Abstract
Invasive plant species threaten native ecosystems, natural resources, and managed lands worldwide. Climate change may increase risk from invasive plant species as favorable climate conditions allow invaders to expand into new ranges. Here, we use bioclimatic envelope modeling to assess current climatic habitat, or lands climatically suitable for invasion, for three of the most dominant and aggressive invasive plants in the southeast United States: kudzu (Pueraria lobata), privet (Ligustrum sinense; L. vulgare), and cogongrass (Imperata cylindrica). We define climatic habitat using both the Maxent and Mahalanobis distance methodologies, and we define the best climatic predictors based on variables that best ‘constrain’ species distributions and variables that ‘release’ the most land area if excluded. We then use an ensemble of 12 atmosphere-ocean general circulation models to project changes in climatic habitat for the three invasive species by 2100. The combined methodologies, predictors, and models produce a robust assessment of invasion risk inclusive of many of the approaches typically used individually to assess climate change impacts. Current invasion risk is widespread in southeastern states for all three species, although cogongrass invasion risk is more restricted to the Gulf Coast. Climate change is likely to enable all three species to greatly expand their ranges. Risk from privet and kudzu expands north into Ohio, Pennsylvania, New York, and New England states by 2100. Risk from cogongrass expands as far north as Kentucky and Virginia. Heightened surveillance and prompt eradication of small pockets of invasion in northern states should be a management priority.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Invasive species and climate change are two of the most pervasive forms of ecosystem disturbance, and both will increasingly threaten biodiversity during the twenty-first century (Millenium Ecosystem Assessment 2003; Thomas et al. 2004; Vitousek et al. 1996). Invasive plant species threaten native ecosystems by outcompeting native species and by altering disturbance regimes (e.g., wildfire) (D’Antonio and Vitousek 1992; Zavaleta 2000). Invasive plants also threaten human-managed systems such as agriculture, rangelands, and forests. Annual economic losses on managed lands are estimated in the billions of dollars for the US alone (Pimentel et al. 2000).
These threats may be exacerbated by climate change (Dukes and Mooney 1999; IPCC 2007; Moore 2004; Thuiller et al. 2007; Weltzin et al. 2003). Higher ambient CO2 concentrations have been shown to increase the competitiveness of invasive plants relative to native species (Nagel et al. 2004; Sasek and Strain 1989, 1991; Smith et al. 2000; Ziska 2003; Ziska et al. 2007). Moreover, many invasive plants are capable of spreading rapidly into disturbed areas, such as recently burned sites, which may become more prevalent with climate change (Dukes and Mooney 1999). Finally, changing precipitation and temperature conditions are likely to alter the distribution of land area at risk of invasion. Effective, long-term invasive plant management needs to include spatially explicit projections of how global change alters invasion risk.
In some locations, climate change could lead to expanded invasion risk from particular species. For example, higher precipitation may enable non-native grasses to spread in the western US (D’Antonio and Vitousek 1992; Martin-R et al. 1995), and higher temperatures may expand the range of invasive plants northward in the southeastern US (Rogers and McCarty 2000). Yet for other invasive species, climatic habitat could be reduced with climate change, creating unprecedented restoration opportunities (Bradley et al. 2009). It is imperative that both risks and opportunities are quantified in order to inform land management decisions in the face of a changing climate.
Changes in land area at risk can be assessed by modeling species’ climatic habitat. Climatic habitat is defined as land area characterized by climate conditions important to a species’ distribution and abundance (Kearney 2006). For invasive plants, an area could be at risk of invasion if climate conditions are suitable for the invasive species. While other factors besides climate also affect invasion risk, climate is one of the most important limiting factors at regional and continental scales (spatial areas of hundreds to thousands of km2) (Guisan and Thuiller 2005; Pearson and Dawson 2003; Walker et al. 1993). Bioclimatic envelope models (BEMs) are frequently used to define climatic habitats of native and invasive species based on their geographic distribution (see for examples, Franklin 1995; Guisan and Thuiller 2005; Guisan and Zimmermann 2000; Pearson and Dawson 2003). However, BEMs are limited because they only consider species-climate relationships and cannot account for other factors such as competition, nutrient use, or dispersal. Unfortunately, to date, very few process-based models exist for any species (Jeltsch et al. 2008), let alone these particular invasive species. Hence, BEMs provide a spatially explicit assessment of invasion risk at regional scales, which can later be refined for local and landscape level assessments using risk factors appropriate to those scales (e.g., land use, dispersal ability) (Guisan and Thuiller 2005; Pearson and Dawson 2003).
A typical limitation of bioclimatic envelope modeling, particularly when applied to climate change conditions, is the high level of uncertainty associated with both the type of envelope model selected and projections of climate change. Although some bioclimatic envelope models have been found to perform better than others (Elith et al. 2006; Tsoar et al. 2007), there is no consensus on the “best” model to use. Further, there is a high level of uncertainty and disagreement between climate change projections from different atmosphere-ocean general circulation models (AOGCMs) (Milly et al. 2005; IPCC 2007). Araujo and New (2007) suggest that the best approach to dealing with this uncertainty is through a model ensemble. That is, researchers should use multiple methodologies and climate change projections to forecast climatic habitats, then combine these results to quantify agreement (Araujo and New 2007). Areas consistently predicted to be at risk by multiple methodologies and climate projections are assumed to be at higher risk than areas predicted to be at risk by only one or two models. Although model ensembles are likely to lead to a more robust forecast of distribution change than any single AOGCM, this type of approach has not been widely implemented.
Several studies have used BEMs to model climatic habitats of invasive plant species in order to assess current invasion risk (Mau-Crimmins et al. 2006; Nielsen et al. 2008; Rouget et al. 2004; Schussman et al. 2006; Thuiller et al. 2005; Welk et al. 2002; Zhu et al. 2007). Others have used climate change projections to forecast future invasion risk (Beerling 1993; Bradley 2009b; Bradley et al. 2009; Kriticos et al. 2003; Mika et al. 2008; Peterson et al. 2008; Sutherst 1995; Zavaleta and Royval 2002). This type of work is critical for management planning aimed at reducing threats from plant invasion. However, despite numerous highly problematic invasive plants worldwide, biogeographical assessments of changing risk with climate change have been performed on only a few species, with scant focus on some of the most widespread and destructive species.
Here, we model invasion risk with climate change for three of the most widespread, aggressive and ecosystem-transforming invasive plants in the southeastern United States: kudzu (Pueraria lobata), Chinese/European privet (Ligustrum sinense; L. vulgare), and cogongrass (Imperata cylindrica). We elected to use the non-native range of these plants to model invasion risk because climatic envelopes for the native and non-native ranges of invasive species can be distinctly different (Broennimann et al. 2007), suggesting that the invaded distribution is most appropriate for assessing invasion risk. Although envelope models underestimate climatic habitat if invasive plants have not invaded to their full potential (Welk 2004), the widespread introduction of these three species makes it reasonable to assume that their distribution approximates equilibrium with current climate. For example, Welk (2004) showed that the distribution of the invasive plant Lythrum salicaria in the US after 100–150 years was appropriate for modeling current invasion risk. The three target species all meet the criterion of long-term establishment (see “Background”) and further were widely planted intentionally. The current distribution of all three invasive species in the United States is largely concentrated within the southeastern states where they were mapped.
To assess current invasion risk, we use an ensemble of two bioclimatic envelope models (Maxent and Mahalanobis Distance) and two sets of climatic variable predictors (based on ‘constraint’ of species distribution and land area ‘released’ with variable exclusion). We then use an ensemble of envelope models, climate predictors, and 12 AOGCMs to forecast spatially explicit invasion risk in the United Stated under year 2100 climate conditions. This work develops spatially explicit projections of invasion risk for three highly problematic plant species. Ecological forecasting of individual invasive species is increasingly important for developing long-term management strategies inclusive of climate change.
Background
Kudzu is a perennial vine pervasive across the southern United States. Kudzu is known as ‘the vine that ate the south’ due to its rapid growth rate and tendency to smother whatever it grows on (saplings, mature trees, utility poles, houses) (Blaustein 2001; Mitich 2000). A native of East Asia, kudzu was initially introduced to the US in 1876 and widely planted for erosion control until it was designated an invasive plant in 1950. Not surprisingly, it poses a substantial threat to native ecosystems (Forseth and Innis 2004), agricultural lands, and forests. Furthermore, economic damages to managed forests and agricultural land are estimated at $100–500 million per year (Blaustein 2001). Expansion of kudzu northwards with climate change is a great concern for mid-Atlantic, and even northern states (Forseth and Innis 2004; Rogers and McCarty 2000). Recent observations of kudzu’s northward expansion have been linked to higher winter temperatures, which reduce the duration of kudzu-killing frosts.
Chinese (L. sinense) and European (L. vulgare) privet are highly invasive flowering shrubs. Privet was planted extensively in the United States beginning in 1852 as an ornamental hedge; however, it quickly escaped cultivation. Privet tends to form thickets, which crowd out other species and threaten native trees and shrubs (Batcher 2000; Wilcox and Beck 2007). Eradication of large infestations is difficult, although some control is possible at early stages of colonization (Batcher 2000). Despite the threat of invasion, privet species are still widely planted for landscaping throughout the United States.
Cogongrass is an invasive perennial grass (Holm et al. 1977) that infests hundreds of millions of hectares globally. It is a growing problem in the southeast United States (Bryson and Carter 1993; MacDonald 2004), where it crowds out native plant species (MacDonald 2004). Cogongrass alters fire cycles, causing wildfires to burn at higher maximum temperatures and killing even normally fire-tolerant species (Lippincott 2000). Following fires, cogongrass quickly invades burned areas to create grassland monocultures (Lippincott 2000). Cogongrass, a native of Southeast Asia, was introduced to the US in the 1930s and widely planted for livestock forage and erosion control. Cogongrass does not tolerate low temperatures, which has led some to initially suggest that states outside the Gulf Coast are not at risk of invasion (Patterson et al. 1980; Wilcut et al. 1988). Warming temperatures with climate change could greatly expand invasion risk.
Methods
Datasets
Regional distributions of cogongrass, kudzu, and privet are based on abundance estimates made by invasive plant experts (Marvin et al. 2009). European and Chinese privet were combined in this mapping effort due to the difficulty of distinguishing between the two species. Distribution data were collected through an online mapping interface (http://invasive.princeton.edu) where users submitted estimates of invasive plant abundance within one-quarter USGS quadrangles (~36 km2) across the Southeastern United States. Data were collected electronically from 187 regional experts, as well as from dozens of other experts on paper maps that were later digitized. Invasive plant experts who participated included individuals from federal and state agencies (e.g., U.S. Forest Service, U.S. Fish & Wildlife Service), non-governmental organizations, and academic institutions. For further information on mapping methodology, see (Marvin et al. 2009).
We reclassified abundance estimates to presence/absence using a threshold of >1% land cover of each invasive species within each one-quarter USGS quadrangle (~36 km2) mapping area. Abundance estimates were not collected comprehensively for the entire southeast due to lack of response, resources, or expertise in many areas (Marvin et al. 2009). As a result, some states were better sampled than others. However, presence locations for all three species span latitudinal, longitudinal, and elevational gradients at a regional scale. Hence, the distributional data are widespread enough to be appropriate for bioclimatic envelope modeling.
Current climate data are based on an interpolation of data from US. weather stations created by the PRISM group (Daly et al. 2002). This gridded interpolation includes average annual and monthly precipitation, minimum temperature, and maximum temperature from 1971 to 2000 for a total of 39 candidate predictor variables. These “current” climate conditions were used to create bioclimatic envelopes for each invasive species based on the spatial relationships between species distribution and average climatic conditions.
Future climate conditions are based on precipitation and temperature projections for the year 2100 from 12 atmosphere-ocean general circulation models (AOGCMs). These global models were standardized and compiled for the Intergovernmental Panel on Climate Change (IPCC) fourth assessment report (IPCC 2007).Footnote 1 We used models based on the SRES A1B scenario, or the “middle of the road” scenario in which CO2 concentrations reach 720 ppm by 2100 (Nakicenovic and Swart 2000). For each model, average monthly precipitation, minimum temperature, and maximum temperature conditions from 1971 to 2000 were subtracted from average monthly climate conditions from 2090 to 2100 to estimate change between the present and 2100. The 10-year 2090–2100 time period was the longest for which the appropriate climate data were available in all of the target AOGCMs. Climate change was added to the higher-resolution PRISM dataset to create a high resolution estimate of 2100 climate conditions.
Invasion risk under current climate
Two models were used to construct bioclimatic envelopes: Maximum entropy (Maxent) (Phillips et al. 2006) and Mahalanobis distance (MD). Maxent uses presence points to define species distributions based on simple functions related to each climate variable (Phillips et al. 2006). MD defines an ellipsoidal envelope around presence points relative to each climate variable (Farber and Kadmon 2003). Both of these techniques have been found to have high predictive accuracy (Elith et al. 2006; Phillips and Dudik 2008; Tsoar et al. 2007).
Two methods were used to select the most appropriate climate variables for the envelope models from the original set of 39 candidate predictor variables. The first method defines the best predictor variables as those that most constrain species distribution (Bradley 2009b; Hirzel et al. 2002). That is, the best climate variables are those that encompass the species distribution with the least additional land area. The 39 candidate predictor variables were ranked by level of constraint, where constraint was defined as the median distance from the mean of all southeast pixels divided by the median distance from the sample mean. The sample mean consists of all pixels where the target species is present. A larger constraint ratio indicates a better climatic predictor (Bradley 2009b; Hirzel et al. 2002). The top predictors based on constraint were identified through forward selection until model fits no longer improved with additional predictor variables.
The second method defines the best predictor variables as those which significantly decrease the fit of the BEM if they are omitted. That is, without these variables the greatest amount of additional land area is released (Arundel 2005). The 39 candidate predictor variables were ranked by release using a jackknife test of variable importance. The number of predictors based on release were identified through forward selection until model fits no longer improved with additional predictor variables.
Due to the inconsistent selection of predictor variables between the two methodologies, we tested the climate variables for autocorrelation. We selected 2,000 random pixels from 11,000 pixels with privet present in the southeast region and measured correlation coefficients for annual and monthly precipitation, minimum temperature, and maximum temperature variables. Privet presence points were selected because they were the most prevalent of the three species.
Both the constraint and release sets of variables were used to construct bioclimatic envelopes using the Maxent and MD methodologies, for a total of four possible envelopes for each species. In each case, the envelope was defined as the Maxent value or Mahalanobis distance that encompassed 95% of the species distribution. The four envelopes were summed to create an ensemble of methodologies to better define current climatic habitat for each invasive species.
We evaluated the relative fits of the four models for each species by comparing predicted frequency to expected frequency for the range of Maxent and MD values (Hirzel et al. 2006). Predicted frequency (P) is defined as the number of occurrences within the model threshold divided by the total number of occurrences in the study area, also known as the fraction of occurrences. Expected frequency (E) is defined as the number of pixels within the model threshold divided by the total number of pixels in the study area, also known as the fraction of total pixels. Expected frequency is the ratio that would be selected by a random model. If the model has no predictive capacity, predicted frequency should equal expected frequency (Hirzel et al. 2006). If the model has high predictive capacity, predicted frequency will be much greater than expected frequency. Higher ratios of predicted to expected frequency (P/E) indicate better model fit. In addition to calculating P/E ratios for the model fits, we also created receiver operator characteristic (ROC) curves for each model. Based on the ROC curves, we calculated area under the curve (AUC) to provide a further estimate of model fit.
Invasion risk with climate change
Future distributions of climatic habitat for the three invasive plants were modeled by applying the four bioclimatic envelopes to AOGCM projected year 2100 climate conditions. The Mahalanobis distance and Maxent values that encompassed 95% of current distribution in the original BEM were used to project potential future distribution for each of the 12 AOGCMs. These 48 total projections (2 models × 2 sets of variables × 12 AOGCMs) were summed to quantify model agreement. Areas with the most agreement between models are assumed more likely to be at risk of invasion with climate change.
Results
Climate variables selected as the best predictors of distribution for the three invasive species based on release and constraint methodologies are shown in Table 1. Both precipitation and temperature variables are important predictors of climatic habitat. Fall and winter variables are typically better predictors than spring and summer variables. For kudzu and privet, there is very little consistency between climate variables identified based on the release and constraint methodologies. For congongrass, there is a higher degree of consistency between the two sets of variables.
In order to evaluate the degree of auto-correlation amongst climate predictor variables, a correlation matrix for minimum temperature in the southeast United States for locations with privet present is shown in Table 2. For both minimum and maximum (not shown) temperature variables, there is a high degree of correlation within seasons (e.g., December–February; winter) and between adjacent seasons (e.g., winter and spring). There is also a high degree of correlation between spring and fall months. The lowest correlations (R = 0.78 for minimum temperature) occur between winter and summer months. Monthly precipitation variables (not shown) do not show the same degree of autocorrelation as temperature variables. Generally, months within the same season are well correlated (e.g., for December–February, R = 0.70–0.92), but months within different seasons are poorly correlated. The one exception is precipitation from winter and spring months, which are well correlated.
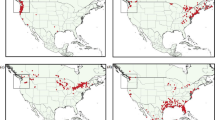
The spatial distribution of kudzu climatic habitat under current conditions is shown in Fig. 1. The risk of kudzu invasion is widespread in the southeast. Envelopes based on constraint variables encompass more land area than envelopes base on release variables. Using the constraint variables, kudzu invasion risk extends west to Oklahoma and Texas, and north to southern New England. The MD and Maxent methodologies produce similar results when given the same predictor variables. The combination of the four sets of methods/variables shows the greatest overlap of current invasion risk in the states of Arkansas, Louisiana, Kentucky, Tennessee, Mississippi, Alabama, Georgia, S. Carolina, N. Carolina, and Virginia.
Kudzu climatic habitat under current climate conditions. a Bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released. b Bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution. c Bioclimatic envelope based on the MD model using predictor variables selected based on land area released. d Bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution. e ¼ USGS quadrangles with greater than 1% cover of Kudzu in the southeast US based on expert opinion (Marvin et al. 2009). f Sum of bioclimatic envelope models a–d. Areas identified as climatic habitat by all four BEMs are more likely at risk from kudzu invasion
The spatial distribution of privet climatic habitat under current conditions is shown in Fig. 2. Like kudzu, risk of privet invasion is also widespread in the southeast. Projections based on constraint variables extend farther to the west than those based on release variable. There are also clear differences between envelopes projected by the MD and Maxent methodologies. MD projections show invasion risk extending farther north into northern Kentucky and West Virginia than do Maxent projections. The combination of the four sets of methods/variables shows the greatest overlap of invasion risk in the states of Arkansas, Louisiana, Kentucky, Tennessee, Mississippi, Alabama, Georgia, S. Carolina, N. Carolina, and Virginia. Privet and kudzu have very similar spatial extents of current invasion risk.
Privet climatic habitat under current climate conditions. a Bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released. b Bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution. c Bioclimatic envelope based on the MD model using predictor variables selected based on land area released. d Bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution. e ¼ USGS quadrangles with greater than 1% cover of privet in the southeast US based on expert opinion (Marvin et al. 2009). f Sum of bioclimatic envelope models a–d. Areas identified as climatic habitat by all four BEMs are more likely at risk from privet invasion
The spatial distribution of cogongrass climatic habitat under current conditions is shown in Fig. 3. Cogongrass invasion risk is highest in Florida and the Gulf coast states. Projections made with release variables are very similar to those made with constraint variables; however, there is also substantial overlap between the two sets of variables (Table 1). There are clear differences between the MD and Maxent model projections, with MD climatic habitat extending further west into Arkansas and Maxent climatic habitat extending northeast along the Carolina coasts. The combination of the four sets of methods/variables shows the greatest overlap of invasion risk in the states of Louisiana, Mississippi, Alabama, Georgia, and Florida. Current risk from cogongrass invasion is concentrated farther south than is the case for either kudzu or privet.
Cogongrass climatic habitat under current climate conditions. a Bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released. b Bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution. c Bioclimatic envelope based on the MD model using predictor variables selected based on land area released. d Bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution. e ¼ USGS quadrangles with greater than 1% cover of cogongrass in the southeast US based on expert opinion (Marvin et al. 2009). f Sum of bioclimatic envelope models a–d. Areas identified as climatic habitat by all four BEMs are more likely at risk from cogongrass invasion
Model fits for all three species have higher predictive accuracy within the southeast states than a random model (Fig. 4). Cogongrass has the best fit, with predicted to expected (P/E) ratios between 2.16 and 2.76 at the 95% occurrence threshold, and AUC values ranging from 0.75 to 0.86 (Fig. 4a). Kudzu has P/E ratios between 1.21 and 1.44 and AUC values ranging from 0.71 to 0.82 (Fig. 4b). Privet has P/E ratios between 1.17 and 1.34 at the 95% occurrence level and AUC values ranging from 0.70 to 0.80 (Fig. 4c). One of the Maxent models performs better than the MD models in each case, however, the relative fits of the predictor variable sets differs by species.
Evaluation of model fits under current climate conditions. Curves for each model show the fraction of species occurrences vs. the fraction of total pixels for incremental increases in MAXENT value or Mahalanobis distance (not pictured). All models have a higher predictive accuracy than a random model (shown by the 1:1 line). A model threshold which encompasses 95% of occurrences was used to construct both current and future range projections
Average projections of climate change for the southeast region are shown in Fig. 5. By year 2100, temperatures are projected to rise by between 1 and 10°C depending on the month and AOGCM used. Average annual warming of 3–4°C for both minimum and maximum temperatures is projected by all the AOGCMs. Precipitation projections do not show a high degree of consistency between AOGCMs. Annual precipitation change is projected anywhere from a loss of 200 mm/year to a gain of 200 mm/year, although more models project precipitation gains than losses. Similarly, monthly precipitation projections are highly variable; however, winter months are fairly consistently projected to receive higher precipitation.
Monthly climate change projections for the southeast region. a Current average maximum temperature between 1971 and 2000 based on PRISM data (bold black line) (Daly et al. 2002) and projected average maximum temperature between 2090 and 2100 from 12 AOGCMs. b Current average minimum temperature between 1971 and 2000 based on PRISM data (bold black line) (Daly et al. 2002) and projected average minimum temperature between 2090 and 2100 from 12 AOGCMs. c Current average precipitation between 1971 and 2000 based on PRISM data (bold black line) (Daly et al. 2002) and projected average precipitation between 2090 and 2100 from 12 AOGCMs
The spatial distribution of kudzu climatic habitat under 2100 climate conditions is shown in Fig. 6. Under the Maxent release and constraint models, climatic habitat expands northward and considerable land area is at high risk. A similar expansion is observed in the MD release model, although overall risk is lower. Under the MD constraint model, climatic habitat shifts northward, but contracts from the current range. An ensemble of both models, variable sets, and all AOGCM projections shows the highest risk in southeast and mid-Atlantic states. Future invasion risk is shifted northwards by about 200 km from current risk, and is projected to expand well into Pennsylvania, New York, and New England states by 2100. Some risk of kudzu invasion is also manifest on the west coast, most notably in coastal Washington and Oregon.
Projected climatic habitat for kudzu in the year 2100. a The bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. b The bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. c The bioclimatic envelope based on the MD model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. d The bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. e Sum of bioclimatic envelope projections a–d. Areas identified as climatic habitat by multiple BEMs and AOGCM conditions are more likely at risk from kudzu invasion
The spatial distribution of privet climatic habitat under 2100 climate conditions is shown in Fig. 7. Similar to kudzu, all models and variable sets show a northward shift, although MD projections show less risk than Maxent projections. An ensemble of both models, variable sets, and all AOGCM projections shows the highest risk in central states of Kentucky, Tennessee, North Carolina, and Virginia. Future invasion risk is shifted northwards by over 300 km from current risk, but is substantially reduced in Gulf Coast states in the majority of projections. Invasion risk increases in Pennsylvania, New York, and New England states by 2100, and also expands westward into Missouri. Some risk of privet invasion also extends to the northwest states of Washington and Oregon.
Projected climatic habitat for privet in the year 2100. a The bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. b The bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. c The bioclimatic envelope based on the MD model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. d The bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. e Sum of bioclimatic envelope projections a–d. Areas identified as climatic habitat by multiple BEMs and AOGCM conditions are more likely at risk from privet invasion
The spatial distribution of cogongrass climatic habitat under 2100 climate conditions is shown in Fig. 8. Once again, all models show a northward expansion, but the Maxent models predict invasion risk under more AOGCM conditions than the MD models. An ensemble of both models, variable sets, and all AOGCM projections shows that current at-risk areas along the Gulf Coast will continue to be vulnerable to invasion by cogongrass, while the invasion risk is likely to expand considerably northward into Kentucky, West Virginia, and Virginia.
Projected climatic habitat for cogongrass in the year 2100. a The bioclimatic envelope based on the MAXENT model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. b The bioclimatic envelope based on the MAXENT model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. c The bioclimatic envelope based on the MD model using predictor variables selected based on land area released is applied to 12 AOGCM projections of 2100 climate conditions. d The bioclimatic envelope based on the MD model using predictor variables selected based on those that most constrain distribution is applied to 12 AOGCM projections of 2100 climate conditions. e Sum of bioclimatic envelope projections a–d. Areas identified as climatic habitat by multiple BEMs and AOGCM conditions are more likely at risk from cogongrass invasion
Discussion
The bioclimatic envelope model results for kudzu, privet, and cogongrass show that climate change is likely to substantially increase land area at risk of invasion. These results provide spatially explicit evidence that supports the previous hypothesis that warming temperatures will expand risk northwards (Rogers and McCarty 2000), and previous projections and observations of expansion with rising temperatures (Jarnevich and Stohlgren 2009). These species-specific results are important for informing long-term management strategies aimed at reducing the impact of three of the most destructive invasive plants in the United States.
For all three species, invasion risk with climate change expands northward by several hundred kilometers by the year 2100. Expansions of climatic habitat are likely to occur gradually for kudzu and cogongrass, with invasive species moving north through short (e.g., wind, water, or animal-borne) and long-distance (e.g., accidentally via transportation corridors) seed dispersal. In the case of privet, seed sources in northern states may already exist due to privet’s use in landscaping, making rapid invasion of forested areas more likely with climate change. For all three species, expanded risk with climate change underscores the need for active control of invasive plant populations along their northern borders now and long-term management planning to prevent invasion into new areas. Particular emphasis should be placed on eradicating small populations of invasive species in northern states that are likely to become sources for future expansion due to climate change (Moody and Mack 1988).
Previous studies have found that some invasive species are likely to contract in range due to climate change (Bradley 2009b; Mika et al. 2008), creating unprecedented opportunities for restoration (Bradley et al. 2009). However, climate-induced restoration opportunities for areas currently occupied by the three species modeled in this paper appear unlikely. For cogongrass, currently invaded land areas in Florida and Gulf Coast states remain at risk under a majority of models (Fig. 8e). For kudzu and privet, the southern edges of their current range may become less climatically suitable (Figs. 6e, 7e). However, several models in the ensemble continue to project these areas as suitable climatic habitat, making it doubtful that large-scale opportunities for restoration will arise.
While projections provided by this analysis are useful for understanding regional risk resulting from climate change, risk assessments using landscape and local datasets are needed for specific localities. For example, Guisan and Thuiller (2005) recommend the hierarchical use of seed dispersal models, land-use disturbance factors, and/or resource models in conjunction with bioclimatic envelope models. The addition of landscape and local-scale risk factors to the climate change risk assessments would improve local and landscape-scale management (Bradley 2009a).
Further, any risk assessments of potential future invasion should be treated as guides rather than predictions because, while we can test model fit relative to current species distribution, we cannot validate the accuracy of projections. However, these and other BEM results provide testable hypotheses of future risk that should be evaluated observationally or experimentally (e.g., Pattison and Mack 2008).
In the southeast, envelope modeling provides few clues as to the most important climate predictors of invasion because monthly climate variables, particularly temperature, have a high degree of autocorrelation (Table 2). Hence, it is difficult to identify the “best” climatic predictors of invasive plant distribution. The high levels of autocorrelation cause the inconsistencies between the climate predictor variables selected based on the constraint and release methodologies. The high autocorrelation also means that the model predictor variables are not independent. However, selecting relatively independent predictor variables (e.g., those with R 2 < 0.60) produces a poor model fit. As a result, we elected to give priority to model fit over predictor variable independence. In a region where climate variables are less correlated, the constraint and release methods should identify similar sets of predictor variables.
One difficulty caused by the high autocorrelation is that we cannot use the predictor variables to interpret invasion ecology for the three species. In the western United States, for example, Bradley (2009b) used climatic habitat predictor variables to hypothesize that spring and summer precipitation affect invasive cheatgrass (Bromus tectorum) competitiveness relative to native shrubs. In the southeast ecoregion, variable autocorrelation makes it hazardous at best to develop hypotheses about invasive plant’s physiological relationships with specific climate variables. For kudzu, privet, and cogongrass, assessments of how climate affects competition and physiology are best done experimentally or observationally (e.g., Ziska 2008) rather than geographically.
This study uses an ensemble of envelope models, predictor variables, and climate change projections to assess current and future invasion risk. Numerous methods and models have been presented for envelope modeling (see for examples, Elith et al. 2006; Tsoar et al. 2007), selecting predictor variables (Arundel 2005; Bradley 2009b; Hirzel et al. 2002), and projecting climate change (IPCC 2007). Unfortunately, in ecological forecasting it is impossible to identify the “best” method for a given species because we cannot validate future occurrences. Similarly, AOGCM projections frequently disagree, and likewise are difficult to validate (but see Milly et al. 2005). However, by combining multiple methods and models as shown here, we are less likely to be misled by any single, erroneous prediction (Araujo and New 2007). Such broad-brush approaches may be necessary to encompass the level of uncertainty inherent in ecological forecasting related to climate change.
There is no consensus among the three case studies as to whether Maxent or MD typically predicts a smaller land area to be climatic habitat. Similarly, there is no consensus on the relative performance of the constraint and release predictor variables. Both of these envelope models and sets of predictor variables project current climatic habitat equally well (Figs. 1, 2 and 3). There is, however, a consistent difference in projections of climatic habitat with climate change. Maxent models typically project a larger land area at risk than MD models. Differences in climatic habitat projections from the two models suggest that multiple envelope-modeling methodologies are necessary to reduce uncertainty in ecological forecasting.
Despite some methodological differences, the model consensus for these three species is that climate change will greatly expand invasion risk. Species-specific risk assessments are a critical component of land management and conservation planning. With spatially explicit projections, conservation practitioners can develop more informed long-term management plans designed to reduce invasion risk proactively, before invasions occur. These types of species-specific projections will become increasingly important for informing management of invasive and native species in light of climate change.
Notes
The following AOGCMs were used in this study: Canadian Centre for Climate Modelling & Analysis (CCCMA) CM3.1, Centre National de Recherches Meteorologiques (CNRM) CM3, Commonwealth Scientific & Industrial Research Organisation (CSIRO) MK3.5, Geophysical Fluid Dynamics Laboratory (GFDL) 2.1, Goddard Institute for Space Studies (GISS), Hadley Centre for Climate Prediction (HAD) CM3, Institute for Numerical Mathematics (INM) CM3, Institut Pierre Simon Laplace (IPSL) CM4, Model for Interdisciplinary Research on Climate (MIROC) 3.2, Max Planck Institute for Meteorology (MPI) ECHAM5, National Center for Atmospheric Research (NCAR) CCSM3.0, National Center for Atmospheric Research (NCAR) PCM1.
References
Araujo MB, New M (2007) Ensemble forecasting of species distributions. Trends Ecol Evol 22:42–47
Arundel ST (2005) Using spatial models to establish climatic limiters of plant species’ distributions. Ecol Model 182:159–181
Batcher MS (2000) Elemental stewardship abstract for Ligustrum spp. Privet. The Nature Conservancy, http://tncweeds.ucdavis.edu/esadocs/documnts/ligu_sp.pdf
Beerling DJ (1993) The impact of temperature on the northern distribution-limits of the introduced species Fallopia japonica and Impatiens glandulifera in north-west Europe. J Biogeogr 20:45–53
Blaustein RJ (2001) Kudzu’s invasion into Southern United States life and culture. In: McNeeley JA (ed) The great reshuffling: human dimensions of invasive species. IUCN, The World Conservation Union, Gland, Switzerland and Cambridge, UK, pp 55–62
Bradley BA (2009a) Assessing ecosystem threats from global and regional change: hierarchical modeling of risk to sagebrush ecosystems from climate change, land use and invasive species in Nevada, USA. Ecography. doi:10.1111/j.1600-0587.2009.05684.x
Bradley BA (2009b) Regional analysis of impacts of climate change on cheatgrass invasion shows potential risk and opportunity. Glob Chang Biol 15:196–208
Bradley BA, Oppenheimer M, Wilcove DS (2009) Climate change and plant invasion: restoration opportunities ahead? Glob Change Biol 15:1511–1521
Broennimann O, Treier UA, Muller-Scharer H, Thuiller W, Peterson AT, Guisan A (2007) Evidence of climatic niche shift during biological invasion. Ecol Lett 10:701–709
Bryson CT, Carter R (1993) Cogongrass, Imperata cylindrica, in the United States. Weed Technol 7:1005–1009
D’Antonio CM, Vitousek PM (1992) Biological invasions by exotic grasses, the grass fire cycle, and global change. Annu Rev Ecol Syst 23:63–87
Daly C, Gibson WP, Taylor GH, Johnson GL, Pasteris P (2002) A knowledge-based approach to the statistical mapping of climate. Clim Res 22:99–113
Dukes JS, Mooney HA (1999) Does global change increase the success of biological invaders? Trends Ecol Evol 14:135–139
Elith J, Graham CH, Anderson RP, Dudik M, Ferrier S, Guisan A, Hijmans RJ, Huettmann F, Leathwick JR, Lehmann A, Li J, Lohmann LG, Loiselle BA, Manion G, Moritz C, Nakamura M, Nakazawa Y, Overton JM, Peterson AT, Phillips SJ, Richardson K, Scachetti-Pereira R, Schapire RE, Soberon J, Williams S, Wisz MS, Zimmermann NE (2006) Novel methods improve prediction of species’ distributions from occurrence data. Ecography 29:129–151
Farber O, Kadmon R (2003) Assessment of alternative approaches for bioclimatic modeling with special emphasis on the Mahalanobis distance. Ecol Model 160:115–130
Forseth IN, Innis AF (2004) Kudzu (Pueraria montana): history, physiology, and ecology combine to make a major ecosystem threat. Crit Rev Plant Sci 23:401–413
Franklin J (1995) Predictive vegetation mapping: geographic modelling of biospatial patterns in relation to environmental gradients. Prog Phys Geogr 19:474–499
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8:993–1009
Guisan A, Zimmermann NE (2000) Predictive habitat distribution models in ecology. Ecol Model 135:147–186
Hirzel AH, Hausser J, Chessel D, Perrin N (2002) Ecological-niche factor analysis: how to compute habitat-suitability maps without absence data? Ecology 83:2027–2036
Hirzel AH, Le Lay G, Helfer V, Randin C, Guisan A (2006) Evaluating the ability of habitat suitability models to predict species presences. Ecol Model 199:142–152
Holm LG, Plucknett DL, Pancho JV, Herberger JP (1977) The world’s worst weeds: distribution and biology. University Press of Hawaii, Honolulu, HI 609 pp
IPCC (2007) Climate change 2007: the physical science basis. Contribution of working group I to the fourth assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, UK and New York, USA
Jarnevich CS, Stohlgren TJ (2009) Near term climate projections for invasive species distributions. Biol Invasions 11:1373–1379
Jeltsch F, Moloney KA, Schurr FM, Kochy M, Schwager M (2008) The state of plant population modelling in light of environmental change. Perspect Plant Ecol Evol Syst 9:171–189
Kearney M (2006) Habitat, environment and niche: what are we modelling? Oikos 115:186–191
Kriticos DJ, Sutherst RW, Brown JR, Adkins SW, Maywald GF (2003) Climate change and the potential distribution of an invasive alien plant: Acacia nilotica ssp. indica in Australia. J Appl Ecol 40:111–124
Lippincott CL (2000) Effects of Imperata cylindrica (L.) Beauv. (Cogongrass) invasion on fire regime in Florida sandhill (USA). Nat Areas J 20:140–149
MacDonald GE (2004) Cogongrass (Imperata cylindrica)—biology, ecology, and management. Crit Rev Plant Sci 23:367–380
Martin-R MH, Cox JR, Ibarraf F (1995) Climatic effects on buffelgrass productivity in the Sonoran Desert. J Range Manag 48:60–63
Marvin DC, Bradley BA, Wilcove DS (2009) A novel, web-based ecosystem mapping tool using expert opinion. Nat Areas J 29:281–292
Mau-Crimmins TM, Schussman HR, Geiger EL (2006) Can the invaded range of a species be predicted sufficiently using only native-range data? Lehmann lovegrass (Eragrostis lehmanniana) in the southwestern United States. Ecol Model 193:736–746
Mika AM, Weiss RM, Olfert O, Hallett RH, Newman JA (2008) Will climate change be beneficial or detrimental to the invasive swede midge in North America? Contrasting predictions using climate projections from different general circulation models. Glob Chang Biol 14:1721–1733
Millenium Ecosystem Assessment (2003) Ecosystems and human wellbeing: a framework for assessment. Island Press, Washington, DC
Milly PCD, Dunne KA, Vecchia AV (2005) Global pattern of trends in streamflow and water availability in a changing climate. Nature 438:347–350
Mitich LW (2000) Intriguing world of weeds series 67—kudzu [Pueraria lobata (Willd.) Ohwi]. Weed Technol 14:231–235
Moody ME, Mack RN (1988) Controlling the spread of plant invasions—the importance of nascent foci. J Appl Ecol 25:1009–1021
Moore PD (2004) Plant ecology—favoured aliens for the future. Nature 427:594–594
Nagel JM, Huxman TE, Griffin KL, Smith SD (2004) CO2 enrichment reduces the energetic cost of biomass construction in an invasive desert grass. Ecology 85:100–106
Nakicenovic N, Swart R (2000) Special report on emissions scenarios. Cambridge University Press, Cambridge, England 570 pp
Nielsen C, Hartvig P, Kollmann J (2008) Predicting the distribution of the invasive alien Heracleum mantegazzianum at two different spatial scales. Divers Distrib 14:307–317
Patterson DT, Flint EP, Dickens R (1980) Effects of temperature, photoperiod, and population source on the growth of cogongrass (Imperata cylindrica). Weed Sci 28:505–509
Pattison RR, Mack RN (2008) Potential distribution of the invasive tree Triadica sebifera (Euphorbiaceae) in the United States: evaluating CLIMEX predictions with field trials. Glob Chang Biol 14:813–826
Pearson RG, Dawson TP (2003) Predicting the impacts of climate change on the distribution of species: are bioclimate envelope models useful? Glob Ecol Biogeogr 12:361–371
Peterson AT, Stewart A, Mohamed KI, Araujo MB (2008) Shifting global invasive potential of European plants with climate change. PLoS ONE 3:e2441
Phillips SJ, Dudik M (2008) Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography 31:161–175
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259
Pimentel D, Lach L, Zuniga R, Morrison D (2000) Environmental and economic costs of nonindigenous species in the United States. Bioscience 50:53–65
Rogers CE, McCarty JP (2000) Climate change and ecosystems of the mid-Atlantic region. Clim Res 14:235–244
Rouget M, Richardson DM, Nel JL, Le Maitre DC, Egoh B, Mgidi T (2004) Mapping the potential ranges of major plant invaders in South Africa, Lesotho and Swaziland using climatic suitability. Divers Distrib 10:475–484
Sasek TW, Strain BR (1989) Effects of carbon dioxide enrichment on the expansion and size of kudzu (Pueraria lobata) leaves. Weed Sci 37:23–28
Sasek TW, Strain BR (1991) Effects of CO2 enrichment on the growth and morphology of a native and an introduced honeysuckle vine. Am J Bot 78:69–75
Schussman H, Geiger E, Mau-Crimmins T, Ward J (2006) Spread and current potential distribution of an alien grass, Eragrostis lehmanniana Nees, in the southwestern USA: comparing historical data and ecological niche models. Divers Distrib 12:582–592
Smith SD, Huxman TE, Zitzer SF, Charlet TN, Housman DC, Coleman JS, Fenstermaker LK, Seemann JR, Nowak RS (2000) Elevated CO2 increases productivity and invasive species success in an arid ecosystem. Nature 408:79–82
Sutherst RW (1995) The potential advance of pests in natural ecosystems under climate change: implications for planning and management. In: Pernetta J, Leemans R, Elder D, Humphrey S (eds) The impact of climate change on ecosystems and species: terrestrial ecosystems. IUCN, Gland, Switzerland, pp 88–98
Thomas CD, Cameron A, Green RE, Bakkenes M, Beaumont LJ, Collingham YC, Erasmus BFN, de Siqueira MF, Grainger A, Hannah L, Hughes L, Huntley B, van Jaarsveld AS, Midgley GF, Miles L, Ortega-Huerta MA, Peterson AT, Phillips OL, Williams SE (2004) Extinction risk from climate change. Nature 427:145–148
Thuiller W, Richardson DM, Pysek P, Midgley GF, Hughes GO, Rouget M (2005) Niche-based modelling as a tool for predicting the risk of alien plant invasions at a global scale. Glob Chang Biol 11:2234–2250
Thuiller W, Richardson DM, Midgley GF (2007) Will climate change promote alien plant invasions? In: Nentwig W (ed) Ecological Studies. Springer, Berlin Heidelberg, pp 197–211
Tsoar A, Allouche O, Steinitz O, Rotem D, Kadmon R (2007) A comparative evaluation of presence-only methods for modelling species distribution. Divers Distrib 13:397–405
Vitousek PM, D’Antonio CM, Loope LL, Westbrooks R (1996) Biological invasions as global environmental change. Am Sci 84:468–478
Walker DA, Halfpenny JC, Walker MD, Wessman CA (1993) Long-term studies of snow-vegetation interactions. Bioscience 43:287–301
Welk E (2004) Constraints in range predictions of invasive plant species due to non-equilibrium distribution patterns: purple loosestrife (Lythrum salicaria) in North America. Ecol Model 179:551–567
Welk E, Schubert K, Hoffmann MH (2002) Present and potential distribution of invasive garlic mustard (Alliaria petiolata) in North America. Divers Distrib 8:219–233
Weltzin JF, Belote RT, Sanders NJ (2003) Biological invaders in a greenhouse world: will elevated CO2 fuel plant invasions? Front Ecol Environ 1:146–153
Wilcox J, Beck CW (2007) Effects of Ligustrum sinense Lour. (Chinese privet) on abundance and diversity of songbirds and native plants in a southeastern nature preserve. Southeast Nat 6:535–550
Wilcut JW, Truelove B, Davis DE, Williams JC (1988) Temperature factors limiting the spread of cogongrass (Imperata cylindrica) and torpedograss (Panicum repens). Weed Sci 36:49–55
Zavaleta E (2000) The economic value of controlling an invasive shrub. Ambio 29:462–467
Zavaleta ES, Royval JL (2002) Climate change and the susceptibility of U.S. ecosystems to biological invasions: two cases of expected range expansion. In: Schneider SH, Root TL (eds) Wildlife responses to climate change: North American case studies. Island Press, Washington, DC, pp 277–341
Zhu L, Sun OJ, Sang WG, Li ZY, Ma KP (2007) Predicting the spatial distribution of an invasive plant species (Eupatorium adenophorum) in China. Landsc Ecol 22:1143–1154
Ziska LH (2003) Evaluation of the growth response of six invasive species to past, present and future atmospheric carbon dioxide. J Exp Bot 54:395–404
Ziska LH (2008) Rising atmospheric carbon dioxide and plant biology: the overlooked paradigm. DNA Cell Biol 27:165–172
Ziska LH, Sicher RC, George K, Mohan JE (2007) Rising atmospheric carbon dioxide and potential impacts on the growth and toxicity of poison ivy (Toxicodendron radicans). Weed Sci 55:288–292
Acknowledgments
This study was supported by the High Meadows Foundation. Dave Marvin was instrumental in collecting the data upon which this study depends. We thank Lew Ziska, Diane Debinski, Nathan Gregory, and Margarita Ramos for their thoughtful comments. We gratefully acknowledge the modeling groups for providing their data for analysis, the Program for Climate Model Diagnosis and Intercomparison (PCMDI) for collecting and archiving the model output, and the JSC/CLIVAR Working Group on Coupled Modeling (WGCM) for organizing the model data analysis activity. The IPCC Data Archive is supported by the Office of Science, U.S. Department of Energy.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Bradley, B.A., Wilcove, D.S. & Oppenheimer, M. Climate change increases risk of plant invasion in the Eastern United States. Biol Invasions 12, 1855–1872 (2010). https://doi.org/10.1007/s10530-009-9597-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-009-9597-y