009). Additionally, extrapolated haplotype richness (estimated through bias-corrected Chao2) and individual-based rarefaction curves of haplotype richness for native and exotic regions of both studied species were estimated with vegan v. 2.2.1 (Oksanen et al. 2015). Chao2 is a non-parametric estimator of the overall regional haplotype richness, extrapolated from the data, based on the assumption that the number of unsampled haplotypes is related to the number of rare haplotypes (Gotelli and Colwell 2010). Haplotype networks were built with pegas v. 0.6 (Paradis 2010).
The best nucleotide substitution model for each species was estimated with jModelTest v. 2.1 (Darriba et al. 2012), based on Akaike Information Criterion (AIC). All model variants available in the program were considered, including invariable sites, rate variation among sites, or both (a total of 88 models). The selected model was TN93 (Tamura and Nei 1993) with gamma correction of the inter-site variation in substitution rates (i.e. TN93+G). This model assumes distinct rates for transitions and transversions. TN93+G was selected because it is widely available in other software, with a simple formula for the distance transformation, and it was ranked within the 15 best models for each species (AICCR = 3212.26; AICEN = 5087.27, subscript CR for crayfish and EN for entocytherids), with a low AIC increase from the top model (less than 10 units). Shape parameters (α) of the gamma distribution of among-site variation for the TN93+G model were estimated by jModelTest (α CR = 0.03; α EN = 0.122). Mean pairwise individual genetic distance based on TN93+G was obtained for native and exotic populations of both species, with ape v. 3.1.4 (Paradis et al. 2004). Hierarchical analysis of molecular variance (AMOVA) was performed with pegas to test for patterns of genetic structure (based on TN93+G distances) between regions (native and exotic), and between populations within a region (i.e. sampling localities). Significance was evaluated from 1000 permutations. A preliminary AMOVA to test for genetic dependence of symbionts inhabiting the same host individual showed no significant effect. Thus, this additional level was not included in the final AMOVA analyses.
Pairwise genetic distances between populations were estimated with Meirmans ΦST (Φ′ST), based on the TN93+G model (R function available in Script S1 in ESM), and removing sites with a unique individual sampled. Φ′ST is a standardised population distance measure with bias correction for different effective populations sizes and different mutation rates (Meirmans 2006). Patterns of isolation by distance (IBD) between populations in native and exotic regions for both species were tested with two approaches. First, correlation between genetic and geographic distances was tested with a Mantel test, implemented in ade4 v. 1.6.2 (Dray and Dufour 2007), with significance determined from 10, 000 permutations. Second, IBD was also tested through ordinary least square (OLS) regressions, based on 1000 permutations, applied with lmodel2 v. 1.7.2 (Legendre 2014). Geographic distances were estimated with fossil v. 0.3.7 (Vavrek 2011). The native crayfish locality LUS15 (Fig. 1a) was removed from the IBD analyses due to its distance from the other localities, to avoid high leverage. After checking all variants with and without log transformations of genetic and geographic distances, the non-transformed variant was selected, based on distribution and model performance criteria. IBD differences between native and exotic ranges were assessed by comparing the slopes (a) of OLS regressions and their confidence intervals (95 % CI). Existence of spatial patterns in genetic variation between populations in native and exotic regions was tested through a redundancy analysis (RDA) based on Moran’s eigenvector maps (MEMs; Galpern et al. 2014). The MEMs were obtained with spdep v. 0.5.88 (Bivand and Piras 2015). Autocorrelated MEMs were identified through a 99-times permutation procedure implemented by spacemakeR v. 0.0.5 (Dray 2013), and removed from the analyses. A principal coordinates analysis (PCoA) was performed on pairwise genetic distances between populations (Φ′ST), and positive axes of the PCoA were used as response variables for the RDA, with MEMs as predictors. Significant MEMs were identified via forward selection based on 1000 permutations, implemented by packfor v. 0.0.8 (Dray et al. 2013), with p values and the adjusted R 2 (R 2adj ) of the full model used as selection criteria.
Correlation between individual genetic distances (TN93+G) of symbionts and their associated host individuals was checked via Mantel test (10,000 permutations) and OLS regression for both native and exotic regions.
Demographic analysis
Mismatch distribution and folded site frequency spectra were used to check for signals of hypothetical demographic events that would have altered expected patterns under a scenario of stable populations. The hypothesis of neutrality of mutations with constant population sizes was tested against alternative scenarios including non-neutral processes (population growth, hitchhiking and background selection) for native and exotic regions with Tajima’s D, Fu and Li’s D* and F*, and Ramos-Onsins and Rozas’s R 2 statistic (10,000 replicates used for simulation) estimated with DNAsp and pegas.
Demographic parameters of P. clarkii and A. sinuosa populations were estimated with fastsimcoal2 (Excoffier et al. 2013) from coalescent simulations based on a simple scenario where the exotic population arose from a bottleneck event in the invasion region. The nucleotide substitution model implemented by this program is k80. The scenario consisted of two populations (native and exotic) with the exotic population appearing from a unique invasion event with migration of some individuals (N FUN ) from the native to the exotic range a number of generations ago (T INV ), followed by a population growth in the exotic range until the present, when the population sizes of native and exotic ranges would be N NAT and N EXO , respectively (Fig. S2 in ESM). Search ranges of demographic parameters were bounded by biological considerations, well-documented events (i.e. first introduction of P. clarkii into Europe occurred in 1973), and the distribution of initial parameter estimates from preliminary trials (for fastsimcoal2 estimation and template files see Appendix S1). Initial parameter estimates were obtained from the observed joint minor allele site frequency spectra (joint-MAF-SFS), with an R function built ad hoc (Available in Script S2 in ESM). The criterion for parameter selection was the composite maximum likelihood (CML). A total of 40 runs were performed, each one consisting of 100,000 simulations per likelihood estimation and 10–40 conditional maximization algorithm (ECM) cycles (stopping criterion of 0.001). Subsequently, support limits of initial CML estimates were obtained through parametric bootstrap. To do this, 100 joint-MAF-SFS were simulated under the evolutionary scenario established in our model and based on the initial CML estimates. A general mutation rate for invertebrates of 2 % divergence per million years was applied. Transition rates were obtained from the optimal parameters estimated by jModelTest for the k80 model (i.e. 0.9723 for P. clarkii and 0.9608 for A. sinuosa). Next, 100 parametric bootstrap estimates were obtained by parameter estimation from the simulated data sets (one run per data set); their distribution defined the support limits.
Results
The final lengths of the COI fragments analysed from P. clarkii and A. sinuosa were 810 and 522 bp, respectively. The total numbers of sequences used were 153 for P. clarkii: 73 native (median and interquartile range per locality 4, 3–5), and 80 exotic (3, 2–4); and 144 for A. sinuosa: 78 native (2, 0.5–5) and 66 exotic (3, 1–3) (Fig. 1; Table S1 in ESM). Both native and exotic populations of A. sinuosa showed very high levels of prevalence (Table S1 in ESM). The median numbers of adult symbionts per crayfish were: 6.5 [0–19] for native populations and 8 [1–24] for exotic populations.
Haplotype diversity and structure
Observed global haplotype richness was higher in the symbiont (118 haplotypes) than the host (25 haplotypes). Exotic populations of both species experienced a loss in single nucleotide polymorphisms, haplotype richness and haplotype diversity (Table 1), which was more pronounced in the host. The overall percentage of singletons (i.e. haplotypes that appear in only one individual) was 56 % in the host and 89.83 % in the symbiont. The higher haplotype diversity in the symbiont and the diversity drop in the exotic region for both species, which was greater in the host, were evident in the rarefaction curves of haplotype richness (Fig. 2).
Individual-based rarefaction curves of haplotype richness for the crayfish host Procambarus clarkii and its ostracod ectosymbiont Ankylocythere sinuosa in the native (Louisiana, USA) and exotic (Iberian Peninsula, Europe) regions. The confidence intervals are drawn as coloured areas around the predicted lines
Haplotype structure of the host (Figs. 1a, b, 3a) was characterised by a very homogeneous pattern in exotic populations (Fig. 1b), with the presence of a highly dominant haplotype (PC01), representing 73.75 % of exotic host individuals, and only one private haplotype (i.e. only appearing in one locality). In the host native range (Fig. 1a), there were several common haplotypes (PC01, PC03 and PC04), and private haplotypes were more frequent. Haplotype PC01 had a wide distribution range in both native and exotic regions, although it was less dominant in the native region, where it represented 17.81 % of native host individuals. There was a second host haplotype widely spread in the exotic region (PC02), which only appeared in a Northern locality of the native range (LUS14). Some less frequent haplotypes of the exotic host (i.e. PC03, PC10 and PC11) were found in exotic populations that were distant from one another.
Haplotype networks of native (Louisiana, USA) and exotic (Iberian Peninsula, Europe) populations of the crayfish host Procambarus clarkii (a), and its ectosymbiotic ostracod, Ankylocythere sinuosa (b), based on a COI mitochondrial DNA fragment (seq. lengths of 810 bp for P. clarkii and 522 bp for A. sinuosa). Circle sizes are proportional to the number of individuals sampled with this haplotype (indicated by the circle legends). The codes of non-private haplotypes are provided by labels (haplotype codes coincide with those used in Fig. 1). The classification of native entocytherid region into two subregions (N and S Louisiana) was based on the first Moran’s eigenvector map (MEM) selected for the native entocytherid in the spatial analysis using MEMs (Fig. 5e)
Almost all native haplotypes of the symbiont (Figs. 1c, d, 3b) were private (Fig. 1c), with only a few haplotypes from S Louisiana (i.e. AS02, AS06, AS07 and AS09) found also in the exotic region and a single haplotype (AS11) shared by two closer localities in N Louisiana. In contrast, exotic populations of the symbiont (Fig. 1d) had a higher number of non-private haplotypes, although the proportion of private haplotypes was still high, compared to the host. The most frequent haplotype of the symbiont (AS01) only appeared in the exotic region, where it represented 12.12 % of symbiont individuals, showing a wide distribution range. As in the host, some other non-private haplotypes of the exotic symbiont were shared by populations that were distant from one another (AS03, AS04 and AS05).
Nucleotide-based diversity and structure
Nucleotide diversity indices (л and лTN93+G; Table 1) were lower in the host than in the symbiont, and in host native populations than in exotic ones. Native and exotic populations of the symbiont had similar nucleotide diversities. Results of AMOVA indicated that there was no significant genetic differentiation between regions for either species (Table S2 in ESM). Native populations of both species showed significant genetic structure (ΦST = 0.25; dfn/dfd = 12/60; p = 0.002 for the host; ΦST = 0.40; dfn/dfd = 10/67; p < 0.001 for the symbiont), whereas there was no signal of genetic structuring in the exotic region (ΦST = 0.27; dfn/dfd = 28/51; p = 0.096 for the host; ΦST = 0; dfn/dfd = 22/43; p = 0.830 for the symbiont).
The IBD analyses (Fig. 4), based on Φ′ST population distances (Fig. S3 in ESM), revealed a significant pattern in the native range for both species (R 2 = 0.12, p mantel = 0.009 for the host; R 2 = 0.60, p mantel = 0.001 for the symbiont), with a positive slope that was higher in the symbiont (a = 0.0011, 95 % CI 0.0003–0.0018 for the host; a = 0.0028, 95 % CI 0.0020–0.0036 for the symbiont). No evidence of IBD pattern was found in the exotic range for either species (R 2 < 0.01, p mantel = 0.78 for the host; R 2 < 0.01, p mantel = 0.44 for the symbiont).
Isolation by distance (IBD) patterns for Procambarus clarkii (a, b) and its ectosymbiont Ankylocythere sinuosa (c, d) for native (a, c) and exotic (b, d) ranges. Pairwise genetic distances between populations were estimated with Meirmans ΦST (Φ′ST) calculated from pairwise individual distances based on the TN93 + G nucleotide substitution model. The continuous line is the ordinary least square regression line, and the discontinuous lines show the confidence intervals of the slopes, obtained from 1000 permutations
Selected MEMs in RDA analysis (Fig. 5) showed the existence of regional-scale patterns of genetic similarity between populations in native range for both species (Fig. 5a, b, e, f). In particular, MEM1 showed a pattern that distinguished between N and S populations (Fig. 5b, e), a trend that was especially strong in the symbiont (R 2adj = 0.63; Fig. 5e). Local-scale patterns showing genetic admixture between populations were detected in exotic range for the host, with low adjusted R 2 (Fig. 5c, d), and no significant patterns were observed for exotic populations of the symbiont.
Spatial patterns in genetic dissimilarity between populations (in both native and exotic regions), detected for the crayfish host Procambarus clarkii (a, b, native region; c, d, exotic region), and for the ostracod ectosymbiont Ankylocythere sinuosa (e, f, native region). The significant Moran’s Eigenvector Maps (MEMs), represented here, explaining the variability of Meirmans ΦST pairwise genetic distances for each species and region, were identified through a redundancy analysis and a forward selection procedure. Circles of similar size and same colour indicate populations with similar scores (large black and white circles describe opposite extremes on the MEM axes). No significant MEMs were selected for A. sinuosa in the exotic region
Genetic distances among symbiont individuals and among their associated host individuals were not correlated in the native range (R 2 < 0.01, p mantel = 0.40). In contrast, they showed a significant positive correlation in the exotic region, although with a very low R 2 (R 2 = 0.05, p mantel < 0.001, a = 0.61, 95 % CI 0.48–0.75).
Demographic analyses
Folded site frequency spectra (SFS) and mismatch distributions (MMD, Fig. 6) showed an unexpected bimodal shape in the exotic host (Fig. 6b). In addition, both native and exotic symbiont populations (Fig. 6c, d), showed the following anomalies compared to a neutral scenario: (1) SFS with an excess of rare alleles at low frequencies (i.e. freq. 1–2); (2) MMD with a scarcity of shorter pairwise genetic distances producing a belly shaped distribution. Almost all neutrality tests applied to the symbiont (with the exception of D for exotic populations) indicated a departure from neutrality towards an excess of rare alleles (Table 1). All tests for the host were non-significant. In addition, by comparing Tajima’s related test statistics (D, D* and F*) between native and exotic populations, the same trend towards higher values (less negative) in the exotic region was observed in both species.
Folded site frequency spectra (outer graphs) and mismatch distributions (inner graphs) of native (a, c) and exotic (b, d) populations of Procambarus clarkii (a, b) and its ectosymbiont Ankylocythere sinuosa (c, d). The pairwise genetic distances for the mismatch distributions are based on the TN93+G nucleotide substitution model. The expected shapes under a neutral scenario are represented with a red line. SNPs are single nucleotide polymorphisms
In general, CML estimates of the simulated demographic scenario of the invasion process had wide support limits (Table S3 in ESM), especially for the case of N EXO . In spite of this, two patterns appear by comparing the species: i) a similar CML estimate for T INV ≈ 80; and ii) generally higher effective population size estimations for the symbiont. Particularly, N NAT was 5.5 times higher in the symbiont, and the distribution of the bootstrap estimates of N FUN for the host was strongly biased towards very low values, whilst the plausible range of N FUN for the symbiont spread more towards higher values (Fig. 7).
Discussion
This work is based on a symbiont with direct life-cycle involving exclusive use of one kind of host. Consequently, there is a stronger link between the symbiont and its host at both demographic and dispersive levels compared to symbionts with indirect cycles. Introduction of P. clarkii in Europe showed a stepping-stone pattern with two consecutive propagule events into SW Iberian Peninsula from New Orleans (Louisiana) in 1973–1974, followed by a human-mediated expansion across Europe by translocation events (Gutiérrez-Yurrita et al. 1999). The wide distribution of A. sinuosa among European P. clarkii populations of the Iberian Peninsula, the absence of the symbiont in other European crayfish (native or exotic; Mestre et al. 2013) and the common association of this symbiont species with P. clarkii in the native range (Hart and Hart 1974) strongly suggest that A. sinuosa and P. clarkii shared the same invasion history. We make this assumption in trying to understand how observed genetic patterns were built during this invasion process.
Genetic diversity patterns
Genetic diversity of both symbiont and host was generally high (Goodall-Copestake et al. 2012). Previous studies of genetic diversity and structure of exotic populations of P. clarkii in Europe (Barbaresi et al. 2007) and China (Li et al. 2012) show similar values of genetic diversity to the exotic P. clarkii populations used in this study. They also highlight the high diversity observed in exotic P. clarkii, based on other genetic markers, but both studies had insufficient sample from native populations for a proper comparison of genetic patterns. Bottleneck effects were detected in Chinese populations of P. clarkii based on microsatellite loci (Yue et al. 2010). The main genetic diversity patterns shown by our symbiont-host system are: (1) a higher genetic diversity of the symbiont than its host; (2) genetic diversity loss in the exotic range for both species; but (3) a less pronounced loss in the symbiont. Changes in genetic diversity can be driven by demographic (e.g. founder effects or population expansion) or selective (genetic draft) events (Jensen et al. 2005). Regarding the demographic hypothesis to explain the observed patterns, effective population size of symbionts with a direct cycle is expected to have a simple relationship with the census number of adult symbionts on their hosts (Criscione et al. 2005). Exotic populations of A. sinuosa have a mean prevalence of 91.15 % (N = 373; Mestre et al. 2014b). The present survey shows that native populations show similarly high prevalence values (Table S1 in ESM). Moreover, the median number of A. sinuosa adults per host was similar for both ranges (6.5 for native and 8 for exotic). These values agree with effective populations sizes estimated for native populations (N NAT ) from the coalescent simulations that were 5.5 times higher in the symbiont. So, the higher effective population size of native populations would explain the higher overall genetic diversity of the symbiont. An alternative hypothesis to explain this pattern is that A. sinuosa has higher mutation rates than P. clarkii due to shorter generation times. However, the short generation time of four and half months estimated for P. clarkii (Huner and Barr 1991), the long survival period of at least two and half months observed in A. sinuosa out of the host (Baker 1969), and the similar univoltine cycles observed in both species (Castillo-Escrivà et al. 2013) indicate that they do not have large differences in generation time.
Genetic diversity loss in the exotic range for both species is explained by their shared invasion history that evidences existence of a bottleneck after the introduction, as the initial crayfish stock was comprised of 200 kg from native populations of S Louisiana (New Orleans; Gutiérrez-Yurrita et al. 1999). So, based on historical data, the maximum value of N FUN for the host was no more than 10,000 individuals (mean weight of a P. clarkii adult in Europe is 20 g; Mestre et al. 2014b), that was likely reduced due to mortality rates during transport and initial acclimatisation. The simulation results suggest a much lower N FUN for the host of less than 500 individuals (Fig. 7a). However, use of a single genetic marker in coalescent simulation results in considerable uncertainty around these parameter estimates. In spite of this, distributions of the bootstrap estimates for N FUN also suggest that the symbiont had an effective population size higher than the host at the initial stage of the invasion (Fig. 7), in agreement with the high values of the population parameters (prevalences and abundances) observed for the symbiont. Thus, the less severe drop in genetic diversity of the exotic symbiont than its host could result from: (1) the higher genetic diversity of source populations of the symbiont and (2) its higher N FUN . This hypothesis, supported by three different data sources, i.e. historical, demographic and genetic, emphasises the importance of propagule size in determining the strength of founder effects in an invasion process.
An alternative hypothesis that could explain the observed diversity patterns is genetic draft, common in organisms with large population sizes and high reproduction rates (Mulligan et al. 2006), like in our model species. According to this hypothesis, selective sweeps in the new exotic environment could have produced the severe drop in genetic diversity of the exotic host. However, this would predict a higher or, at least, similar drop in diversity for the exotic symbiont, that was also exposed to a new environment, and has higher population sizes than its host. A possible reason why this was not observed is that the species are affected by different selective pressures: differences in these pressures between native and exotic ranges are higher in the host, because the environment of the symbiont, i.e. the host body, remains more constant. However, new potential sources of selection on the host in the exotic range can also affect the symbiont, as discussed below.
Overall, the neutrality tests reflect a general trend of singleton losses in the exotic range for both species (i.e. higher values of Tajima’s related statistics), as a consequence of founder effects. The bimodal shape of the SFS and MMD for the exotic host makes sense in the scenario of stronger founder effects suffered by the host, as multimodal shapes (including bimodal) can be indicative of historical population contractions (Burbrink et al. 2008). Apparent signals of population expansion observed in the symbiont by the SFS, MMD (i.e. belly shaped) and neutrality tests may be an effect of the considerably high genetic variability observed in the symbiont. Particularly for these tests, strong conservation of some sites (mainly second codon positions) causes more departure from the infinite sites mutation model as the overall diversity gets greater.
Patterns of genetic structure
In spite of the limited sampling sizes per site of this survey, consistent patterns at population level were observed that are congruent with the biological traits and biogeographic history of the study organisms. Native populations of both partners were genetically structured, showing IBD and regional spatial patterns. The long evolutionary history of the symbiont-host system in the native region along with dispersal limitations of the host, most probably promoted these patterns. Freshwater ecosystems are often spatially fragmented and crayfish, unlike other freshwater crustaceans, do not use natural vectors like birds or mammals for increasing their dispersal. Particularly, the natural dispersal range of P. clarkii is up to 2.5 km (Siesa et al. 2011), but they are strongly affected by hydrologic connectivity (Paulson and Martin 2014). Furthermore, the observed spatial patterns for the native symbiont were more marked than for the native host (i.e. higher ΦST of AMOVA, higher slope of IBD and higher R 2adj of the main MEM). These results agree with the limited off-host dispersal of the symbiont (Mestre et al. 2015). The general loss of genetic structure in exotic populations of both species is congruent with the documented invasion history, where human-mediated translocations promoted admixture of European populations, with minimal time for subsequent population differentiation. Presence of local patterns in the host (Fig. 5c, d), close to a panmixia model (Galpern et al. 2014), strengthens this idea. Other genetic studies of exotic P. clarkii in Europe and China corroborate the role of human-mediated translocations as mechanisms that eliminate dispersal barriers and preclude appearance of IBD patterns (Barbaresi et al. 2007; Yue et al. 2010; Li et al. 2012). In contrast, these studies show some evidence of among-population structure in exotic P. clarkii, although they included samples from native populations in the analysis (Barbaresi et al. 2007), or were based on a different invasion process with an older invasion history, i.e. China (Yue et al. 2010; Li et al. 2012), where P. clarkii was introduced in 1929.
Evolutionary implications and future perspectives
We have presented the case of a symbiont invader that has overcome founder effects apparently better than its host co-invader, due to a high diversity in source populations and a high propagule size. In an invasion context, the symbiont is therefore provided with a high adaptive potential in the new environment. However, what role do novel selective pressures from the exotic host or off-host environment play in our symbiont model? The host environment is susceptible to change by two mechanisms. First, the host invader can be subjected to rapid evolution or phenotypic variation (Huey et al. 2000). Second, the symbiont can colonise novel hosts through a spill-over process (Roy and Handley 2012). Our symbiont model, A. sinuosa, is especially prone to the second phenomenon as it has a wide host specificity (i.e. generalist symbiont) and native European crayfish represent an empty niche for entocytherids (Mestre et al. 2013). Mestre et al. (2015) successfully tested the experimental spill-over of A. sinuosa onto a native European crayfish. Moreover, Europe is also rich in other exotic crayfish species (at least 10) that could also be colonised by the symbiont (Kouba et al. 2014). Selection of a generalist symbiont to colonise different host species has been documented, and may be an important diversifying mechanism in symbionts through sympatric speciation (McCoy et al. 2001). Our symbiont model is also particularly sensitive to the off-host environment, due to its ectosymbiotic, non-parasitic life-style (i.e. dependence on external food sources), and the environmental variability inherent in freshwater habitats (Mestre et al. 2014b). Thus, variation in off-host environmental variables (physico-chemical or climatic) as a result of the range expansion of the host co-invader can promote new selective pressures on the symbiont. The adaptive potential, limited dispersal and off-host environmental sensitivity evidenced in this symbiont invader could favour future allopatric differentiation as a result of adaptive processes to local environmental conditions in Europe, if human-mediated translocations are not enough to maintain sufficient gene flow to preclude the processes.
Considering that invasive species and symbiont-host systems have served, separately, as very powerful models in ecology and evolutionary biology, the combination of both approaches with models of symbiont-host co-invaders, comparing patterns of population genetics between native and exotic populations of both associated species, has great potential. Insights into important topics from different fields such as evolutionary, conservation and invasion biology can be obtained with this little-explored approach. Understanding the factors related to genetic responses to founder effects, drivers of genetic diversity (demographic vs. evolutionary), the actual role of genetic diversity in evolutionary change, or differences in sympatric vs. allopatric speciation processes are some examples. The rich variety of symbiont life styles offers a great opportunity to test effects of different life-history traits (e.g. generation time, reproductive mode or dispersal capacity) on genetic responses to an invasion process. However, a very limited range of symbiont-host models has been studied hitherto, most involving parasites with complex life cycles. We encourage the use of a wider spectrum of symbiont-host systems. Particularly, we have shown the usefulness of a very simple model, not used previously, of a non-parasitic freshwater ectosymbiont with direct life-cycle inhabiting a host with a known invasion history. We emphasize the utility of this kind of model to better interpret patterns, test hypotheses and understand more complex systems.
References
Allendorf FW, Lundquist LL (2003) Introduction: population biology, evolution, and control of invasive species. Conserv Biol 17:24–30
Avise JC (1994) Molecular markers, natural history and evolution, 2nd edn. Chapman & Hall, London
Baker JH (1969) On the relationship of Ankylocythere sinuosa (Rioja 1942) (Ostracoda, Entocytheridae) to the crayfish Procambarus simulans simulans (Faxon 1884). Trans Am Microsc Soc 88:293–294
Barbaresi S, Gherardi F, Mengoni A, Souty-Grosset C (2007) Genetics and invasion biology in fresh waters: a pilot study of Procambarus clarkii in Europe. In: Gherardi F (ed) Biological invaders in inland waters: profiles, distribution, and threats. Springer, Dordrecht, pp 381–400
Bazin E, Glémin S, Galtier N (2006) Population size does not influence mitochondrial genetic diversity in animals. Science 312:570–572
Bivand R, Piras G (2015) Comparing implementations of estimation methods for spatial econometrics. J Stat Softw 63:1–36
Blakeslee AM, Byers JE, Lesser MP (2008) Solving cryptogenic histories using host and parasite molecular genetics: the resolution of Littorina littorea’s North American origin. Mol Ecol 17:3684–3696
Blouin MS, Yowell CA, Courtney CH, Dame JB (1995) Host movement and the genetic structure of populations of parasitic nematodes. Genetics 141:1007–1014
Burbrink FT, Fontanella F, Pyron RA, Guiher TJ, Jimenez C (2008) Phylogeography across a continent: the evolutionary and demographic history of the North American racer (Serpentes: Colubridae: Coluber constrictor). Mol Phylogenet Evol 47:274–288
Castillo-Escrivà A, Mestre A, Monrós JS, Mesquita-Joanes F (2013) Population dynamics of an epibiont Ostracoda on the invasive red swamp crayfish Procambarus clarkii in a western Mediterranean wetland. Hydrobiologia 714:217–228
Charlesworth D (2003) Effects of inbreeding on the genetic diversity of populations. Philos Trans R Soc Lond Ser B 258:1051–1070
Combes C (2004) Parasitism: the ecology and evolution of intimate interactions. University of Chicago Press, Chicago
Cox GW (2004) Founder effects and exotic variability. In: Cox GW (ed) Alien species and evolution: the evolutionary ecology of exotic plants, animals, microbes, and interacting native species. Island Press, Washington, pp 32–46
Criscione CD, Blouin MS (2004) Life cycles shape parasite evolution: comparative population genetics of salmond trematodes. Evolution 58:198–202
Criscione CD, Poulin R, Blouin MS (2005) Molecular ecology of parasites: elucidating ecological and microevolutionary processes. Mol Ecol 14:2247–2257
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Dray S (2013) spacemakeR: spatial modelling. R package version 0.0-5/r113
Dray S, Dufour AB (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22:1–20
Dray S, Legendre P, Blanchet G (2013) packfor: forward selection with permutation (Canoco p.46). R package version 0.0-8/r109
Excoffier L, Dupanloup I, Huerta-Sánchez E, Foll M (2013) Robust demographic inference from genomic and SNP data. PLoS Genet 9:e1003905
Gaither MR, Aeby G, Vignon M, Meguro Y, Rigby M, Runyon C et al (2013) An invasive fish and the time-lagged spread of its parasite across the Hawaiian archipelago. PLoS ONE 8:e56940
Galpern P, Peres-Neto PR, Polfus J, Manseau M (2014) MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol Evol 5:1116–1120
Goodall-Copestake W, Tarling G, Murphy E (2012) On the comparison of population-level estimates of haplotype and nucleotide diversity: a case study using the gene cox1 in animals. Heredity 109:50–56
Gotelli NJ, Colwell RK (2010) Estimating species richness. In: Magurran AE, McGill BJ (eds) Biological diversity: frontiers in measurement and assessment. Oxford University Press, Oxford, pp 39–54
Gutiérrez-Yurrita PJ, Martínez JM, Bravo-Utrera MA, Montes C, Ilhéu M, Bernardo JM (1999) The status of crayfish populations in Spain and Portugal. In: Gherardi F, Holdich DM (eds) Crayfish in Europe as alien species: How to make the best of a bad situation?. A. A. Balkema, Rotterdam, pp 161–192
Hart DG, Hart CW Jr (1974) The ostracod family Entocytheridae. Fulton Press Inc., Lancaster
Hobbs HH Jr (1972) Biota of freshwater ecosystems, identification manual 9: Crayfishes (Astacidae) of North and Middle America. US Environmental Protection Agency, Washington
Huey RB, Gilchrist GW, Carlson ML, Berrigan D, Serra L (2000) Rapid evolution of a geographic cline in size in an introduced fly. Science 287:308–309
Huner JV, Barr JAE (1991) Red swamp crayfish, biology and exploitation, 3rd edn. Louisiana State University, Baton Rouge
Jarne P (1995) Mating system, bottlenecks and genetic polymorphism in hermaphroditic animals. Genet Res 65:193–207
Jensen JD, Kim Y, DuMont VB, Aquadro CF, Bustamante CD (2005) Distinguishing between selective sweeps and demography using DNA polymorphism data. Genetics 170:1401–1410
Kolbe JJ, Larson A, Losos JB, de Queiroz K (2008) Admixture determines genetic diversity and population differentiation in the biological invasion of a lizard species. Biol Lett 4:434–437
Kouba A, Petrusek A, Kozák P (2014) Continental-wide distribution of crayfish species in Europe: update and maps. Knowl Manag Aquat Ecosyst 413:05
Legendre P (2014) lmodel2: model II regression. R package version 1.7-2
Li Y, Guo X, Cao X, Deng W, Luo W, Wang W (2012) Population genetic structure and post-establishment dispersal patterns of the red swamp crayfish Procambarus Clarkii in China. PLoS ONE 7:e40652
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lockwood JL, Cassey P, Blackburn T (2005) The role of propagule pressure in explaining species invasions. Trends Ecol Evol 20:223–228
McCoy KD, Boulinier T, Tirard C, Michalakis Y (2001) Host specificity of a generalist parasite: genetic evidence of sympatric host races in the seabird tick Ixodes uriae. J Evol Biol 14:395–405
Meirmans PG (2006) Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution 60:2399–2402
Mestre A, Monrós JS, Mesquita-Joanes F (2011) Comparison of two chemicals for removing an entocytherid (Ostracoda: Crustacea) species from its host crayfish (Cambaridae: Crustacea). Int Rev Hydrobiol 96:347–355
Mestre A, Aguilar-Alberola JA, Baldry D, Balkis H, Ellis A, Gil-Delgado JA et al (2013) Invasion biology in non-free-living species: interactions between abiotic (climatic) and biotic (host availability) factors in geographical space in crayfish commensals (Ostracoda, Entocytheridae). Ecol Evol 3:5237–5253
Mestre A, Monrós JS, Mesquita-Joanes F (2014a) A review of the Entocytheridae (Ostracoda) of the world: updated bibliographic and species checklists and global georeferenced database, with insights into host specificity and latitudinal patterns of species richness. Crustaceana 87:923–951
Mestre A, Monrós JS, Mesquita-Joanes F (2014b) The influence of environmental factors on abundance and prevalence of a commensal ostracod hosted by an invasive crayfish: are ‘parasite rules’ relevant to non-parasitic symbionts? Freshw Biol 59:2107–2121
Mestre A, Castillo-Escrivà A, Rueda J, Monrós JS, Mesquita-Joanes F (2015) Experimental spillover of an exotic ectosymbiont on an European native crayfish: the importance of having a chance. Hydrobiologia 755:225–237
Miura O, Torchin ME, Kuris AM, Hechinger RF, Chiba S (2006) Introduced cryptic species of parasites exhibit different invasion pathways. Proc Natl Acad Sci USA 103:19818–19823
Mulligan CJ, Kitchen A, Miyamoto MM (2006) Comment on “Population size does not influence mitochondrial genetic diversity in animals”. Science 314:1390
Novak SJ, Mack RN (2005) Genetic bottlenecks in alien plant species: influence of mating systems. In: Sax DF, Stachowicz JJ, Gaines SD (eds) Species invasions: insights into ecology, evolution, and biogeography. Sinauer Associates Inc, Sunderland, pp 201–228
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB et al (2015) vegan: community ecology package. R package version 2.2-1
Paradis E (2010) pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290
Paulson EL, Martin AP (2014) Discerning invasion history in an ephemerally connected system: landscape genetics of Procambarus clarkii in Ash Meadows, Nevada. Biol Invasions 16:1719–1734
Poulin R (2007) Evolutionary ecology of parasites, 2nd edn. Princeton University Press, Princeton
Poulin R, Morand S (2005) Parasite biodiversity. Smithsonian Institution Scholarly Press, Washington
Roy HE, Handley LJL (2012) Networking: a community approach to invaders and their parasites. Funct Ecol 26:1238–1248
Sax DF, Stachowicz JJ, Brown JH, Bruno JF, Dawson MN, Gaines SD et al (2007) Ecological and evolutionary insights from species invasions. Trends Ecol Evol 22:465–471
Siesa ME, Manenti R, Padoa-Schioppa E, Bernardi FD, Ficetola GF (2011) Spatial autocorrelation and the analysis of invasion processes from distribution data: a study with the crayfish Procambarus clarkii. Biol Invasions 13:2147–2160
Stefani F, Aquaro G, Azzurro E, Colorni A, Galli P (2012) Patterns of genetic variation of a Lessepsian parasite. Biol Invasions 14:1725–1736
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM (2003) Introduced species and their missing parasites. Nature 421:628–630
Untergrasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115
Vavrek MJ (2011) Fossil: palaeoecological and palaeogeographical analysis tools. Palaeontol Electron 14:1T
Wares JP, Hughes AR, Grosberg RK (2005) Mechanisms that drive evolutionary change: insights from species introductions and invasions. In: Sax DF, Stachowicz JJ, Gaines SD (eds) Species invasions: insights into ecology, evolution, and biogeography. Sinauer Associates Inc, Sunderland, pp 229–258
Young W (1971) Ecological studies of Entocytheridae (Ostracoda). Am Midl Nat 85:399–409
Yue GH, Li J, Bai Z, Wang CM, Feng F (2010) Genetic diversity and population structure of the invasive alien red swamp crayfish. Biol Invasions 12:2697–2706
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A greedy algorithm for aligning DNA sequences. J Comput Biol 7:203–214
Acknowledgments
We thank Josep Antoni Aguilar-Alberola, Maria Antón, Marco Arruej, Andreu Castillo, Andreu Escrivà, Jose Antonio Gil-Delgado, Cristina Molina, Adrian Ponz, Josep R. Roca, Olivier Schmit, Luis Valls, Laia Zamora, Noel Novelo, Jonathan L. West, Will Sheftal, Julie Delabbio, Taren Manley, Mike Kennedy, Shirley and Shai for their help in the fieldwork; Andy Kruppa, Gavin Horsburgh and Barbara Morrissey for their assistance in the lab; Laurent Excoffier and Ivan Scotti for their help in coalescent simulation analyses. This research was funded by the Spanish Ministry of Science and Innovation Project ECOINVADER (CGL2008-01296/BOS) and the University of Valencia (“V-Segles” predoctoral grant to A. Mestre).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mestre, A., Butlin, R.K., Kelso, W.E. et al. Contrasting patterns of genetic diversity and spatial structure in an invasive symbiont-host association. Biol Invasions 18, 3175–3191 (2016). https://doi.org/10.1007/s10530-016-1207-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-016-1207-1