Abstract
The recent recognition of invasive hybrid watermilfoil (Myriophyllum spicatum × M. sibiricum) in North America has necessitated a more thorough evaluation of its overall distribution and occurrence in natural populations. A comprehensive survey of watermilfoil populations was conducted in five Minnesota lakes, three of which were suspected a priori to contain hybrid watermilfoil. DNA sequence data verified that hybrid plants between the nonindigenous M. spicatum L. and indigenous M. sibiricum Kom. occurred in three of the five lakes sampled. Myriophyllum spicatum was not detected in lakes where hybrids were prevalent. Further sampling of lakes in Idaho, Michigan, Minnesota, Wisconsin and Washington identified 30 additional hybrid watermilfoil populations. In only three of these populations the hybrid watermilfoil was found to co-occur with M. spicatum. To facilitate the field identification of the two parental species and their hybrid, morphological data from watermilfoil specimens collected across the United States were evaluated. We determined that leaf segment/leaf length measurements can effectively distinguish M. spicatum and M. sibiricum; however, hybrids are intermediate for these characters and such measurements frequently overlap with respect to their parental taxa. By incorporating a combined molecular and morphological approach to identifying watermilfoils, the hybrids can be identified readily and their distributions elucidated both within and between lakes. Because hybrids may respond differently to local ecological conditions than their parents, information on their presence and distribution should be of particular importance to management and conservation programs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The nonindigenous Eurasian watermilfoil (Myriophyllum spicatum L.) is widely recognized as a problematic invasive plant in North America, primarily due to the rampant spread of this aquatic species throughout waterbodies of 46 states and three Canadian provinces (Jacono and Richerson 2003; Kim 2005). Cronk and Fennessy (2001) categorized Eurasian watermilfoil among the five most noxious wetland plants and currently it is the most widely managed aquatic weed in the United States (Bartodziej and Ludlow 1998). During the course of a comparative molecular systematic study of Myriophyllum, Moody and Les (2002) discovered that some invasive, North American watermilfoil populations that had been attributed previously to M. spicatum actually consisted of hybrids between that nonindigenous species and the closely related, but indigenous M. sibiricum Kom. In that study, DNA sequence data were used to confirm the existence of three hybrid populations in Minnesota and Wisconsin.
The documentation of hybrid watermilfoils is significant because hybridity has been linked to aggressive and invasive traits in plants (Galatowitsch et al. 1999; Ellstrand and Schierenbeck 2000). The frequency of this consequence is evidenced by Ellstrand and Schierenbeck (2000) who provided 28 examples where invasiveness was preceded by hybridization and attributed in many cases to hybrid vigor. Hybridity also can produce novel genotypes (thus phenotypes) with ecological tolerances that differ from those of the parents (Anderson 1948). For example, recent studies have demonstrated that three Helianthus species of hybrid origin survive in extreme habitats not suitable for their parental taxa while retaining unique combinations of genes acquired in linkage groups typifying each of their parental species (Rieseberg et al. 2003; Rieseberg 2001; Schwarzbach et al. 2001).
There also is the potential for hybrids to respond differently to herbivory. Although hybrid plants generally are more susceptible to herbivory than their parental species, some studies have shown that in some instances they may be more resistant as well (Whitham et al. 1999; Floate and Whitham 1994; Fritz et al. 1994). Accordingly, it seems imperative that hybrid genotypes be taken into account when biocontrol is considered. For example the aquatic milfoil weevil (Euhrychiopsis lecontei) can reduce some M. spicatum populations effectively (Creed and Sheldon 1995) while having little effect on the native M. sibiricum (Newman et al. 1997). However, the effectiveness and specificity of these weevils with respect to the known hybrid between these species remains poorly known. There are similar concerns with the use of herbicides, as the susceptibility to herbicide application also remains uncertain for hybrid watermilfoil. Thus, as more biological and ecological information on hybrid milfoils is amassed, a better understanding of their distribution and population composition will be necessary to effectively direct management programs.
Molecular markers are useful in the identification of taxa that are difficult to distinguish using morphology alone. This factor is particularly pertinent to differentiating between M. spicatum and M. sibiricum, which traditionally have been identified by the presence (M. spicatum) or absence (M. sibiricum) of turions (a feature observable only in autumn) or by the number of compound leaf segments (pinnae), which has come to be regarded as inconsistent (Patten 1954; Orchard 1981). Furnier et al. (1995) first attempted the use of molecular (RAPD) markers to distinguish native watermilfoil species from the nonindigenous M. spicatum, but results were inconsistent. Moody and Les (2002) subsequently found that nrDNA sequence data (internal transcribed spacer region; ITS) could consistently and accurately differentiate almost all recognized North American watermilfoil species. Because these nuclear sequences are inherited biparentally it was possible to readily identify interspecific hybrids by recovering alleles specific to each parental species by cloning the ITS sequences obtained from hybrid individuals. This approach facilitated the unambiguous taxonomic assignment of even morphologically intractable specimens (Moody and Les 2002).
Although invasive hybrid populations of Myriophyllum spicatum × M. sibiricum have been documented, their overall distribution in any given waterbody has not yet been investigated. It remains to be determined whether watermilfoil hybrids simply are aberrations that occasionally co-occur with the parental taxa, or if hybrid populations could envelop water-bodies as expansive, homogeneous stands. Such information could provide some insight into whether hybrid watermilfoils are capable of persisting in habitats occupied by the parental species, or even possibly exhibit greater adaptiveness to specific lake environments. Furthermore, our current knowledge of overall distribution for watermilfoil hybrids in all of North America is limited to a survey of only three lakes (Moody and Les 2002). Thus, the main objectives of this research were to incorporate molecular markers and a strategic sampling strategy to: (1) determine whether M. spicatum and M. sibiricum commonly co-occur in the Minnesota and Wisconsin lakes where the invasive hybrid populations had been identified previously; (2) survey a broad sample of North American lakes for the presence of invasive hybrid watermilfoil populations; and (3) re-examine morphological characters that have been used traditionally to distinguish M. spicatum from M. sibiricum in light of the influence of hybrid individuals on these potentially distinguishing features.
Materials and methods
Population homogeneity sampling
Watermilfoil specimens were collected from five lakes with invasive watermilfoil populations in southeastern Minnesota for the purpose of DNA extraction and morphological analyses. Lakes examined included: Bald Eagle Lake (BE), Otter Lake (OT) and White Bear Lake (WB) in Ramsey County; and Cedar Lake (CED) and Lake Minnetonka (MK) in Hennepin County (Table 1; GPS location for each accession available upon request from authors). Ten individual plant accessions from each lake were collected from locations around the perimeter of these lakes, except Otter Lake (11) and Lake Minnetonka where nine accessions were collected from various bays. The sampling strategy was designed to survey populations from an assortment of distinct vegetated areas occurring within each lake. Sampled lakes were selected using different criteria. Lake Minnetonka and Cedar Lake were shown previously to contain plants with RAPD phenotypes similar to those shared by many M. spicatum populations (Furnier et al. 1995). Furthermore, these plants were identified confidently as M. spicatum by their morphology (Minnesota DNR, personal communication). Bald Eagle Lake, Otter Lake, and White Bear Lake were selected because they contained watermilfoil plants whose assignment to M. spicatum or M. sibiricum on the basis of morphology alone was equivocal (Minnesota DNR, personal communication). In addition, White Bear Lake previously was known to contain specimens that had been identified as hybrids between M. spicatum and M. sibiricum (Moody and Les 2002). Also, Bald Eagle Lake, Otter Lake and White Bear Lake contained at least some watermilfoil specimens with RAPD phenotypes distinct from those most commonly sampled by Furnier et al. (1995), thus suggesting that these anomalous individuals may represent hybrids.
In addition 2–4 individual plant accessions (Table 1) were collected from lakes identified in Wisconsin by Jester et al. (2000) as having low watermilfoil weevil density on invasive watermilfoil population and from several Minnesota lakes with invasive watermilfoil that occurred within the same geographic region as those suspected of harboring hybrids. The sampling for these lakes was less comprehensive than for the previously discussed Minnesota lakes, but was meant to obtain additional general information on invasive watermilfoil distribution.
Sampling for the distribution of hybrid watermilfoil
Plants suspected a priori as hybrid watermilfoil were collected from lakes in several states (Idaho, Michigan, Minnesota, Washington, Wisconsin) by one of the authors or by aquatic plant management agencies (Table 1). This sampling scheme did not comprehensively survey lakes but specifically targeted invasive watermilfoil plants (i.e., those forming dense, homogeneous stands) that possessed an ambiguous morphology.
DNA isolation, subcloning, and sequencing
Total DNAs were extracted from either NaCl-CTAB preserved (Rogstad 1992) or fresh leaf material using a miniprep procedure (Doyle and Doyle 1987). The nuclear DNA ITS region was amplified and sequenced to determine plant identity and/or hybrid origin. Double-stranded DNAs were amplified using the polymerase chain reaction (PCR) to amplify the ITS-1, ITS-2 and 5.8S region of nuclear ribosomal DNA using the ITS4 and ITS5 primers (White et al. 1990). The chloroplast DNA trnL/F region was amplified and sequenced in order to determine the maternal parentage of hybrid watermilfoils in Minnesota lakes given that in plants the chloroplast DNA is generally inherited maternally. The cpDNA trnL/F spacer was amplified using the primers “C” and “F” (Taberlet et al. 1991) for accessions collected from the more intensively sampled Minnesota lakes (Table 1). For specimens collected in these lakes, the PCR products were cloned into plasmids using a TOPO TA cloning kit (Invitrogen). Between four and 10 clones were screened from each individual that showed polymorphic sites in ITS by reamplifying the entire ITS region. Clones were sequenced for the entire ITS-2 and 5.8S region as well as a portion of ITS-1 which included 9 point mutations and 1 indel (1 bp) that varied consistently between M. spicatum and M. sibiricum. Cloned specimens were cycle sequenced using the ITS4 (White et al. 1990) primer. The trnL region was sequenced using the primer “D” (Taberlet et al. 1991). Subsequent to confirmation that ITS sequence polymorphisms positively identified hybridization through cloning of the aforementioned specimens, DNA sequence polymorphisms alone were used in determining hybrids for all other analyses (Table 1). Sequences were obtained using an ABI 3100 DNA automated sequencer. Sequence data have been deposited in the GenBank database as accession numbers AF513839, AF513849, AF513850, DQ786012- DQ786029.
Analyses of sequence data
ITS sequence data from all watermilfoil specimens initially were compiled into a single data set. These data were compared to data obtained previously by Moody and Les (2002) in order to verify that the polymorphisms occurred at those specific nucleotide sites that differed consistently between M. spicatum and M. sibiricum. Subsequently we cloned ITS sequences from the Minnesota material (Table 1) that exhibited ITS polymorphisms, as well as some pure (i.e., monomorphic) M. sibiricum and M. spicatum accessions. Cloned ITS sequence data from each accession were compared to ITS sequence data from M. spicatum and M. sibiricum to verify the presence of the nucleotide sites in these putative hybrids that matched each cloned parental sequence. Cloned sequences from the hybrid watermilfoils, M. spicatum and M. sibiricum were then compiled into individual data sets representing each lake. Sequences were aligned manually and analyzed for polymorphisms and/or variable sites using Sequencher 4.1.2 (Gene Code Corp.) and MacClade 4 (Maddison and Maddison 2001). Parsimony analyses were performed on sequence data originating from those lakes that contained hybrids using PAUP 4.0b8 (Swofford 2000). Heuristic searches (with 10 random taxon addition sequences) and tree bisection-reconnection with unordered, equally weighted characters were implemented. Indels (sequence gaps) were treated as binary characters. Parsimony analysis was conducted including single copies of cloned sequences (repetitive sequences not included) from hybrid watermilfoils compiled from the three more intensively sampled Minnesota lakes with hybrid watermilfoil. Subsequently a parsimony analysis was performed on a data set containing all sampled specimens from Lake Minnetonka and Cedar Lake and a representative sample of sequence clones from the previous analysis. Sequence data from the chloroplast trnL region were compiled into a single data set and then compared (as above) to determine their direct correspondence to the sequence of either parent (i.e., M. spicatum or M. sibiricum). This data set also was analyzed by parsimony as described above.
Morphological analysis
Leaf length, pinna number, and basal pinna length (unless unavailable) were measured from 10 leaves from each collected and sequenced accession from each Minnesota lake (Table 1). Subsequently, leaf length and pinna number (3–4 leaves) were obtained for specimens from multiple accessions of hybrids (Table 1) and their parental species that had been identified positively using DNA markers from several states and populations (CA [2 populations], CO [1], CT [4], FL [1], IN [1], MN [5], NH [1], WA [1], WI [13]; contact authors for location data). Leaf measurements were made from pressed dried specimens. These specific leaf characteristics were chosen because of the traditional use of pinna number to differentiate M. spicatum from M. sibiricum. Leaves were chosen for measurement by their position on the plant. Measurements were made of mature leaves positioned on submerged stems greater than three whorls above the base of the stem but lower than two whorls below the apex of the stem or base of the inflorescence. The position of measured leaves is particularly important given that leaves at the base of the stem or near the inflorescence, due likely to developmental constraints, have fewer pinnae than elsewhere on the mature plant. Bract shape and length measurements were considered initially; however, due to the frequent lack of reproductive structures and, when present, the inconsistency of reproductive stage (e.g. fruit or flower) these characters were not analyzed.
Results
Distribution
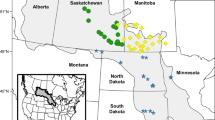
Hybrid watermilfoil populations were identified from 30 water-bodies in five states (Table 1; Fig. 1). In three cases both M. spicatum and hybrid watermilfoil were identified in the same lake (Fish Lake [2 hybrid, 3 M. spicatum] and Mud Lake [1 hybrid, 1 M. spicatum], Dane Co., WI; Round Lake [1 hybrid, 5 M. spicatum] Bonner, ID). Fish Lake and Mud Lake were inundated with dense populations of invasive watermilfoil and both hybrids and M. spicatum were collected from these populations whereas, Round Lake had only one sample identified as a hybrid among the more prevalent M. spicatum.
Distribution maps of the 30 known hybrid watermilfoil populations in Michigan, Minnesota and Wisconsin. Locations are based on lat./long. coordinates. ● identifies hybrid watermilfoil population verified with molecular markers. Some populations are close to each other and may appear as a single marker on the map due to overlap
Minnesota populations
The ITS data matrices analyzed consisted of 543 base pairs (bp) of which nine point mutations and one indel varied consistently between M. spicatum and M. sibiricum. All 10 watermilfoil accessions sampled from Otter Lake, 8/10 from White Bear Lake and 5/10 from Bald Eagle Lake were hybrid individuals that displayed ITS polymorphisms at all nine of the nucleotide sites that are known to differ between M. spicatum and M. sibiricum (Table 1). Two accessions from White Bear Lake and five accessions from Bald Eagle Lake had sequence data corresponding to M. sibiricum (Table 1). All accessions from Cedar Lake and Lake Minnetonka had ITS sequences corresponding only to M. spicatum (Table 1; Fig. 2).
Single most parsimonious tree of 28 steps from ITS sequences of accessions collected from Cedar Lake and Lake Minnetonka and a representative selection of cloned ITS sequences from hybrid and M. sibiricum taxa. Bold branches represent M. spicatum genotypes; all other branches represent M. sibiricum genotypes. B = Bald Eagle Lake, C = Cedar Lake, M = Lake Minnetonka, O = Otter Lake, W = White Bear Lake. Numbers following lake abbreviations represent accession # (followed by clone # if applicable). Numbers next to branches represent branch lengths. Cloned ITS sequences from M. sibiricum accessions are in boxes; cloned ITS sequences data from hybrid taxa are circled
All cloned ITS sequences (i.e., alleles) recovered from accessions with nucleotide polymorphisms corresponded either to M. spicatum or M. sibiricum. In addition, multiple copies of cloned M. sibiricum ITS sequences were recovered that varied consistently at specific nucleotide sites within and among populations, thus resulting in clustering of similar genotypes that shared common nucleotide changes in ITS (Figs. 2 and 3). This extent of nucleotide variation among M. sibiricum ITS sequences occurred in addition to those sites that varied consistently between M. spicatum and M. sibiricum. Other minor variations were observed. Some cloned ITS sequences showed unique substitutions at one or more sites (Figs. 2 and 3) and several ITS sequences cloned from hybrid accessions were mosaics (sequences with a combination of characters from both parental species) in addition to possessing copies of each individual parental sequence (i.e., Moody and Les 2002) likely a result of in vitro chimera formation (i.e., Cronn et al. 2002).
Single most parsimonious tree of 44 steps using cloned ITS sequences of accessions collected from Bald Eagle Lake, Otter Lake and White Bear Lake. Bold branches represent taxa with cloned ITS sequence data representing the M. spicatum genotype, all other branches represent cloned ITS sequence data representing M. sibiricum genotypes. Watermilfoil accessions are represented by the lake: B = Bald Eagle Lake, O = Otter Lake, W = White Bear Lake; followed by numbers representing accession # followed by clone #. Numbers next to branches represent branch lengths. ITS sequences cloned from M. sibiricum accessions are in boxes. All other sequences originated from cloned hybrid plants
The trnL region of cpDNA (422 bp) contained one point mutation and two indels (16 bp and 15 bp, respectively) that differed between all sampled accessions of M. spicatum and M. sibiricum. All Minnesota hybrid accessions showed inheritance of the plastid genome from M. spicatum, as indicated by the match of their cpDNA trnL sequences (not shown).
Morphology
Morphological data effectively differentiate M. spicatum and M. sibiricum when both leaf length and total pinna number are considered (Fig. 4). Hybrids were not distinctive in this measurement, but overlapped with both M. spicatum and M. sibiricum. At higher leaf lengths the hybrids trended towards intermediacy between the parental species in their leaf length/pinna number (Fig. 4). Basal pinna length was not distinctive (not shown).
Scatter diagram representing leaf length/pinnae number measurements taken from midstem leaves of M. sibiricum, M. spicatum and hybrid watermilfoil taxa. Although these leaf characters clearly distinguish M. sibiricum from M. spicatum (which do not overlap), the broad overlap of hybrid plants with both of the parents makes morphological identification difficult when hybrids are present. Specimens examined are listed in Table 1 and methods section
Discussion
The documentation of invasive watermilfoil hybrids in lakes ranging from Michigan to Washington indicates that they are widely dispersed across the northern portion of the United States and do not simply represent local, isolated occurrences (Table 1, Fig. 1). The presence of hybrids is now extended to include five states. Although most hybrid populations have been identified in Wisconsin, this result reflects a bias towards the active sampling conducted by the Wisconsin Department of Natural Resources. Sampling in the states where hybrid watermilfoil already has been identified is still being conducted, which likely will increase the number of sites with documented hybrid populations.
We are uncertain whether the fairly broad distribution pattern observed reflects the vegetative dispersal of plants resulting initially from a single or few hybrid events, or if repeated hybridization has occurred throughout areas where ranges of the parental species overlap. Aiken (1981) showed that F1 hybrids can be produced in the greenhouse, but further studies on hybrid fecundity are necessary. The presence of cpDNA sequence data matching either M. sibiricum or M. spicatum in different hybrid populations (Moody and Les 2002) does indicate at least two separate hybridization events, although all the Minnesota populations from the current study show maternal inheritance from M. spicatum. Further observations on the fertility and dispersal of hybrid watermilfoil should be undertaken to help clarify questions pertaining to hybrid origins.
The finding that watermilfoil accessions sampled from Otter Lake and White Bear Lake were primarily of hybrid origin; whereas, those from Lake Minnetonka and Cedar Lake all were M. spicatum (Figs. 2 and 3) corresponds with our phenotypic observations and interpretations of population level molecular (RAPD) data presented by Furnier et al. (1995). We identified five watermilfoil accessions from Bald Eagle Lake as M. sibiricum and five accessions to be of hybrid origin. The lower frequency of hybrids in this lake may indicate either that hybrids were introduced recently to Bald Eagle Lake or that there is less potential for their spread in this lake due to some as yet unknown ecological limitations. We did not find M. spicatum to co-occur in Minnesota lakes where hybrid populations were identified. In the five Minnesota lakes sampled most intensively, those in which hybrids were positively identified M. spicatum was not observed and lakes where M. spicatum was positively identified hybrids were not observed. Because that sampling was limited to 10 specimens per lake this result does not preclude the potential for co-occurrence, but given the collection from apparently homogenous stands of watermilfoil it does strongly suggest that either the hybrid or M. spicatum are dominant in these lakes. Including the less intensively surveyed lakes in our study, we identified only three lakes (Fish Lake and Mud Lake, WI; Round Lake, ID) out of 30 where both the hybrid and M. spicatum co-occurred (Table 1). A more comprehensive sampling will be necessary to better determine the distribution of each genotype in these lakes.
The abundance of invasive populations of both M. spicatum and hybrids (M. spicatum × M. sibiricum) does not strictly conform to the hypotheses of Ellstrand and Schierenbeck (2000) regarding hybridization as the primary cause for this invasion. Based on other ecological studies of hybrids (Emms and Arnold 1997; Rieseberg et al. 2003; Figueroa et al. 2003) it certainly is conceivable that hybrid watermilfoils might have a selective advantage over their parent M. spicatum in some environments while M. spicatum is more competitive in others. Because the hybrids combine traits of the indigenous M. sibiricum (which arguably would be well-adapted to local conditions) and the nonindigenous invasive M. spicatum, they may be more capable of thriving under specific environmental circumstances where M. spicatum would be less successful. It also is possible that the distribution we observe for hybrid watermilfoil could be a consequence of historical factors. The first invasive taxon to reach a lake may spread so quickly as to preclude the establishment of the other. Naturally, additional studies and experiments will be necessary to test these hypotheses.
Regardless, the fairly widespread distribution of invasive hybrid watermilfoils presents novel problems for management and conservation efforts. The spread of hybrids into lakes inhabited by M. sibiricum potentially may displace indigenous watermilfoil populations as evidenced by the loss of the species from Otter Lake. The long-term integrity of the indigenous M. sibiricum genome is another concern. It remains unclear whether introgression is occurring in this system, but the ability of M. spicatum to hybridize with M. sibiricum could lead to a reduction of pure M. sibiricum lineages through competition and/or genomic contamination resulting from repeated backcrossing. Our current molecular approach is unable to evaluate the occurrence of introgression. Although ITS markers are definitive in the recognition of recent hybridization events, it is likely that given enough time for potential backcrossing, the ITS region could become homogenized to a single copy (i.e. of an individual parent). The incorporation of additional population variable molecular markers (e.g., microsatellites) and greenhouse studies could be used to determine the extent, if any, of this potentially complicating factor.
The fairly widespread occurrence of hybrid watermilfoils also necessitates the reevaluation of biocontrol programs that involve the watermilfoil weevil (Euhrychiopsis lecontei). A long-term experiment by Newman (2004) has shown that Otter Lake invasive watermilfoil populations (identified here as hybrid plants) have experienced a greater and more consistent decline (presumably due to weevil treatment) than did those in several lakes containing M. spicatum. These results indicate that weevil biocontrol could be more effective on hybrid populations than on M. spicatum. Prior to the knowledge that hybrid watermilfoils existed, this result was explained primarily as a consequence of insectivory on the milfoil weevil. In light of the discovery of hybrids, these findings should be reinterpreted, taking into account potential genotypic effects. A recent greenhouse study was conducted by Roley and Newman (2006) specifying genotype, which suggests that watermilfoil weevils actually may have an intermediate effect on hybrid watermilfoil plants (as measured by watermilfoil weevil survival) with respect to M. spicatum (higher weevil survival) and M. sibiricum (lower weevil survival). Evidently there are confounding factors including the abundance of insectivores and sediment-genotype interactions that may affect the performance of watermilfoil weevils (Newman 2004). Yet, with the differential results between populations of hybrid watermilfoil and M. spicatum, future studies of this nature should consider genotype. Furthermore, the disparity of results between hybrid and parent also indicates that the taxa do respond differently to at least some ecological variables.
This study is the first to report the presence of multiple copies of the ITS region in M. sibiricum. All of these multiple copies were present in the hybrid watermilfoils examined as determined using DNA cloning techniques (Figs. 2 and 3). These results would be expected if the mechanisms of concerted evolution were not functioning properly and/or if polyploidy has occurred in M. sibiricum (Hughes et al. 2002; Hershkovitz et al. 1999; Campbell et al. 1997). Chromosome counts (Love and Love 1958; Love and Ritchie 1966; Aiken 1978) indicate that both M. sibiricum (2n = 42) and M. spicatum (2n = 28, 42) are polyploid in comparison to M. alterniflorum (2n = 14), their sister species (Moody and Les 2002). If copies of ITS reside on different chromosomes (as the result of polyploidy) the divergence of ITS within a single species could be expected; thus, the mechanisms of concerted evolution (homogenization of the ITS region) would be disrupted. Whether M. spicatum × M. sibiricum is a diploid or polyploid hybrid has yet to be determined.
Although molecular data can distinguish between watermilfoil species and their hybrids effectively (Moody and Les 2002), their procurement can be time-consuming and relatively expensive in comparison to field identification. Thus, it is desirable to identify reliable morphological markers that can distinguish the various taxa in the field. Two morphological characteristics have been used most commonly to differentiate the two similar species M. spicatum and M. sibiricum: (1) presence or absence of turions and (2) differences in leaf pinna number (Crow and Hellquist 2000; Orchard 1981, Fernald 1919). Turion presence or absence is not diagnostic because hybrids resemble M. spicatum in lacking turions. Thus far, no field collections of hybrid specimens have possessed turions. Furthermore, when hybrid watermilfoils were grown in greenhouse tanks together with their parental species, they did not form turions even though turions were produced in M. sibiricum (unpublished data).
Because turions are not present during the principal growing season of these plants, differences in leaf pinna number have been used predominantly to differentiate these species. The pinnately compound leaves of M. spicatum originally were characterized by having >13 pinna pairs; whereas, those of M. sibiricum had <12 pinna pairs (Fernald 1919). However, specimens more recently collected in the field often have been found to exhibit considerable overlap in leaf segment number (Patten 1954; Orchard 1981), thereby making their confident identification virtually impossible using this trait. We believe that the current lack of taxonomic confidence in leaf segment characters may reflect the collection of hybrid specimens whose intermediate leaf segment features blur the distinction that once existed when only the parental species were encountered and compared. Indeed, our results show that pinna number does appear to overlap only minimally between the two parental species, and also that any overlap is lost if leaf length is taken into consideration (Fig. 4) and traits are evaluated on leaves collected from the midstem region of mature, submerged plants as recommended by Crow and Hellquist (2000). Yet, even the consideration of pinna number and leaf length together does not consistently differentiate hybrids from either of the parents (Fig. 4). There is a trend towards pinna number intermediacy in most hybrid populations but the inconsistency of this trait suggests molecular characterization still remains the most reliable method of conclusively identifying hybrid watermilfoil and its parental species.
Hybrid watermilfoil has emerged as a serious invasive threat to North American waters and additional studies that further document its distribution, evaluate its ecological tolerances and interactions, clarify its biology and describe its responses to various control measures are much needed before any management strategy can be implemented effectively.
References
Aiken SG (1981) A conspectus of Myriophyllum (Haloragaceae) in North America. Brittonia 33:57–69
Aiken SG (1978) [Counts on Haloragaceae] p. 522. In: A. Love, IOPB Chromosome number reports LXII. Taxon 27:519–535
Anderson E (1948) Hybridization of the habitat. Evolution 2:1–9
Bartodziej W, Ludlow L (1998) Aquatic vegetation monitoring by natural resources agencies in the United States. Aquatics 20:15–18
Campbell CS, Wojciechowski MA, Baldwin BG, Alice LA, Donoghue MJ (1997) Persistent nuclear ribosomal DNA sequence polymorphism in the Amelanchier agamic complex. Mol Biol Evol 14:81–90
Creed RP Jr, Sheldon SP (1995) Use of a native insect as a biological control for an introduced weed. Ecol Appl 5:1122–1132
Cronk JK, Fennessy MS (2001) Wetland plants: biology and ecology. Lewis Publishers, Boca Raton, Florida
Cronn R, Cedroni M, Haselkorn T, Grover C, Wendel JF (2002) PCR-mediated recombination in amplification products derived from polyploid cotton. Theor Appl Genet 104:482–489
Crow GE, Hellquist CB (2000) Aquatic and wetland plants of Northeastern North America Vol. 1. The University of Wisconsin Press, Madison, Wisconsin
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bul Bot Soc Am 19:11–15
Ellstrand NC, Schierenbeck KA (2000) Hybridization as a stimulus for the evolution of invasiveness in plants? Proc Nat Acad Sci USA 97:7043–7050
Emms SK, Arnold ML (1997) The effect of habitat on parental and hybrid fitness: Transplant experiments with Louisiana irises. Evolution 51:1111–1119
Fernald ML (1919) Two new Myriophyllum and a species new to the United States. Rhodora 21:121–124
Figueroa ME, Castillo JM, Redondo S, Luque T, Castellanos EM, Nieva FJ, Luque CJ, Rubio-Casal AE, Davy AJ (2003) Facilitated invasion by hybridization of Sarcocornia species in a salt-marsh succession. J Ecol 91:616–626
Floate KD, Whitham TG (1994) Aphid–ant interaction reduces chrysomelid herbivory in a cottonwood hybrid zone. Oecologia 97:215–221
Fritz RS, Nichols-Orians CM, Brunsfeld SJ (1994) Interspecific hybridization of plants and resistance to herbivores: hypotheses, genetics, and variable responses in adverse herbivore community. Oecologia 97:106–117
Furnier GR, Olfelt JP, Stolz AM (1995) Genetic variation in Eurasian watermilfoil. Report submitted as deliverables C2.4.1 and C3.1.1. Minnesota Department of Natural Resources, Ecological Services, St. Paul, MN
Galatowitsch SM, Anderson NO, Ascher PD (1999) Invasiveness in wetland plants in temperate North America. Wetlands 19:733–755
Hershkovitz MA, Zimmer EA, Hahn WJ (1999) Ribosomal DNA sequences and angiosperm systematics. In: Hollingsworth PM, Bateman RM, Gornall RJ (eds) Molecular systematics and plant evolution. Taylor & Francis, London, UK, pp 268–326
Hughes CE, Bailey CD, Harris SA (2002) Divergent and reticulate species relationships in Leucaena (Fabaceae) inferred from multiple data sources: insights into polyploid origins and nrDNA polymorphism. Am J Bot 89:1057–1073
Jacono CC, Richerson MM (2003) http://nas.er.usgs.gov/taxgroup/plants/docs/my_spica.html
Jester LL, Bozek MA, Helsel DR, Sheldon SP (2000) Euhrychiopsis lecontei distribution abundance and experimental augmentations for Eurasian watermilfoil control in Wisconsin lakes. J Aquat Plant Manag 38:88–97
Kim AS (2005) Milfoil campaign spreading. Portland Press Herald, Monday, February 14, 2005
Love A, Love D (1958) The American element in the flora of the British Isles. Bot Notes 3:373–388
Love A, Ritchie JG (1966) Chromosome numbers from central and northern Canada. Can J Bot 44:429–439
Maddison DR, Maddison WP (2001) MacClade. Analysis of phylogeny and character evolution. 4.01. Sinauer, Sunderland, MA
Moody ML, Les DH (2002) Evidence of hybridity in invasive watermilfoil (Myriophyllum) populations. Proc Nat Acad Sci USA 99:14867–14871
Newman RM (2004) Biological control of Eurasian Watermilfoil completion report for 2001–2004 submitted to the Minnesota Department of Natural Resources. Ecological Services, St. Paul, MN
Newman RM, Borman ME, Castro SW (1997) Developmental performance of the weevil Euhrychiopsis lecontei on native and exotic watermilfoil host plants. J North Am Benthol Soc 16:627–634
Orchard AE (1981) A revision of South American Myriophyllum (Haloragaceae), and its repercussion on some Australian and North American species. Brunonia 4:27–65
Patten BC (1954) The status of some American species of Myriophyllum as revealed by the discovery of intergrade material between M. exalbescens and M. spicatum in New Jersey. Rhodora 56:213–225
Rieseberg LH (2001) Chromosomal rearrangements and speciation. Trends Ecol Evol 16:351–358
Rieseberg LH, Raymond O, Rosenthal DM, Lai Z, Livingstone K, Nakazato T, Durphy JL, Schwarzbach AE, Donovan LA, Lexer C (2003) Major ecological transitions in wild sunflowers facilitated by hybridization. Science 301:1211–1216
Roley SS, Newman RM (2006) Developmental performance of the milfoil weevil, Euhrychiopsis lecontei (Coleoptera: Curculionidae), on northern watermilfoil, Eurasian watermilfoil and hybrid (northern × eurasian) watermilfoil. Environ Entomol 35:121–126
Rogstad SH (1992) Saturated NaCl-CTAB solution as a means of field preservation of leaves for DNA analyses. Taxon 41:701–708
Schwarzbach AE, Donovan LA, Rieseberg LH (2001) Transgressive character expression in a hybrid sunflower species. Am J Bot 88:270–277
Swofford DL (2000) Phylogenetic analysis using parsimony. Version 4.0. Sinauer Associates, Sunderland
Taberlet P, Gielly L, Pautou G, Bouvet J (1991) Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol 17:1105–1109
White TJ, Bruns SL, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, San Diego, California, USA, pp 315–322
Whitham TG, Martinsen GD, Floate KD, Dungey HS, Potts BM, Keim P (1999) Plant hybrid zones affect biodiversity: tools for a genetic-based understanding of community structure. Ecology 80:416–428
Acknowledgements
We acknowledge the Department of Natural Resources in Minnesota (esp. Chip Welling), Wisconsin (esp. Laura Herman), and Washington (esp. Jenifer Parsons), and also B. Pullman for their assistance in collecting specimens for this study. Funding for this project has been provided by NEBC Graduate Student Research Award Program, CAWS Michael Lefor Wetland Science Research Grant, Karling Graduate Student Research award (BSA), William R. Anderson Student Research Grant (ASPT), University of Connecticut Bamford Endowment Fund, Minnesota Dept. of Natural Resources and NSF DDIG 0309123.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Moody, M.L., Les, D.H. Geographic distribution and genotypic composition of invasive hybrid watermilfoil (Myriophyllum spicatum × M. sibiricum) populations in North America. Biol Invasions 9, 559–570 (2007). https://doi.org/10.1007/s10530-006-9058-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10530-006-9058-9