Abstract
The range of the barred owl (Strix varia) has expanded westward over the past century and now entirely overlaps the range of the federally threatened northern spotted owl (S. occidentalis caurina) in the Pacific Northwest. We compared Haemoproteus blood parasite assemblages among northern spotted owls in their native range and barred owls in both their native and invasive ranges to evaluate predictions of five hypotheses about parasites and biological invasions: (1) Enemy Release, where hosts benefit from a loss of parasites in their invasive range, (2) Enemy of My Enemy, where invasive hosts introduce parasites to naïve native hosts, (3) Parasite Spillback, where invasive hosts act as a new reservoir to native parasites, (4) Increased Susceptibility, where native hosts introduce parasites to naïve invasive hosts, and (5) Dilution Effect, where invasive species act as poor hosts to native parasites and decrease the density of potential hosts in their invasive range. We used haplotype network analyses to identify one haplotype common to both owl species throughout North America, three more haplotypes that appeared to be isolated to the barred owl’s historic range, and a fifth haplotype that was only found in California. Based on infection status and parasite diversity in eastern and western barred owl populations, we found strong support for the Enemy Release Hypothesis. Northern spotted owls had higher parasite diversity and probability of infection than sympatric barred owls, offering some support for the Parasite Spillback and Dilution Effect Hypotheses. Overall, this study demonstrates the complexity of host-parasite relationships and highlights some of the ways in which species’ range expansions may alter such relationships among both invasive and native hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Parasites can have profound impacts on natural systems by affecting both the ecology and evolution of host populations. When parasite communities change, cascade effects can lead to population-wide changes among host species present in the community through the loss of native parasites (e.g., Torchin et al. 2001), the introduction of novel parasites (e.g., van Riper et al. 1986), or a shift in the abundance of parasites (e.g., Fiorello et al. 2004). As anthropogenic impacts on natural environments increasingly alter parasite and disease dynamics among wildlife populations, parasites have become an important concern for conservation and management (Brearley et al. 2013).
Invasive species can impact parasite community dynamics in their non-native ranges, which in turn affects competitive interactions among invasive and native host species (Perkins et al. 2008). Ecological and phylogenetic similarities of hosts and parasites likely facilitate parasite transmission between invasive and native hosts (Lebarbenchon et al. 2007), yet the impacts caused by changes in parasite communities are not well known. Five hypotheses (Enemy Release, Parasite Spillover, Enemy of My Enemy, Increased Susceptibility, and Dilution Effect Hypotheses) have been proposed to explain how biological invasions can affect communities of “natural” parasites (those found in an invasive host species’ native range), and “native” parasites (those already present in the invasive host species’ introduced or expanded range) (Colautti et al. 2004).
The Enemy Release Hypothesis (ERH) predicts that invasive host populations will be infected with fewer parasite species in their introduced or expanded ranges compared to their native ranges (Torchin et al. 2003; Lebarbenchon et al. 2007). Invasive host populations are generally founded by a few individuals that are likely infected with only a subset of the parasites from larger source populations (Colautti et al. 2004). In addition, abiotic (e.g., climate) and biotic (e.g., vector abundance) differences between native and introduced environments can disrupt the life cycles of natural parasites, leading hosts to escape infections from such parasites in introduced ranges (Phillips et al. 2010). This freedom from parasites provides an invasive host both energetic and competitive advantages in introduced areas (Williamson 1996; Crawley 1997).
Once an invasive host is established in a novel area, four general scenarios can occur among invasive hosts, native hosts, and their parasites, each of which has different implications for the conservation and management of native host populations. Under the first scenario, parasites that accompany invasive host species through the invasion process may be transmitted to new hosts in the invader’s new environment (parasite “spillover” Power and Mitchell 2004). These parasites subsequently may be more virulent in naïve native hosts (Lymbery et al. 2014). Invasive hosts may benefit through apparent competition when the negative impacts of an introduced parasite are considerably higher among an area’s native hosts than for the invasive hosts (Colautti et al. 2004). These concepts have been encapsulated in the Enemy of My Enemy Hypothesis (EEH) (Sabelis et al. 2001), which predicts relatively higher infection prevalence, higher mean infection intensity, and a greater number of parasite lineages in native versus invasive hosts.
Under the second and third scenarios, invasive species may serve as novel, competent hosts for native parasites. The Parasite Spillback Hypothesis (PSH) predicts that invasive species will serve as new reservoirs for native parasites, subsequently increasing native hosts’ exposure to these parasites and the overall proportion of infected native hosts (Kelly et al. 2009). Alternatively, the Increased Susceptibility Hypothesis (ISH) predicts that invasive species will act as naïve hosts to native parasites and be more vulnerable to parasites in new areas they invade relative to native hosts, resulting in higher infection prevalence, higher mean infection intensity, and a greater number of parasite species in invasive host populations (Colautti et al. 2004). Given the high cost of infection, invasive hosts may be placed at a competitive disadvantage to native hosts under the ISH.
Finally, if invasive species serve as poor rather than competent hosts to parasites present in their new environment, the addition of invasive species may dilute the density of potential hosts in an area, which, in turn, can lower the number of native hosts infected with parasites over time (Paterson et al. 2011; Poulin et al. 2011). The Dilution Effect Hypothesis (DEH) predicts low parasite prevalence, diversity, and infection intensity in invasive host populations, as well as a decrease in parasite prevalence in native host populations over time.
We tested predictions of these hypotheses with respect to infection with avian blood parasites of the genus Haemoproteus among native northern spotted owls (Strix occidentalis caurina) and sympatric, invasive barred owls (S. varia) in northwestern California. Distributed throughout the Pacific Northwest, the northern spotted owl is considered threatened under the Endangered Species Act (US Fish and Wildlife Service 1990). The barred owl historically occurred from south-central Mexico north through the southern US and into the eastern US and Canada (Johnsgard 1988). In the early 1900s, the barred owl range began expanding westward to British Columbia and then south through the Pacific Northwest, reaching northern California by 1976 (Livezey 2009).
Today, barred owl presence has been linked to reductions in northern spotted owl site occupancy, reproduction, and survival; thus, barred owls pose a “significant and complex threat” to northern spotted owl populations (US Fish and Wildlife Service 2011). Additionally, the potential for barred owl range expansion to alter parasite assemblages and disease dynamics of both owl species has raised concerns over the parasite-mediated conservation implications of this range expansion for northern spotted owls (US Fish and Wildlife Service 2011). Among the parasites that have been listed as a potential threat to northern spotted owls are avian blood parasites of the genus Haemoproteus (Ishak et al. 2008). Haemoproteus comprise a diverse group of vector-borne parasites that have been used extensively to study host-parasite interactions in birds (e.g., Atkinson and van Riper 1991). Clinical signs of infection with Haemoproteus are typically mild in most bird species but severe anemia has been reported in raptors (Remple 2004) including several owl species (Mutlow and Forbes 2000).
Our overall objective was to gain a better understanding of the general patterns and principles shaping parasite transmission between invasive and native hosts in the context of host range expansion. Motivated by the competitive threat to northern spotted owls posed by invasive barred owls in northern California, we compared metrics of Haemoproteus parasite diversity and infection status between the two species to test predictions of the five hypotheses. We also included relevant ecological and demographic variables to gain additional insights into patterns of parasite distribution in these host species.
Materials and methods
Parasite metrics
We estimated parasite assemblage similarity, haplotype diversity, probability of infection, and infection intensity of Haemoproteus in three populations of owl hosts: northern spotted owls in their native geographic range in northwestern California, sympatric barred owls in their non-native range in northwestern California (“western barred owls”), and barred owls in their historic range in the eastern US (“eastern barred owls”). We used these metrics to test specific predictions under each of the five hypotheses (Table 1).
Haemoproteus assemblage similarity and haplotype diversity were analogous to species metrics (Poulin and Morand 2004), where a haplotype was considered a unique DNA sequence (Posada and Crandall 2001) that differed by at least one base pair in the cytochrome b region. We defined assemblage similarity as the total number of haplotypes present in both host populations (“shared haplotypes”) and haplotype diversity as a combination of haplotype richness (the number of unique haplotypes present in a single host population) and evenness (the number of birds infected with each haplotype) in a single host population (Poulin and Morand 2004). We estimated the probability of infection as (1) the probability that an owl was infected with a Haemoproteus spp., regardless of haplotype, and (2) the probability that an owl was infected with haplotypes shared by the populations of interest. In both cases, we focused on whether host population was important in predicting probability of infection. Finally, we estimated infection intensity as the proportion of infected blood cells divided by the total number of blood cells examined in a blood smear from an individual host.
Sample collection
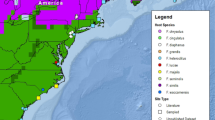
From April 2008 to September 2012, blood samples from northern spotted and western barred owls were collected from private and federal lands throughout Siskiyou, Trinity, Humboldt, and Mendocino counties in northwest California (Online Resource 1). This area was located near the barred owl’s invasion front, where the effects of host range expansion on parasite assemblages are expected to be most pronounced (Phillips et al. 2010). Blood was collected via brachial venipuncture from live captured individuals or within 10 min of death from barred owls shot as part of a removal experiment. Sex and age were recorded for each sampled bird, as well as UTM coordinates of the capture location using a GPS. Location information was subsequently imported into ArcMap 10 (ESRI, Redlands, CA) to calculate the distance (kilometers) of each owl’s sampling location to the coast.
For eastern barred owls, blood samples were collected year-round from March 2011 through May 2012 by eight raptor rehabilitation centers located throughout the barred owl’s historic range (Online Resource 1). While we recognize that this sampling strategy may bias estimates of prevalence if immune-compromised birds are overrepresented in rehabilitation centers, previous studies found negligible differences in prevalence between wild-caught and rehabilitation birds (Tella et al. 1999; Krone et al. 2001). In most cases (n = 155), samples were collected during routine examinations performed at the time of a bird’s admission. In some cases (n = 20), samples were collected from resident birds that had been in captivity for up to 3 years. Sex, age, and capture location (denoted as the closest city to which the bird was found) were recorded at the time of sample collection. Samples included: (1) thin blood smears using 1 drop (8 µL) of blood, (2) whole blood stored on lysis buffer, and (3) blood on Whatman filter paper or FTA cards (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) (Lewicki 2013). We stored all samples at room temperature until laboratory processing.
Laboratory analyses
Infection status
To test for the presence of Haemoproteus spp. in blood samples, we extracted genomic DNA from paper or buffer samples using a DNeasy extraction kit (Qiagen, Valencia, CA). We then followed a nested PCR protocol described by Hellgren et al. (2004). In each PCR run, we used aliquots of DNA from samples with known avian Haemoproteus spp. infections as positive controls and aliquots of purified water as negative controls. We ran 2µL of the PCR amplification product on a 2 % agarose gel with a 100 bp ladder (New England Biolabs, Ipswich, MA) followed by ethidium bromide staining, UV visualization, and digital imaging. Positive samples were identified by the presence of a band of moderate to bright intensity at approximately 480 base pairs in size on gels.
Because the nested PCR protocol amplified portions of the cytochrome b region of both Haemoproteus and Plasmodium parasite DNA, we sequenced samples considered positive using the initial screening protocol to differentiate infections with the two genera. Excess primers and unincorporated nucleotides were removed from 20µL aliquots of PCR product by adding 1µL of Exosap-IT® (USB Corporation, Cleveland, OH) and incubating at 37 °C for 15 min, followed by 80 °C for 15 min. Cleaned PCR products were cycle sequenced using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems Inc., Foster City, CA). Unincorporated dye terminators and salts from sequence products were then removed using a PrepEase® DNA Clean-Up Kit (USB Corporation, Cleveland, OH). Final sequencing products were visualized on an ABI Prism 3130 genetic analyzer (Applied Biosystems Inc., Foster City, CA). The sequences were deposited in GenBank, Accession Numbers KF747368–KF747372.
Haplotype identity
Sequences were aligned and edited using Sequencher v4.10.1 (Gene Codes Corporation, Ann Arbor, MI). We excluded sequences with one or more ambiguous peaks (e.g., polymorphisms or weak peaks) from any analyses involving haplotype assignment because only one base pair difference was required to classify a sequence as a unique haplotype. We compared our sequences with published sequences in GenBank (Benson et al. 2006) using the National Center for Biotechnology Information nucleotide Basic Local Alignment Search Tool (BLAST) and the MalAvi database (Bensch et al. 2009) to (1) assign each sequence to a parasite genus, (2) evaluate whether any of our haplotypes had been detected in other birds, and (3) evaluate how genetically similar our haplotypes were to published sequences.
We collated parasite sequences from GenBank that overlapped ours by at least 468 bp and that matched our sequences with 98 % or higher identity. We aligned all sequences using Sequencher v4.10.1 and constructed a haplotype table showing polymorphic sites using DNADiffer (Ritland 2012) (Online Resource 2). We then used a haplotype network approach to infer relationships among Haemoproteus haplotypes because it allowed for better resolution of relationships from intraspecific datasets with low genetic divergence and possibly non-hierarchical relationships among haplotypes than a phylogenetic tree (Posada and Crandall 2001). We used NETWORK v4.6 (Fluxus Technology, Suffolk, United Kingdom) to generate a median-joining (MJ) network (Bandelt et al. 1995) using the Haemoproteus haplotypes included in our haplotype table but limited to wild birds of North America, given our interest in parasite haplotypes present in barred and northern spotted owl ranges. Following Ricklefs et al. (2005), we defined “evolutionary lineages” as groups including ≥2 haplotypes that were separated from each other by ≤2 mutations and we included these lineage demarcations in the final haplotype network.
Infection intensity
To measure infection intensity, we examined blood smears from PCR-positive samples using an Olympus BX43 microscope with a DP72 digital camera and i-Solution Lite image analysis software (IMT i-Solution Inc., Burnaby, British Columbia, Canada). We photographed 25 fields within a 2 cm2 area of the slide at 1,000× magnification under oil immersion following the field selection protocol outlined in Lewicki (2013). Using the Manual Tag tool in i-Solution Lite, we counted the total number of erythrocytes in each field, as well as the number of erythrocytes infected with Haemoproteus spp. Infection intensity was estimated as the proportion of total erythrocytes enumerated in the 25 fields that were classified as infected.
Statistical analyses
Competing hypotheses were expressed as statistical models where Haemoproteus haplotype diversity, infection status, and infection intensity were response variables (Table 2). The number of individuals (i.e., sample size) included in an analysis of a single metric varied depending on (1) the availability of clean sequence data or blood smears, (2) the owl population of interest under a given hypothesis, (3) whether juveniles were included, and (4) whether an analysis focused on all sampled individuals or only infected individuals (Online Resource 3). Birds with ambiguous or multiple Haemoproteus haplotype infections were excluded from all analyses.
Parasite haplotype diversity
We estimated Haemoproteus haplotype diversity for each population using the software EstimateS (Colwell 2005). We estimated (1) the Shannon Index (Shannon and Weaver 1962; Sanders 1968), which emphasizes the haplotype richness component of diversity, and (2) the inverse of the Simpson Index (1/D) (Simpson 1949; Magurran 2004), which emphasizes the evenness component of diversity. For both indices, we estimated a bootstrap standard deviation among 1,000 randomizations. We inferred statistically different diversity values if their 95 % confidence intervals (CI) did not overlap the mean of the other group. We also used Nei and Li’s (1989) formula for nucleotide diversity, which quantifies the average number of nucleotide differences per site between two sequences. We calculated nucleotide diversity within each owl population using the software DNAsp (Librado and Rozas 2009).
Probability of infection and infection intensity
Because of concerns about small sample sizes in estimating probability of infection and infection intensity (Jovani and Tella 2006), we examined the effect that smaller sample sizes could have on our estimates by resampling the populations with larger sample sizes using our smallest sample size. For example, we estimated prevalence for northern spotted (n = 98) and eastern barred (n = 135) owls by bootstrapping these samples using the sample size for western barred owls (n = 49). We used a bootstrapping algorithm in PROC SURVEYSELECT in SAS v.9.3 software (SAS Institute Inc. 2011) where we randomly sampled without replacement for 1,000 replicate bootstrap samples (Cassell 2010). From these bootstrapped samples, we then calculated point estimates and 95 % CI for comparison with our naïve estimates.
Statistical models for probability of infection and infection intensity were analyzed using SAS v.9.3 software (SAS Institute Inc. 2011). We used logistic regression (PROC LOGISTIC) to estimate probability of infection, where infection status (‘infected’ or ‘not infected’) was the binary response variable, and generalized linear models (PROC GENMOD) to estimate infection intensity, where proportion of cells infected was the continuous response variable. We used normal-distribution based procedures in analyzing infection intensity because these methods perform better than do other distributions for over-dispersed count data (McDonald and White 2010).
Model sets included up to five biologically relevant ecological variables as covariates (Table 2; Lewicki 2013). The suite of variables included in a given model set depended on which populations of owls were compared in relation to our hypotheses. We used an information-theoretic approach (Burnham and Anderson 2002) to select appropriate models for inference. We used a bias-corrected version of Akaike’s Information Criteria (AICc) to objectively rank models and ΔAICc and Akaike weights to compare models (Burnham and Anderson 2002). We also used R 2 values as a measure of the proportion of variation in the data explained by each model.
To account for model selection uncertainty, we model averaged parameter estimates \((\tilde{\bar{\beta }})\) and their sampling variances across all models in a given model set (Burnham and Anderson 2002) and used the model-averaged estimates to compute 95 % CI. We also reported estimates of the effects of other important variables when competing models with non-trivial Akaike weights included these variables. The relevance of parameter estimates was assessed based on whether 95 % CI overlapped zero.
Results
We obtained blood samples from 353 owls (127 northern spotted, 51 western barred, and 175 eastern barred). Seventy-one samples were excluded from analyses to avoid bias from apparent misclassifications or ambiguous peaks (Online Resource 3). Of the remaining samples (n = 98 northern spotted, 49 western barred, and 135 eastern barred owls), northern spotted owls had slightly lower prevalence of Haemoproteus infection (76.5 %, 95 % CI 68.1, 84.9 %) than eastern barred owls (88.1 %, 95 % CI 82.6, 93.6 %), while western barred owls had the lowest prevalence of Haemoproteus infection (30.6 %, 95 % CI 17.7, 43.5 %). Mean infection intensity (infected cells/10,000 cells examined) was almost 100 times greater in northern spotted (95.0) than western barred owls (1.0). The smaller sample size for western barred owls had little effect on bootstrapped point estimates for either prevalence (northern spotted owl: 76.6 %; eastern barred owl: 88.1 %) or intensity (northern spotted owl: 94). Thus, we assumed differing samples sizes among the three populations had little effect on our subsequent analyses. Across samples from all populations, 478 base pairs of the cytochrome b gene were sequenced from a total of 209 owls with putative Haemoproteus spp. infections. Of these 209 infections, we detected five Haemoproteus haplotypes (H1–H5) representing four Haemoproteus lineages (Fig. 1). Based on our searches of GenBank and MalAvi using our search criteria, we found five other Haemoproteus haplotypes from other studies of birds in North America (EU627834, EU627836, EU627839, EU627840 from Ishak et al. 2008, and AF465589 from Ricklefs and Fallon 2002) that were sufficiently similar to ours to include in our haplotype network (Fig. 1; Online Resource 2). All of these haplotypes occurred in owls.
Haplotype network for Haemoproteus haplotypes detected in barred and northern spotted owls sampled in this study and all Haemoproteus haplotypes reported in GenBank or MalAvi databases for samples from wild birds of North America that shared 98 % or higher sequence agreement. Empty circles in the network indicate mutations, filled black circles are median vectors, and boxes encompass evolutionary lineages (groups of haplotypes separated by ≤2 mutations)
Enemy Release (ERH) Hypothesis
We detected four haplotypes (H1–H4; Fig. 1) among the eastern barred owls we sampled; two (H1 and H2) did not match any sequences published on GenBank or MalAvi as of 15 October 2013 (Online Resource 2). We detected H1 in 17 barred owls from states in the Midwest and northeastern United States. We found H2 in only one barred owl from Alabama. Because H2 differed from H1 by one base pair, we considered these as belonging to the same putative evolutionary lineage. We found H3 in only one barred owl from Alabama and it matched a haplotype found in a barred owl from Wisconsin in Ishak et al. (2008) (EU627840). The majority of eastern barred owls were infected with H4, which matched a Haemoproteus sequence detected in both a barred and a great horned owl from Florida (AF465589) and differed by one base pair from another sequence detected in several other owl species across the world (EU627834; Fig. 1; Online Resource 2).
We detected two haplotypes among the western barred owls we sampled. The first haplotype was the same haplotype detected in the majority of eastern barred owls (H4). The second haplotype, H5, was detected in only one barred owl from California and matched a Haemoproteus sequence detected in a California spotted owl (S. o. occidentalis) in Ishak et al. (2008) (EU627839; Fig. 1; Online Resource 2).
The Shannon diversity index for Haemoproteus in eastern barred owls (0.47, 95 % CI 0.46, 0.48) was almost twice that for western barred owls (0.25, 95 % CI 0.20, 0.30), and the Simpson diversity index was also higher for eastern barred owls (1.35, 95 % CI 1.30, 1.40) than for western barred owls (1.21, 95 % CI 1.15, 1.27). Nucleotide diversity was approximately twice as high for Haemoproteus in eastern (πx = 0.0067) than western (πx = 0.0037) barred owls.
The top-ranked model (Akaike weight = 0.638) for infection probability [Pr(Inf)] with any haplotype included only the population covariate (\(\tilde{\bar{\beta }}\) = 1.51, 95 % CI 1.07, 1.96; Table 3); eastern barred owls [Pr(Inf) = 0.903, 95 % CI 0.829, 0.947] had almost three times the probability of infection than western barred owls [Pr(Inf) = 0.313, 95 % CI 0.198, 0.456]. The top-ranked model for predicting the probability that a barred owl was infected with the shared H4 haplotype had an Akaike weight of 0.672 and, again, included the population covariate (\(\tilde{\bar{\beta }}\) = 1.03, 95 % CI 0.64, 1.42) as the only effect (Table 3); eastern barred owls [Pr(Inf) = 0.778, 95 % CI 0.685, 0.849] had 2.5 times the probability of infection with H4 than western barred owls [Pr(Inf) = 0.313, 95 % CI 0.200, 0.456].
Post-invasion hypotheses
We detected the same two haplotypes in northern spotted owls that we detected in western barred owls (H4 and H5). However, 10 (10.2 %) northern spotted owls were infected with H5 (the California-specific haplotype) compared to one (2.0 %) western barred owl (Fig. 1; Online Resource 2). Shannon and Simpson diversity indices were both much higher for Haemoproteus in northern spotted owls (0.38, 95 % CI 0.37, 0.39, and 1.30, 95 % CI 1.27, 1.34, respectively) than western barred owls (see above). In addition, parasite nucleotide diversity was approximately twice as large for Haemoproteus from northern spotted (πx = 0.0064) than sympatric barred owls (see above).
The top-ranked model for probability of infection had an Akaike weight of 0.158 and included effects of population and the natural log of distance to coast (LnDC; Table 3). Model-averaged estimates of these effects indicated that western barred owls had a lower probability of infection than northern spotted owls (\(\tilde{\bar{\beta }}\) = −0.90, 95 % CI −1.13, −0.66), while the probability of an owl being infected increased as the natural log of distance from the coast increased (\(\tilde{\bar{\beta }}\) = 0.65, 95 % CI 0.20, 1.10; Fig. 2a). Although the two top-ranked models included distance to the coast covariates (either log transformed or untransformed) with a combined Akaike weight of 0.280, models including the management intensity (MG) covariate also appeared to be competitive, with Akaike weights similar to the top-ranked models (Table 3). However, model-averaged estimates of the management intensity effect were not different than zero (\(\tilde{\bar{\beta }}\) = −1.46, 95 % CI −9.78, 6.87) and management intensity and distance to coast were highly correlated (r = −0.91).
Predicted probabilities of infection (a) and infection intensities (b) of Haemoproteus in northern spotted and western barred owls from northwest California. Probabilities of infection were estimated from a logistic regression model where owl population and the natural log of distance to the coast were covariates [Pr(Inf) ≈ PO + LnDC]; estimates for northern spotted owls are solid lines and for barred owls are dashed lines, and 95 % CI are shown in gray. Infection intensities were estimated from a general linear model in which population, sex, and a population by sex interaction were covariates (Intensity ≈ PO + SX + PO × SX)
On average, 2,431 (95 % CI 2,328, 2,534) erythrocytes were examined per bird for infection intensity analyses. The top-ranked generalized linear model included population, sex, and a population by sex interaction, with an Akaike weight of 0.235 (Table 3). Model-averaged estimates of these effects indicated that male western barred owls had the lowest predicted infection intensities (1 infected cell per 10,000 blood cells examined, 95 % CI −49, 51), while female northern spotted owls had the highest predicted infection intensities (90 infected cells per 10,000 blood cells examined, 95 % CI 43, 136; Fig. 2b). Female western barred owls and male northern spotted owls also had much lower infection intensities than female northern spotted owls (Fig. 2b).
Discussion
We compared five Haemoproteus assemblage metrics of northern spotted, western barred, and eastern barred owls in order to test predictions of five hypotheses that describe how host range expansion can affect parasite assemblages of both native and invasive host populations. Birds from all three populations were infected with Haemoproteus parasites, ranging from 33.3 to 89.9 % sample prevalence. Overall, Haemoproteus prevalence was highest in eastern barred owls and lowest in western barred owls. Our results provided strong support for the ERH, strong to moderate support for the Dilution Effect and Parasite Spillback Hypotheses, and very little support for the Enemy of My Enemy and Increased Susceptibility Hypotheses.
Enemy Release Hypothesis
Western barred owls were much less likely to be infected with Haemoproteus than eastern barred owls, which supports the ERH prediction that infection prevalence is lower in a host species’ invasive range than its native range (Table 1). We also detected lower haplotype diversity in western than eastern barred owls, which supports the ERH prediction that host populations escape native parasites when invading ecological communities in which those native parasites have yet to adapt (Table 1). Furthermore, the ERH predicts that as a host species invades new regions, rare parasites will be lost from invading host populations while common, generalist parasites will persist among host populations (Colautti et al. 2004). Our results support this prediction as well: eastern and western barred owls shared one haplotype (H4), and in both owl populations this haplotype comprised the majority (63 % eastern barred; 94 % western barred) of infections. This same haplotype was also found in the majority (87 %) of infected northern spotted owls, and a closely related (1 bp different) haplotype has been found in owls from North America, Africa, and Europe (Ishak et al. 2008), suggesting that this is a common, cosmopolitan haplotype.
Interestingly, western barred owls had a lower probability of infection with H4, contradicting the prediction that probability of infection should be similar among eastern and western barred owls for shared haplotypes. It is unlikely that the observed difference in probability of infection of H4 was driven by this haplotype being naturally rarer in the Pacific Northwest because northern spotted owls had a high prevalence of this haplotype. Alternative explanations may include (1) western barred owls may have new behavioral adaptations and habitat associations that have decreased their exposure to Haemoproteus vectors, and/or (2) western barred owls may have lower susceptibility to infection with these avian blood parasites.
The observed higher prevalence in eastern versus western barred owls is similar to the pattern observed by Ishak et al. (2008) and supports the notion that suitable habitat for and/or abundance of Haemoproteus vectors is heterogeneous and fragmented across North America. Haemoproteus parasites require warm temperatures for development in biting midge vectors (Valkiūnas 1996). Cold temperatures in the Rocky and Cascade Mountain ranges may hinder rarer Haemoproteus haplotypes from accompanying barred owl hosts with invasion of the west because of the parasites’ dependence on these warmer temperatures; sampling populations from the invasion corridor along southern Canada would allow for a better test of this hypothesis.
Post-invasion hypotheses
Of the two haplotypes detected in northern spotted and western barred owls, one (H4) appears to be common and cosmopolitan, while the other (H5) was detected only in California. We found no evidence that either of these haplotypes originated from eastern North America, which fails to support the EEH prediction that barred owls would introduce novel Haemoproteus haplotypes to northern spotted owls through range expansion. The California-specific haplotype (H5) is noteworthy because it has not been documented in other studies (e.g., Perkins and Schall 2002; Ricklefs and Fallon 2002) and is genetically distant (≥13 bp different) from Haemoproteus haplotypes detected in previous studies. Discovery of this haplotype lends support to a prediction of the ISH because it suggests that barred owls may be acquiring new Haemoproteus haplotypes as they expand their range (Table 1); however, we observed that northern spotted owls had a higher probability of infection and infection intensity than western barred owls, providing strong support against ISH predictions that these parasites would more negatively affect western barred than northern spotted owls (Table 1). Intuitively, these results are not surprising; if Haemoproteus were having a strong, negative impact on western barred owl populations, it is doubtful that these populations would invade the Pacific Northwest as successfully as has been observed in the past several decades (US Fish and Wildlife Service 2011).
Parasite similarity, probability of infection, and infection intensity results all lend support to PSH predictions that barred owls acquire new parasites as they expand their range and that these parasites would more negatively affect northern spotted owls (Table 1); however, an alternative explanation for the low probability of infection, parasite diversity, and infection intensity observed in western barred owls is that western barred owls are poor hosts to parasites in their invasive range, which supports predictions of the DEH (Table 1). Without historic data on Haemoproteus haplotypes of northern spotted owls, we could not evaluate a central component of these hypotheses—how Haemoproteus prevalence has changed in northern spotted owls since the barred owl range expansion. The conservation implications of these two hypotheses differ markedly: under the PSH, the presence of barred owls may negatively affect northern spotted owls by perpetuating high Haemoproteus prevalence in northern spotted owls while, under the DEH, the presence of barred owls may indirectly benefit northern spotted owls by decreasing Haemoproteus prevalence. Given these implications, it will be important to continue monitoring for changes in Haemoproteus assemblages in these two host species to better understand the impacts of barred owl range expansion on northern spotted owl health.
Inferences about the transmission of parasites between native and invasive hosts would also benefit from future studies evaluating the role of additional biological variables in shaping transmission dynamics. In our study, the highest-ranked probability of infection model included the natural log of distance to coast (LnDC, Table 3), which may be related to the regional distribution of Haemoproteus vectors. Generally, the biting midge vectors of Haemoproteus have higher reproductive success at warmer temperatures (Mellor et al. 2000), and, in our study area, inland sites were characterized by cool, wet winters and hot, dry summers, while coastal sites experienced milder temperatures and higher year-round precipitation (Ting 1998; Franklin et al. 2000). It is possible that the warmer temperatures inland during the peak parasite transmission season led to increased vector abundance at these sites and, in turn, higher levels of parasite transmission. Vector abundance has also been used to explain differences in parasite prevalence between avian assemblages of disturbed and undisturbed sites. Chasar et al. (2009) found lower blood parasite prevalence in areas with high disturbance, possibly because areas of high disturbance provide less suitable habitat for vectors than undisturbed areas. Similar reasoning may explain why many of our highest-ranking models included management intensity (Table 3), which predicted a higher probability of infection in lower intensity management areas. However, distance to coast and management intensity were highly correlated because of our opportunistic sampling; most of our samples on low-intensity management areas were further inland than those on high-management areas. Although distance to coast was more strongly supported in our analyses, understanding the differing effects of these two covariates on Haemoproteus transmission would be improved by more even sampling between sites with low and high intensity management at coastal and inland areas. We did not find a similar relationship between infection intensity and distance to coast (Table 3); sex and species appeared to have the strongest effects with female northern spotted owls having a higher infection intensities than males or either sex in barred owls. Other studies have detected sex-biased parasitism of blood parasites in owl hosts (e.g., Korpimaki et al. 1993), which may be driven by differences in life history traits and exposure to environmental stressors between the sexes.
Overall, our results suggest that Haemoproteus assemblage dynamics of northern spotted owls are not solely influenced by the presence or absence of invasive barred owls, and evaluating the role of additional biological variables may help broaden our global understanding of the relationships among invasive and native hosts, their parasites, and the environment.
The true cost of parasitism?
We found that northern spotted owls were more likely to be infected with Haemoproteus haplotypes than sympatric barred owls, but our study did not directly evaluate if and to what extent parasite infection status and intensity influence northern spotted owl and barred owl fitness. Although generally considered to be relatively innocuous in their avian hosts, Haemoproteus parasites can become pathogenic when coupled with additional stressors (Remple 2004), such as competition with barred owls.
We found strong support for the ERH for Haemoproteus parasites of invasive barred owls, but the true cost of Haemoproteus infections also has implications for invasive barred owl fitness. If Haemoproteus parasites are relatively innocuous to their barred owl hosts, the loss of these parasites among western barred owls may not have much biological relevance. Nevertheless, our results demonstrate an important pattern that may be occurring among more cost-demanding parasites that were not examined in this study. Thus, we echo Ishak et al. (2008) suggestion that follow-up studies should evaluate the relationship of infection status with immunocompetence, estimated survival and reproductive rates for infected compared to uninfected birds, and competitive interactions of both northern spotted and barred owls.
Finally, we compared parasite haplotype diversity in this study under the assumption that host populations infected with a low diversity of parasites were more immunologically competent than host populations infected with a high diversity of parasites. However, Hudson et al. (2006) argue that high native parasite diversity may be an indicator of the health of a given ecosystem because it is often a result of long chains of multispecies connections that can only be present in healthy ecosystems. We detected greater haplotype diversity among northern spotted owls than western barred owls, and Ishak et al. (2008) reported a high diversity of Leucocytozoon blood parasite lineages among northern spotted owls relative to Leucocytozoon assemblages of other owl species across the world. If the blood parasite infections we detected among northern spotted owls are a result of host-vector-parasite interactions that have co-evolved over a long period of time, then our study suggests that Haemoproteus infections may be benign if not beneficial in northern spotted owls. Svensson-Coelho et al. (2013) found that avian host species with high Haemoproteus prevalence showed low Plasmodium prevalence and vice versa. One explanation for this observed pattern is that infection of parasites from one genus may inhibit infection of parasites from another. In the context of our study system, it is possible that northern spotted owls have adapted to high Haemoproteus prevalence as part of a defense mechanism against more virulent Plasmodium parasites, which are seemingly rare among northern spotted owls (Gutiérrez 1989; Ishak et al. 2008; Lewicki 2013). Future studies on this concept in northern spotted owls would help elucidate both the role that blood parasites have on northern spotted owl fitness and the complex relationships between blood parasites and avian hosts in the face of invasion in general.
References
Atkinson CT, Van Riper CIII (1991) Pathogenicity and epizootiology of avian haematozoa: Plasmodium, Leucocytozoon, and Haemoproteus. In: Loye JE, Zuk M (eds) Bird-parasite interactions: ecology, evolution, and behavior. Oxford University Press, London, pp 19–48
Bandelt HJ, Forster P, Sykes BC, Richards MB (1995) Mitochondrial portraits of human populations using median networks. Genetics 141:743–753
Bensch S, Hellgren O, Pérez-Tris J (2009) MalAvi: a public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol Ecol Resour 9:1353–1358
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2006) GenBank. Nucleic Acids Res 34:D16–D20
Brearley G, Rhodes J, Bradley A, Baxter G, Seabrook L, Lunney D, Liu Y, McAlpine C (2013) Wildlife disease prevalence in human-modified landscapes. Biol Rev 88:427–442
Burnham KP, Anderson DR (2002) Model selection and multimodel inference: a practical information-theoretic approach, 2nd edn. Springer, New York
Cassell DL (2010) BootstrapMania!: re-sampling the SAS® way. In: SAS Institute (ed) Proceedings of the SAS® Global Forum 2010 conference, pp 1–11
Chasar A, Loiseau C, Valkiūnas G, Iezhova T, Smith TB, Sehgal RNM (2009) Prevalence and diversity patterns of avian blood parasites in degraded African rainforest habitats. Mol Ecol 18:4121–4133
Colautti RI, Ricciardi A, Grigorovich IA, MacIsaac HJ (2004) Is invasion success explained by the enemy release hypothesis? Ecol Lett 7:721–733
Colwell RK (2005) EstimateS: statistical estimation of species richness and shared species from samples, version 7.5. http://purl.oclc.org/estimates. Accessed 20 March 2013
Crawley MJ (ed) (1997) Plant invasions. Blackwell Science, Oxford
Fiorello C, Deem S, Gompper M, Dubovi E (2004) Seroprevalence of pathogens in domestic carnivores on the border of Madidi National Park, Bolivia. Anim Conserv 7:45–54
Franklin AB, Anderson DR, Gutiérrez RJ, Burnham KP (2000) Climate, habitat quality, and fitness in northern spotted owl populations in northwestern California. Ecol Monogr 70:539–590
Gutiérrez RJ (1989) Hematozoa from the spotted owl. J Wildl Dis 25:614–618
Hellgren O, Waldenström J, Bensch S (2004) A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium and Haemoproteus from avian blood. J Parasitol 90:797–802
Hudson PJ, Dobson AP, Lafferty KD (2006) Is a healthy ecosystem one that is rich in parasites? Trends Ecol Evol 21:381–385
Ishak HD, Dumbacher JP, Anderson NL, Keane JJ, Valkiūnas G, Haig SM, Tell LA, Sehgal RNM (2008) Blood parasites in owls with conservation implications for the spotted owl (Strix occidentalis). PLoS One 3:e2304
Johnsgard PA (1988) North American owls: biology and natural history. Smithsonian Institution Press, Washington
Jovani R, Tella JL (2006) Parasite prevalence and sample size: misconceptions and solutions. Trends Parasitol 22:214–218
Kelly DW, Paterson RA, Townsend CR, Poulin R, Tompkins DM (2009) Parasite spillback: a neglected concept in invasion ecology? Ecology 90:2047–2056
Korpimaki E, Hakkarainen H, Bennet GF (1993) Blood parasites and reproductive success of Tengmalm’s owls: detrimental effects on females but not males? Funct Ecol 7:420–426
Krone O, Priemer J, Streich J, Sommer P, Langgemach T, Lessow O (2001) Haemosporida of birds of prey and owls from Germany. Acta Protozool 40:281–289
Lebarbenchon C, Poulin R, Thomas F (2007) Parasitism, biodiversity, and conservation biology. In: Thomas F, Guegan J-F, Renaud F (eds) Ecology and evolution of parasitism. Oxford University Press, Oxford, pp 149–160
Lewicki KE (2013) Haemosporidian parasites of barred owls (Strix varia) and northern spotted owls (S. occidentalis caurina): investigating the effects of an invasive species on parasite transmission and community dynamics. Thesis, Colorado State University
Librado P, Rozas J (2009) DNAsp v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Livezey KB (2009) Range expansion of barred owls, part I: chronology and distribution. Am Midl Nat 161:49–56
Lymbery AJ, Morine M, Kanani HG, Beatty SJ, Morgan DL (2014) Co-invaders: the effects of alien parasites on native hosts. Int J Parasitol 3:171–177
Magurran A (2004) Measuring biological diversity. Blackwell Science Ltd., Malden
McDonald TL, White GC (2010) A comparison of regression models for small counts. J Wildl Manag 74:514–521
Mellor PS, Boorman J, Baylis M (2000) Culicoides biting midges: their role as arbovirus vectors. Annu Rev Entomol 45:307–340
Mutlow A, Forbes N (2000) Haemoproteus in raptors: pathogenicity, treatment, and control. In: Proceedings of the Association of Avian Veterinarians, pp 157–163
Nei M, Li J (1989) Variances of the average number of nucleotide substitutions within and between populations. Mol Biol Evol 6:290–300
Paterson RA, Townsend CR, Poulin R, Tompkins DM (2011) Introduced brown trout alternative acanthocephalan infections in native fish. J Anim Ecol 80:990–998
Perkins SL, Schall JJ (2002) A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol 88:972–978
Perkins SE, Altizer S, Bjornstad O, Burdon JJ, Clay K, Gomez-Aparicio L, Jeschke JM, Johnson PT, Lafferty KD, Malmstrom CM, Martin P, Power A, Strayer DL, Thrall PH, Uriarte M (2008) Invasion biology and parasitic infections. In: Ostfeld RS, Keesing F, Eviner VT (eds) Infectious disease ecology: effects of ecosystems on disease and of disease on ecosystems. Princeton University Press, Princeton, pp 179–204
Phillips BL, Kelehear C, Pizzatto L, Brown GP, Barton D, Shine R (2010) Parasites and pathogens lag behind their host during periods of host range advance. Ecology 91:872–881
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Poulin R, Morand S (2004) Parasite biodiversity. Smithsonian Books, Washington
Poulin R, Paterson RA, Townsend CR, Tomkins DM, Kelly DW (2011) Biological invasions and the dynamics of endemic diseases in freshwater ecosystems. Freshw Biol 56:676–688
Power AG, Mitchell CE (2004) Pathogen spillover in disease epidemics. Am Nat 164:S79–S89
Remple JD (2004) Intracellular hematozoa of raptors: a review and update. J Avian Med Surg 18:75–88
Ricklefs RE, Fallon SM (2002) Diversification and host switching in avian malaria parasites. Proc R Soc Lond B Biol Sci 269:885–892
Ricklefs RE, Swanson BL, Fallon SM, Martinéz-Abraín A, Scheuerlein A, Gray J, Latta SC (2005) Community relationships of avian malaria parasites in southern Missouri. Ecol Monogr 75:543–559
Ritland K (2012) DNADiffer. http://gdc.forestry.ubc.ca/downloads. Accessed 25 July 2013
Sabelis MW, Janssen A, Kant MR (2001) Ecology: the enemy of my enemy is my ally. Science 291:2104–2105
Sanders HL (1968) Marine benthic diversity: a comparative study. Am Nat 102:243–282
SAS Institute Inc (2011) SAS® 9.3 SQL Procedure user’s guide. SAS Institute Inc., Cary, NC
Shannon CE, Weaver W (1962) The mathematical theory of information. University of Illinois Press, Urbana
Simpson EH (1949) Measurement of diversity. Nature 163:688
Svensson-Coelho M, Blake JG, Loiselle BA, Penrose AS, Parker PG, Ricklefs RE (2013) Diversity, prevalence, and host specificity of Avian Plasmodium and Haemoproteus in a western amazon assemblage. Ornithol Monogr 76:1–47
Tella JL, Blanco G, Forero MG, Gajón Á, Donázar JA, Hiraldo F (1999) Habitat, world geographic range, and embryonic development of hosts explain the prevalence of avian hematozoa at small spatial and phylogenetic scales. Proc Natl Acad Sci USA 96:1785–1789
Ting TF (1998) The thermal environment of northern spotted owls in northwestern California: possible explanations for use of interior old growth and coastal early successional stage forest. Thesis, Humboldt State University
Torchin ME, Lafferty KD, Kuris AM (2001) Release from parasites as natural enemies: increased performance of a globally introduced marine crab. Biol Invasions 3:333–345
Torchin ME, Lafferty KD, Dobson AP, McKenzie VJ, Kuris AM (2003) Introduced species and their missing parasites. Nature 421:628–630
US Fish and Wildlife Service (1990) 50 CFR Part 17 Endangered and threatened wildlife and plants; determination of threatened status for the northern spotted owl; final rule. Fed Regist 55:26114–26194
US Fish and Wildlife Service (2011) Revised recovery plan for the Northern Spotted Owl (Strix occidentalis caurina). US Fish and Wildlife Service, Portland
Valkiūnas G (1996) Ecological implications of hematozoa in birds. Bull Scand Soc Parasitol 6:101–103
van Riper C, van Riper SG, Goff ML, Laird M (1986) The epizootiology and ecological significance of malaria in Hawaiian land birds. Ecol Monogr 56:327–344
Williamson M (1996) Biological invasions. Chapman & Hall, London
Acknowledgments
We thank Peter Carlson and Jeremy Rockweit of Klamath Biological Research Station; Mark Higley and the Hoopa Valley Tribe; Green Diamond Resource Company; Robert Feamster and Sierra Pacific Industries; and Laurie Clark and the National Council for Air and Stream Improvement, Inc., for allowing us to conduct research on their lands and for assistance in collecting western owl samples. The Avian Conservation Center (South Carolina), Wildcare Foundation (Oklahoma), Avian Haven (Maine), Carolina Raptor Center (North Carolina), Audubon of Florida (Florida), The Raptor Center (Minnesota), Tri-State Bird Rescue & Research (Delaware), and Alabama Raptor Center (Alabama) collected all the eastern barred owl samples analyzed in this study. Additional field and laboratory assistance came from Constanza Toro, Annie Kellner, Matthew Hopken, Nikki Crider, and Jeremy Dertien. Dr. Thomas Gidlewski provided access to his microscope; Nic Berrong assisted in installing and navigating the i-Solution Lite software; and Drs. Ellen Martinsen and Robert Ricklefs provided positive control samples. Finally, Drs. Liba Pejchar, Brian Foy, Ken Burnham, and Ann Hess assisted in study design and analysis and 2 anonymous reviewers provided helpful comments and suggestions that greatly improved our manuscript. This work was conducted under the auspices of the Colorado State University Institutional Animal Care and Use Committee protocol #10-1818A. Funding and additional support for this project was provided by the U.S.D.A. Forest Service Region 5 contract 11-CS-11052007-319 and Colorado State University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Lewicki, K.E., Huyvaert, K.P., Piaggio, A.J. et al. Effects of barred owl (Strix varia) range expansion on Haemoproteus parasite assemblage dynamics and transmission in barred and northern spotted owls (Strix occidentalis caurina). Biol Invasions 17, 1713–1727 (2015). https://doi.org/10.1007/s10530-014-0828-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-014-0828-5