Abstract
A Southern hemisphere lineage of the blue mussel Mytilus galloprovincialis has been diverging in allopatry from Northern hemisphere conspecifics for 0.84–1.2 million years. Secondary contact between Southern and Northern hemisphere mussels in Chile, New Zealand and Australia provides an opportunity to better understand the extent and consequences of extensive range expansion. Non-native M. galloprovincialis and hybrids, as detected from RFLP assays of nuclear and mitochondrial DNA, are present in all three countries and significant cytonuclear disequilibria exist for native homozygotes in Chile and New Zealand, non-native homozygotes in Chile and non-native heterozygotes in New Zealand. Introductions into Australia are rare events given that no pure non-native mussels were detected. Immigration from one or both taxa into the hybrid zone may underlie disequilibria in New Zealand, whilst gender-directional crossing with limited ongoing hybridization contributes to disequilibria in Chile. Hybridization dynamics do not pose a threat to the Southern lineage in Chile and Australia, but in New Zealand, introgression, continued immigration and slight hybridization gender bias towards non-native maternal parents could lead to the regional extirpation of the native lineage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dispersal, selection and assortative fertilization are driving forces that maintain and shape the spatial genetic composition of hybrid zones (Arnold 1997; Barton and Hewitt 1989; Gardner 1994, 1997; Mallet 2007, and reference therein). Such zones have long been of interest because of what they can tell us about natural processes contributing to speciation. This may include the influence of differential parental versus hybrids fitness or the contribution of environmental factors to the structural origin and maintenance of hybrid zones as a stepped cline between “pure” parental types or as a mosaic of parental and hybrid genotypes (e.g., Harrison and Rand 1989). More recently, there has been increasing recognition of human-mediated deliberate or accidental introductions and the role these play in threatening genetic integrity of native species by promoting speciation through introgressive hybridization (Rhymer and Simberloff 1996; Simberloff 2005). Such events may have profound conservation implications and management objectives have to take such outcomes into consideration (Keller and Taylor 2010; Simberloff 2005).
Cytonuclear disequilibria, analogous to the measure of linkage disequilibria in the context of two nuclear and mitochondrial genes, may be used to examine the extent and direction of introgressive hybridization, infer fertilization success by determining gender direction in crosses and monitor the flow of non-native alleles into a native population (Arnold et al. 1988; Asmussen et al. 1987, 1989; Avise 2000; Avise and Saunders 1984). With respect to the introduction of non-native species, cytonuclear disequilibria statistics are especially useful for determining the degree of consistency among multiple contact regions and evaluating the relative conservation impacts of such introductions (Avise 2000). Patterns of disequilibria documented under experimental conditions (e.g., Scribner and Avise 1993, 1994) provide a reference for evaluating the signatures of introgressive hybridization processes occurring in natural settings (Arnold 1993; Asmussen et al. 1987; Avise 2000) and are used in this context as a posteriori hypotheses.
Taxonomic differentiation within the “Mytilus edulis species complex”—M. edulis (Linne. 1758), M. galloprovincialis (Lam. 1819) and M. trossulus (Gould 1850)—may rely on morphometric, biochemical and molecular methods (Daguin and Borsa 2000; Gardner and Thompson 2001; Gérard et al. 2008; McDonald and Koehn 1988; McDonald et al. 1991; Seed 1992; Westfall and Gardner 2010). These mussels are widely distributed throughout the world and exhibit an anti-tropical distribution (Hilbish et al. 2000). Sexes are separate and gametes are shed directly into the sea where fertilization is external. As such, there is no active mate choice (Gardner 1997; Seed 1992).
Hybridization occurs readily between Mytilus sibling species in areas of sympatry, resulting in variable patterns of genetic introgression attributed to assortative mating via external fertilization, habitat selection and hybrid unfitness (Gardner 1994, 1997; Kijewski et al. 2006; Rawson et al. 1999; Riginos et al. 2004). Extensive interbreeding at numerous spatially well defined zones of contact (e.g., Bierne et al. 2003; Gardner 1996, 1997) has demonstrated that Mytilus spp. hybrid zones are often characterized by asymmetric introgression of genes and semi-permeable barriers to gene flow (Kijewski et al. 2006; Rawson and Hilbish 1998; Rawson et al. 1999). The recognition of human-mediated blue mussel introductions into many countries or regions (e.g., South Africa, Hong Kong, Japan, the Pacific coast of North America and more recently into the Southern hemisphere—Bownes and McQuaid 2006; Brannock et al. 2009; Elliott et al. 2008; Geller 1999; Grant and Cherry 1985; Heath et al. 1995; Lee and Chown 2007; Westfall and Gardner 2010) has highlighted the threat that bioinvasion can play in an ecological and a genetic sense by displacement of native biota and introgression into or swamping of native genotypes. Modeling the introduction and spread of genes into an invaded or newly colonized area demonstrates rapid and massive introgression of neutral genes and that most documented patterns of introgression for plant and animal species are consistent with this model (Currat et al. 2008). Given the propensity of smooth-shelled blue mussels to interbreed, the ease with which these mussels are moved around the globe and the lack of baseline knowledge about the situation in the Southern hemisphere, this threat is now viewed as serious (Westfall and Gardner 2010).
Unique genetic lineages of Mytilus galloprovincialis are present in the Northern and Southern hemispheres and have been diverging in allopatry for approximately 0.84 (Gérard et al. 2008) to 1.2 (Hilbish et al. 2000) million years, although evolutionary relationships among species and lineages are still under investigation. Mitochondrial gene trees define reciprocally monophyletic lineages of Northern and Southern hemisphere M. galloprovincialis (Borsa et al. 2007; Daguin and Borsa 2000; Gérard et al. 2008; Hilbish et al. 2000), leading to the suggestion that these geographically separated lineages be considered as regional subspecies (sensu Moritz 1994) for the purposes of conservation (Westfall and Gardner 2010).
The blue mussel Mytilus galloprovincialis (putatively from the Northern hemisphere) has been classified as one of the top 100 invasive threats in the world (Lowe et al. 2000). The Northern hemisphere lineage has previously been identified in Australia (Gérard et al. 2008), New Zealand (suggested as a possibility in Hilbish et al. 2000) and Chile (Daguin and Borsa 2000; Toro et al. 2005) where human-mediated vectors are the probable causes of these introductions (Westfall and Gardner 2010; Westfall et al. 2010). Another population of non-native blue mussels exists in South Africa but will not be considered further because of the natural absence of native Southern hemisphere blue mussels in this region (Grant and Cherry 1985). The introduction of non-native mussels to the Southern hemisphere is a cause of concern for conservation of the recently identified unique genetic lineage. Extensive hybridization between hemispheric lineages of M. galloprovincialis is predicted in all regions of co-occurrence due to their close taxonomic affinity (Westfall and Gardner 2010) and historical precedent within sympatric populations (e.g., Kijewski et al. 2006; Rawson and Hilbish 1998; Rawson et al. 1999). Comparing the outcomes of hybridization among regions will aid management priorities for incursion prevention and extirpation of non-native mussels or in the development of mariculture programs by identifying the potential outcome of non-native mussel spat importation. In such cases, managers or policy makers may be better informed about genetic impacts of current or future incursions and also mariculture practices. Monitoring the presence of non-native alleles in native populations will be a principal tool for assessing the efficacy of existing maritime biosecurity policies.
The following investigation examines hybridization dynamics between Northern and Southern Mytilus galloprovincialis lineages with the goal of evaluating relative conservation impacts of non-native mussel introductions in newly identified hybrid regions in Chile, New Zealand and Australia (Westfall and Gardner 2010). Cytonuclear associations between native and non-native mussels are quantified and specific disequilibria sign patterns and zone architecture are compared among regions to investigate the effects of anthropogenic introduction on the genetic composition of native Southern hemisphere blue mussels. Results are discussed in the context of the genetic conservation of the unique Southern hemisphere M. galloprovincialis lineage, advancing predictions for outcomes in each region.
Materials and methods
Sampling regions
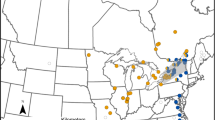
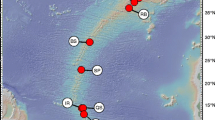
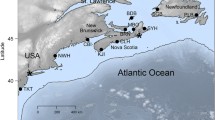
Three Southern hemisphere countries with known non-native mussel presence (Westfall et al. 2010) were sampled for blue mussels (total n = 190) (Table 1). In New Zealand, sample locations ranged from the Bay of Islands (North Island) to Lyttleton Harbour (South Island) (Fig. 1a) (Table 1). Two locations were sampled in each of Australia (Fig. 1b) and Chile (Fig. 1c) (Table 1).
Maps of sample locations in a New Zealand, b Chile and c Australia. In (a) the inset is a detailed picture of the Bay of Islands. Locations are given as 2-letter codes for cytonuclear data (Table 2) and 4-letter codes for phylogenetic accuracy data (Table 1), note that some locations have both codes provided
Taxonomic status
Detailed DNA extraction methods are provided by Westfall and Gardner (2010). A mitochondrial DNA 16s RFLP assay (Westfall et al. 2010) classifies three Mytilus galloprovincialis haplogroups: Southern hemisphere M. galloprovincialis, Northern hemisphere M. galloprovincialis “Mediterranean” haplogroup and a shared “North Atlantic” M. galloprovincialis/M. edulis haplogroup (Hilbish et al. 2000; Rawson and Hilbish 1995). In the latter group, M. galloprovincialis are distinguished from M. edulis using the Me15/16 nuclear DNA diagnostic marker (Inoue et al. 1995). For further details of assay procedures refer to Westfall et al. (2010). The male mitochondrial mitotype is eliminated due to length variation of the 16s rRNA PCR amplicon.
The nuclear DNA RFLP assay targets the Me15/16 PCR amplicon (Inoue et al. 1995) and was developed by Santaclara et al. (2006) to differentiate Mytilus galloprovincialis and M. chilensis. Recent reclassification of M. chilensis to M. galloprovincialis (Westfall and Gardner 2010) supporting the Southern hemisphere lineage divergence of M. galloprovincialis identified by Hilbish et al. (2000) and Gérard et al. (2008) means that the assay differentiates between Northern and Southern hemisphere M. galloprovincialis. The assay from Santaclara et al. (2006) was modified as follows: enzymatic digestion in a 25 μL total reaction volume contained 50 ng DNA, 1 U restriction endonuclease AciI, 1X NEB (New England Biolabs) Buffer #3, and 100 mg bovine serum albumin, digested at 37 °C for 1 h and heat inactivated at 65 °C for 20 min. From Santaclara et al. (2006), the M. galloprovincialis Me15/16 amplicon of 126 bp contains a single restriction site resulting in fragments of 69 and 57 bp, whereas Southern hemisphere M. galloprovincialis mussels have a point mutation that has removed the AciI cut site. In our investigation, two fragments are generated for the Northern M. galloprovincialis at 77 and 49 bp, while the Southern M. galloprovincialis Me15/16 amplicon remains uncut at 126 bp. Me 15/16 amplicon sequences of known Southern (n = 10) and Northern (n = 8) hemisphere M. galloprovincialis lineages (based on geographic origin) were subject to virtual restriction digest to ensure assay integrity with observed and predicted fragment profiles (GenBank Acc. HQ257459 to HQ257475; present paper).
Virtual restriction digests of GenBank sequences submitted by Santaclara et al. (2006) (GenBank Acc. DQ640590 to DQ640610) generate fragments of 53 and 49 bp for Mytilus galloprovincialis and no cut site for “M. chilensis”, constituting a total sequence length of 102 bp, therefore these database sequences were reported without the reverse primer of 20 bp (giving a total length of 126 bp as in Inoue et al. 1995). Virtual restriction digests of Genbank sequences with the reverse primer sequence added (see Inoue et al. 1995 for primer sequence) generate restriction fragments of 77 and 49 bp for Northern M. galloprovincialis, an exact match to results from our virtual (from sequenced individuals) and laboratory digests.
Phylogenetic accuracy of markers
To ensure that the cytoplasmic and nuclear allelic polymorphism identified by the RFLP markers accurately reflects Mytilus galloprovincialis hemispheric lineage taxonomy, subsets of individuals were sequenced for all taxonomic variants (Table 1). Phylogenies were reconstructed for subsets of mussels where initial taxonomic designation was assigned from geographic origin (Fig. 1; Table 2). In the Southern hemisphere where possible, mussels were sampled from regions well removed from hybrid zones (Fig. 1; Table 2). The goal of phylogenetic reconstructions was to establish the taxonomic differentiation between Northern and Southern hemisphere M. galloprovincialis that is captured by both the nuclear and cytoplasmic RFLP assays.
16s
A 527 bp fragment of the mitochondrial 16s rRNA gene was amplified using the universal primers 16sAR/16sBR (Palumbi 1995) under the following cycling conditions: 95 °C, 3′; [95 °C, 30″; 52 °C, 30″; 72 °C, 45″] ×30; 72 °C, 3′. After sequencing, a reliable 420 bp fragment of the 16s rRNA gene was used for phylogenetic reconstruction. An unrooted neighbour-joining tree was generated from 16s rRNA gene M. galloprovincialis sequences (n = 31) in MEGA v4.0.1 (Tamura et al. 2007) with 500 bootstrap iterations.
Me15/16
Northern (n = 11) and Southern (n = 10) hemisphere sequences described by Santaclara et al. (2006) (GenBank Acc. DQ640590 to DQ640610) were aligned with a reference Mytilus galloprovincialis genome sequence (GenBank Acc. AY497292). Additional Northern (n = 8) and Southern (n = 10) hemisphere M. galloprovincialis (as defined by geographic placement) individuals were directly sequenced from Me15/16 PCR products (Inoue et al. 1995) and simply aligned with GenBank sequences. An unrooted maximum parsimony tree was generated from GenBank and newly obtained M. galloprovincialis sequences (n = 18) in MEGA v4.0.2 (Tamura et al. 2007) with 500 bootstrap iterations.
Cytonuclear disequilibria statistics
The program CNDd (Asmussen et al. 1987; Asmussen and Basten 1994) tests for cytonuclear disequilibrium (D) and normalized disequilibrium (D’—within minimal and maximal marginal frequencies specific to each region) (Asmussen and Basten 1996) for all cytonuclear genotypic and allelic combinations grouped by country. The program requires coding the resulting 16s RFLP’s three haplogroups into two while conserving hemispheric lineage identification: nuclear alleles were coded as capital S (Southern lineage) and N (Northern lineage) and mitochondrial alleles as lowercase s and n, respectively. The composite genotype and haplotype are referred to as the cytonuclear genotype; individuals typed as pure Southern lineage are S/S–s and Northern lineage are N/N–n. We note that the use of one mtDNA and one nDNA marker in concert will most probably provide a conservative estimate of cytonuclear dynamics. The addition of further genes will, of course, increase resolution, but is unlikely to substantively affect the results presented here or their interpretation.
Fisher’s exact tests (Basten and Asmussen 1997) quantify associations of overall cytonuclear genotypes and among specific combinations. The contingency method generated expected values of cytonuclear genotypic combinations without assuming linkage disequilibrium between the markers. Chi square tests were performed for single cells (each cytonuclear genotypic combination) and Fisher’s exact tests are performed within each sample country across all cytonuclear combinations. Sequential Bonferroni correction was employed (Rice 1989).
Results
Taxonomic status
A total of 190 individuals typed by two RFLP assays (Table 2) into three genotypes (S/S, S/N, N/N) and two haplotypes (s and n) produce six unique genotypic and haplotypic combinations: all alleles are fixed for each taxon (Table 2). The nuclear Northern Mytilus galloprovincialis allele frequency ranges from 8.4 % in New Zealand to 34.5 % in Chile (Table 3). The cytoplasmic Northern allele frequency ranges from 20 % in Australia to 42.7 % in New Zealand (Table 3). The Australian samples are characterized by low nuclear (11.7 %) and mitochondrial (20 %) Northern lineage M. galloprovincialis allele frequencies (Table 3), but this same pattern of frequencies of both alleles at one end of the spectrum is not observed in other regions.
Evidence of hybridization is observed across all study regions with the presence of S/N genotype with either s or n haplotype (Table 3). Furthermore, backcrossing events are evident in New Zealand and Australia (both from the cytonuclear genotype combinations S/S–n and N/N–s) and not observed in Chile. It is expected the mitochondrial identity of an individual mussel will indicate the mitochondrial identity of the maternal parent. s haplotype indicates a Southern lineage maternal parent and n haplotype indicates a Northern lineage maternal parent (Table 4).
Phylogenetic accuracy of markers
16s RFLP
In total, 16 unique haplotypes are identified from 96 mussels: eight unique haplotypes in 57 Southern hemisphere mussels and eight unique haplotypes in 39 Northern hemisphere mussels. The four haplogroups identified by the 16s RFLP marker cluster into monophyletic clades and/or subclades of the neighbour-joining phylogenetic tree (Online Resource 1). Bootstrap support values are high for divergence of the shared M. edulis/M. galloprovincialis North Atlantic haplogroup but only 53 % for the divergence of Mediterranean and Southern hemisphere M. galloprovincialis. Low bootstrap support for hemispheric divergence is indicated in previously published mitochondrial phylogenies (Hilbish et al. 2000; Gérard et al. 2008) and is most likely due to the low mutation rate of the 16s gene, which does not contain sufficient phylogenetic information to support a lineage divergence estimated in the late Pleistocene (Hilbish et al. 2000; Gérard et al. 2008).
Me15/16 RFLP
Hemispheric lineage divergence of M. galloprovincialis is highly supported in the nuclear Me15/16 phylogeny due to a single point mutation in the amplicon sequence that is the AciI restriction enzyme cut site (Online Resource 2). Not all expected taxonomic designations are obtained for sequence data. Table 2 indicates that one individual from each of Kaikoura (New Zealand), Port Arthur (Australia) and Concepçion (Chile) has a Northern hemisphere M. galloprovincialis genotype (when Southern hemisphere was expected) and resolves onto the Northern clade of the tree. This is due to the presence of non-native mussels in these areas (Westfall and Gardner 2010).
Cytonuclear disequilibria statistics and genotype frequencies
An excess of the S/S–s and a deficit of the N/N–n cytonuclear genotype combinations in mussels from Chile result in overall significant genotypic disequilibrium (Table 5) and significant differences between observed and expected individual cytonuclear genotypes (Table 4), respectively. Overall genotypic disequilibrium that is significantly different from the null hypothesis (Table 5) and a significant departure of observed from expected numbers of individual genotypes (Table 4) in Chile derives from an excess of the S/S–s and a significant deficit of N/N–s cytonuclear genotypes with associated D’ values at their maximal and minimal limits, respectively. Large standard error estimates reveal a deficit of the N/S–s genotypic combination (low sample size) and a median D’ (Table 5). Chile also has a significant excess of N/N–n cytonuclear genotypes and deficit of N/N-s cytonuclear genotypes (Table 4).
Overall genotypic disequilibrium that is significantly different from the null hypothesis in New Zealand mussels derives from an excess of the S/S–s (similar to Chile) and a deficit of the N/S–s cytonuclear genotypic combinations, with associated D’ values at median levels (Table 5). Significantly large S/S–s allelic disequilibrium with D’ at its maximal limit is also detected. Large standard error values reveal a deficiency of the N/N–s genotypic combination and a median D’. Significant excess of the S/N–s and a deficit of the S/N–n cytonuclear genotypes are also observed (Table 4).
Australia is the only region where non-significant disequilibria (Table 5) and differences between observed and expected frequencies (Table 4) across all cytonuclear genotypic combinations are observed. The D’ values are at their maximal and minimal boundaries for N/N–s and S/S–s cytonuclear genotypic combinations, respectively (Table 5).
Discussion
Southern hemisphere overall
Significant cytonuclear associations for individual native/non-native Mytilus galloprovincialishybrid populations in Chile, New Zealand and Australia are the first reported for Mytilus spp. in the Southern hemisphere. Investigations of Northern hemisphere M. trossulus × M. edulis hybridization resulting in cytonuclear disequilibria in and around the Baltic Sea have uncovered significant results only when samples across sites are pooled but not when individual sites or groups of related populations are tested (Kijewski et al. 2006; Smietanka et al. 2004). In this case, significant cytonuclear associations are thought to reflect covariation of allelic and haplotypic frequencies across a geographic range (Kijewski et al. 2006) and are influenced more by geographic factors than taxonomy (Kijewski et al. 2010).
Chilean and New Zealand hybrid mussel populations exhibit significant departures from random associations of native and non-native cytonuclear alleles. The pattern observed in both regions of D S/S,s > 0 and D N/N,s < 0 is also observed in controlled laboratory experiments where either one or both parental taxa are constantly migrating into the hybrid region (Arnold 1993; Avise et al. 1990). This suggests disequilibria in these populations are permanent, at a steady state and a model with migration alone is the best explanation for observed data (Table 5). The difference between Chile and New Zealand lies with the polarity of D S/N,s (positive and negative, respectively): they are expected to be the same if cytonuclear interactions are consistent between regions. The additive effects of assortative fertilization or selection against hybrids until adulthood may also produce the same sign pattern whilst the polarity relates to gender directionality of crosses (Arnold 1993), as discussed below for each region.
Chile
Negative D S/N,s in Chile in addition to zero observed recombinant individuals and normalized disequilibria at their maximum (D’ N/N,s ) and minimum (D’ S/S,s ) values supports an inference of high mating fidelity within the Southern hemisphere lineage due to either assortative fertilization or genetic drift from unequal population sizes. Further support for assortative fertilization comes from the difference between observed and expected frequencies of the cytonuclear genotypes: a non-significant excess of S/N–s cytonuclear genotypes and deficit of S/N–n (Table 4). The number of observed hybrids with the s haplotype being greater in frequency than expected and hybrids with n haplotype less than expected (Table 4) indicate that crosses include a Southern lineage maternal parent and Northern lineage paternal parent more often than the opposite pairing.
The failure to identify recombinant cytonuclear genotypes in Chile coupled with normalized disequilibria at marginal values for all cytonuclear combinations indicates hybridization between M. galloprovincialis lineages in this region is not ongoing and barriers to gene flow (either intrinsic or extrinsic) are preventing genetic introgression between lineages. Lack of recombinant genotypes are consistent with barriers to gene flow that have previously been observed among members of the M. edulis species complex in hybrid zones (Rawson et al. 1999), but not between divergent populations (or lineages) of a single species.
In the face of non-native mussel introduction, conservation of the unique genomic content of native Southern lineage M. galloprovincialis in Chile is managed by the nature of the parental populations themselves, an interpretation that differs from the genetic consequences of non-native mussel introductions in New Zealand and Australia.
New Zealand
Normalized disequilibria values for New Zealand mussels are typically about ±0.5 for all cytonuclear genotypic combinations. This suggests taxonomic boundaries are being compromised through the homogenizing effects of hybridization (Avise 2000) and barriers to gene flow between lineages do not seem to be present within this hybrid region. Furthermore, the significantly excess of hybrid individuals with a Northern lineage maternal parent (S/N–n) (Table 4) and significant deficit of observed hybrid individuals with a Southern lineage maternal parent (S/N–s) provide additional evidence favouring Northern female/Southern male crossings. This is supported by the greatest observed frequency of non-native cytoplasmic alleles (42.7 %), a value that is more than double the frequency observed in other regions (average 20.4 %) (Table 3).
The pervasiveness of Northern hemisphere mitochondrial haplotypes and apparent lack of reproductive barriers to gene flow among mussels in New Zealand indicate that if Northern lineage M. galloprovincialis continue to be introduced or perpetuate naturally throughout the national distribution then the status of the New Zealand lineage is under threat. These results suggest non-native mussel introductions into New Zealand pose a greater threat to the genetic integrity of native mussels here than in Chile or Australia.
Australia
The absence of observed pure Northern Mytilus galloprovincialis indicates that Australian study sites (Melbourne and Tasmania) have not experienced recent non-native mussel introductions. Normalized D’ values at maximal and minimal values (±1.0) associated with non-significant disequilibria indicate that drift and/or selection dynamics are maintaining the taxonomic boundaries between lineages (Avise 2000). All recombinant cytonuclear genotypes are present in frequencies higher than expected (except N/N–n at Melbourne frequency = 0, Table 4). One interpretation is that we are seeing the historical signal of old introductions as European sailing vessels have been visiting parts of Australia since the late eighteenth century, upon which is now superimposed one of the world’s most active maritime biosecurity policies (Department of Agriculture, Fisheries and Forestry— http://www.daff.gov.au/animal-plant-health/pests-diseases-weeds/marine-pests). There are several possible causes for the observed cytonuclear genotypic frequencies; (1) low rates of introduction, possibly associated with Australia’s stringent national policies on marine biosecurity. (2) unfavourable environmental conditions, or (3) a substantial amount of time has elapsed since the last introduction and pure non-native mussels have largely died out. When one species or taxon is rare in a hybrid setting, it runs the risk of being swamped by introgressive hybridization and eventually losing out to the more common taxon (Rhymer and Simberloff 1996). Points (1) and (3) outlined above demonstrate the efficacy of Australia’s maritime biosecurity policies and other regions in the Southern hemisphere should look towards the strategies employed by Australia to prevent further non-native mussel incursions.
Conclusions
Variation of the genetic outcomes of non-native mussel introductions in three Southern hemisphere countries was an unexpected result of the current investigation, requiring post hoc hypotheses based on genetic signatures of hybridization under laboratory conditions to provide the framework for future research. Firstly, genetic variation among source populations of introduced Northern hemisphere mussels may contribute to the observed pattern of apparent reproductive barriers to gene flow between lineages operating in Chile but not in New Zealand or Australia. Non-native mussels from across the Southern hemisphere sample regions exhibit ancestry from both “Mediterranean” and “North Atlantic” haplogroups (Hilbish et al. 2000; Westfall and Gardner 2010; Westfall et al. 2010) and could potentially be sourced from a very large geographical range.
Another possible mechanism underlying these hybridization outcomes is uncharacterized taxonomic variation among Southern hemisphere regions. Southern hemisphere mussels are regarded as having greatest affinity to two Northern hemisphere taxa based on biochemical and morphological variation (Daguin and Borsa 2000; McDonald et al. 1991). The first category includes blue mussels from South American (Pacific and Atlantic coasts) and the Kerguelen Islands with biochemical variation closely resembling M. edulis from the Northern hemisphere and morphological characteristics intermediate between Northern hemisphere M. edulis and M. galloprovincialis (McDonald et al. 1991). The second group consists of Australasian (New Zealand and Australia) blue mussels that most closely resemble M. galloprovincialis from the Northern hemisphere (Daguin and Borsa 2000; McDonald et al. 1991). Furthermore, phylogenetic analysis of mitochondrial DNA indicates Chile and the Kerguelen Islands constitute a monophyletic clade (Gérard et al. 2008). Although random nuclear markers developed for Northern hemisphere species have previously identified all blue mussels in Chile, New Zealand and Australia as M. galloprovincialis (Westfall et al. 2010; Westfall and Gardner 2010), further taxonomic characterization may uncover distinctive South American and Australasian populations with unique genetic attributes. The apparent barriers to gene flow acting within the Chilean hybrid region could reflect either higher level taxonomic differentiation or within-lineage differentiation, but further genomic characterization in necessary to test these hypotheses.
Genetic swamping of Northern Mytilus galloprovincialis alleles in Australia is an example of how extremely low frequencies of non-native alleles can affect the genetic make-up of native populations. The gender bias of first generation crosses to include more Southern maternal parents coupled with no evidence of recombinant genotypes in Chile is a desired outcome when considering the potential for non-native introductions to alter the genomic composition of native mussel populations. In Australia and Chile, the potential exists for minimal impact on the native population from the introduction of Northern hemisphere M. galloprovincialis. The outcome in New Zealand is less optimistic because introgressive hybridization, continued immigration of non-native mussels, slight gender bias to interlineage crosses for more Northern than Southern maternal parents and no detected selection against hybrid mussels may lead to continued cytonuclear disequilibria and genetic mixing of lineages.
Variation of non-native mussel introduction outcomes among the three Southern hemisphere regions included in this study has implications for the non-native introduction of blue mussels worldwide and conservation of native mussels. The genetic consequences of non-native introductions cannot be explicitly predicted based on the presence/absence of non-native mussels, rather, each case of introduction must be examined separately. Further investigation into the underlying cause(s) of the observed variation will help to predict the genetic outcomes of non-native mussel introductions worldwide and guide strategies for conserving the unique genetic content of blue mussel populations and taxa. Managers and policy makers may also consider the genetic impacts of mariculture practice when spat are moved among different regions of a country. Countries may now need to consider their national maritime biosecurity policies so that they develop a focus on both pre-border and post-border controls (Forrest et al. 2009).
References
Arnold J (1993) Cytonuclear disequilibria in hybrid zones. Ann Rev Ecol Syst 24:521–554
Arnold ML (1997) Natural hybridization and evolution. Oxford University Press, Oxford, UK
Arnold J, Asmussen M, Avise JC (1988) An epistatic mating system model can produce permanent cytonuclear disequilibria in a hybrid zone. PNAS 85: 1893–1896
Asmussen M, Basten C (1994) Sampling theory for cytonuclear disequilibria. Genetics 138:351–1363
Asmussen M, Basten C (1996) Constraints and normalised measure for cytonuclear disequilibria. Heredity 76:201–214
Asmussen M, Arnold J, Avise JC (1987) Definition and properties of disequilibrium statistics for associations between nuclear and cytoplasmic genotypes. Genetics 115:755–768
Avise JC (2000) Cytonuclear genetic signatures of hybridization phenomena: rationale, utility and empirical examples from fishes and other aquatic animals. Rev Fish Biol Fish 10:253–261
Avise JC, Saunders NM (1984) Hybridization and introgression among species of sunfish (Lepomis): Analysis by mitochondrial DNA and allozyme markers. Genetics 108:237–255
Avise JC, Nelson WS, Arnold J, Koehn RK, Williams GC, Thorsteinsson V (1990) The evolutionary genetic status of Icelandic eels. Evolution 44:1254–1262
Barton NH, Hewitt GM (1989) Adaptation, speciation and hybrid zones. Nature 341:497–503
Basten C, Asmussen M (1997) The exact test for cytonuclear disequilibria. Genetics 146:1165–1171
Bierne N, Borsa P, Daguin C, Jollivet D, Viard F, Bonhomme F, David P (2003) Introgression patterns in the mosaic hybrid zone between Mytilus edulis and M. galloprovincialis. Mol Ecol 12:447–461
Borsa P, Daguin C, Bierne N (2007) Genomic reticulation indicates mixed ancestry in Southern hemisphere Mytilus spp mussels. Biol J Linn Soc 92:747–754
Bownes SJ, McQuaid CD (2006) Will the invasive mussel Mytilus galloprovincialis Lamarck replace the indigenous Perna perna L. on the south coast of South Africa? J Exp Mar Biol Ecol 338:140–151
Brannock PM, Wethey DS, Hilbish TJ (2009) Extensive hybridization with minimal introgression in Mytilus galloprovincialis and M. trossulus in Hokkaido. Japan. Mar Ecol Prog Ser 383:161–171
Currat M, Ruedi M, Petit RJ, Excoffier L (2008) The hidden side of invasions: massive introgression by local genes. Evolution 62:1908–1920
Daguin C, Borsa P (2000) Genetic relationships of Mytilus galloprovincialis Lmk populations worldwide: evidence from nuclear-DNA markers. In: Crame A, Harper E, Taylor J (eds) Bivalve systematics and evolution, vol 177. Geological Society of London Special Publications volume, London, pp 389–397
Elliott J, Holmes K, Chambers R, Leon K, Wimberger P (2008) Differences in morphology and habitat use among the native mussel Mytilus trossulus, the non-native M. galloprovincialis, and their hybrids in Puget Sound, Washington. Mar Biol 156:39–53
Forrest BM, Gardner JPA, Taylor M (2009) Internal borders for the management of invasive marine species. J Appl Ecol 46:46–54
Gardner JPA (1994) The structure and dynamics of naturally occurring hybrid Mytilus edulis Linnaeus, 1758 and Mytilus galloprovincialis Lamarck, 1819 (Bivalvia, Mollusca) populations: review and interpretation. Arch Hydrobiol 99:37–71
Gardner JPA (1996) The Mytilus edulis species complex in southwest England: the extent of hybridization and introgression and their effects upon interlocus associations and morphometric variation. Mar Biol 125:385–399
Gardner JPA (1997) Hybridization in the Sea. Adv Mar Biol 31:1–78
Gardner JPA (2004) A historical perspective of the genus Mytilus (Bivalvia: Mollusca) in New Zealand: multi-variate morphometric analyses of fossil, midden and contemporary blue mussels. Biol J Linn Soc 82:329–344
Gardner JPA, Thompson RJ (2001) The effects of coastal and estuarine conditions on the physiology and survivorship of the mussels Mytilus edulis, M trossulus and their hybrids. J Exp Mar Biol Ecol 265:119–140
Geller JB (1999) Decline of a native mussel masked by sibling species invasion. Cons Biol 13:661–664
Gérard K, Bierne N, Borsa P, Chenuil A, Feral J-P (2008) Pleistocene separation of mitochondrial lineages of Mytilus spp mussels from Northern and Southern hemispheres and strong genetic differentiation among southern populations. Mol Phylogenet Evol 49:84–91
Grant S, Cherry I (1985) Mytilus galloprovincialis Lmk in Southern Africa. J Exp Mar Biol Ecol 90:179–191
Harrison RG, Rand D (1989) Mosaic hybrid zones and the nature of species boundaries. In: Otte D, Endler J (eds) Speciation and its consequences. Sinauer Associates, Sunderland, Massachusetts, pp 111–133
Heath DD, Rawson PD, Hilbish TJ (1995) PCR-based nuclear markers identify alien blue mussel (Mytilus spp.) genotypes on the west coast of Canada. Can J Fish Aquat Sci 52:2621–2627
Hilbish T, Mullinax A, Dolven S, Meyer A, Koehn R, Rawson P (2000) Origin of the antitropical distribution pattern in marine mussels (Mytilus spp): routes and timing of transequatorial migration. Mar Biol 136:69–77
Inoue K, Waite J, Matsouka M, Odo S, Harayama S (1995) Interspecific variations in adhesive protein sequences of Mytilus edulis, M galloprovincialis, and M trossulus. Biol Bull 189:370–375
Keller SR, Taylor DR (2010) Genomic admixture increases fitness during a biological invasion. J Evol Biol 23:1720–1731
Kijewski T, Zbawicka M, Väinölä R, Wenne R (2006) Introgression and mitochondrial DNA heteroplasmy in the Baltic populations of mussels Mytilus trossulus and M edulis. Mar Biol 149:1371–1385
Kijewski T, Smietanka B, Zbawicka M, Gosling E, Hummel H, Wenne R (2010) Distribution of Mytilus taxa in European coastal areas as inferred from molecular markers. J Sea Res 65:224–234
Lee JE, Chown SL (2007) Mytilus on the move: transport of an invasive bivalve to the Antarctic. Mar Ecol Pro Ser 339:307–310
Lowe S, Browne M, Boudjelais S, De Poorter M (2000) 100 of the world’s worst invasive alien species: a selection from the global invasive species database Published by The Invasive Species Specialist Group (ISSG), a specialist group of the Species Survival Commission (SSC) of the World Conservation Union (IUCN), pp 12
Mallet J (2007) Hybrid speciation. Nature 446:279–283
McDonald J, Koehn R (1988) The mussels Mytilus galloprovincialis and M trossulus on the Pacific coast of North America. Mar Biol 99:111–118
McDonald J, Seed R, Koehn R (1991) Allozymes and morphometric characters of three species of Mytilus in the Northern and Southern hemispheres. Mar Biol 111:323–333
Palumbi SR (1995) Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mabel BK (eds) Molecular Systematics. Sinauer Associates, Sunderland, Massachusetts, pp 205–247
Rawson PD, Hilbish TJ (1995) Evolutionary relationships among the male and female mitochondrial DNA lineages in the Mytilus edulis species complex. Mol Biol Evol 12:893–901
Rawson PD, Hilbish TJ (1998) Asymmetric introgression of mitochondrial DNA among European populations of blue mussels (Mytilus spp). Evolution 52:100–108
Rawson P, Agrawal V, Hilbish T (1999) Hybridization between the blue mussels Mytilus galloprovincialis and M trossulus along the Pacific coast of North America: evidence for limited introgression. Mar Biol 134:201–211
Rhymer JM, Simberloff D (1996) Extinction by hybridization and introgression. Ann R Ecol Syst 27:83–109
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Riginos C, Hickerson MJ, Henzler CM, Cunningham CW (2004) Differential patterns of male and female mtDNA exchange across the Atlantic Ocean in the blue mussel, Mytilus edulis. Evolution 58:2438–2451
Santaclara FJ, Espineira M, Cabado AG, Aldasoro A, Gonzalez-Lavin N, Vieites JM (2006) Development of a method for the genetic identification of mussel species belonging to Mytilus, Perna, Aulacomya and other genera. J Agric Food Chem 54:8461–8470
Scribner KT, Avise JC (1993) Cytonuclear genetic architecture in mosquitofish populations and the possible roles of introgressive hybridization. Mol Ecol 2:139–149
Scribner KT, Avise JC (1994) Cytonuclear genetics of experimental fish hybrid zones inside biosphere 2. Proc Nat Acad Sci USA 91:5066–5069
Seed R (1992) Systematics, evolution and distribution of mussels belonging to the genus Mytilus: an overview. Am Malacol Bull 9:123–137
Simberloff D (2005) Non-native species DO threaten the environment. J Agric Environ Ethic 18:595–607
Smietanka B, Zbawicka M, Wolowicz M, Wenne R (2004) Mitochondrial DNA lineages in the European populations of mussels. Mar Biol 146:79–92
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Toro JE, Ojeda JA, Vergara AM, Castro GC, Alcapán AC (2005) Molecular characterization of the Chilean blue mussel (Mytilus chilensis Hupe 1854) demonstrates evidence for the occurrence of Mytilus galloprovincialis in southern Chile. J Shellfish Res 24:1117–1121
Westfall KM, Gardner JPA (2010) Genetic diversity of Southern hemisphere blue mussels (Mytilidae; Bivalvia) and the identification of non-indigenous taxa. Biol J Linn Soc 101:898–909
Westfall KM, Wimberger PH, Gardner JPA (2010) An RFLP mtDNA assay to determine if Mytilus galloprovincialis (Mytilidae; Bivalvia) is of Northern or Southern hemisphere origin. Mol Ecol Resour 10:573–575
Acknowledgments
This research was funded by Biosecurity New Zealand (to JPAG) and by the Centre for Marine Environmental & Economic Research, Victoria University of Wellington. KMW was supported by a Victoria University of Wellington PhD scholarship. We thank (in alphabetical order) the Australian National Museum, M. Curwen, C. & H. Hay, N. Moltschaniwskyj, E. Tarifeno and A. Wood for providing samples. We would also like to thank two anonymous reviewers for helpful comments and suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Westfall, K.M., Gardner, J.P.A. Interlineage Mytilus galloprovincialis Lmk. 1819 hybridization yields inconsistent genetic outcomes in the Southern hemisphere. Biol Invasions 15, 1493–1506 (2013). https://doi.org/10.1007/s10530-012-0385-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-012-0385-8