Abstract
Objective
This study was aimed to assess the effect of a novel postbiotic on bacterial community composition and structure within the intestinal ecosystem of rainbow trout (Oncorhynchus mykiss), as well as evaluate its capacity to protect rainbow trout from Lactococcus garvieae infection.
Results
After 30 days of dietary postbiotic supplementation, high-throughput 16S rRNA gene sequencing revealed that bacterial community composition, diversity and richness were significantly higher in treated fish than in control fish. The proportion of sequences affiliated to the phylum Tenericutes, and to a lesser extent, the phyla Spirochaetes and Bacteroidetes was increased in fish fed a postbiotic-enriched diet compared to control fish, whereas the abundance of Fusobacteria was higher in control fish. Moreover, the treated fish showed significantly (p < 0.05) improved protection against L. garvieae compared to control fish.
Conclusions
These findings suggest that dietary postbiotic supplementation may represent an environmentally friendly strategy for preventing and controlling diseases in aquaculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactococcosis is a disease caused by Lactococcus garvieae, which is responsible for severe economic losses in farmed marine and freshwater fish species (Vendrell et al. 2006; Gibello et al. 2016; Meyburgh et al. 2017). Disease outbreaks are usually treated with vaccines or antibiotics. Although vaccination has proven effective in protecting fish from lactococcosis, this immunity lasts a short period of time or the process is often ineffective when applied to immunologically immature fish (Ravelo et al. 2006; Embregts and Forlenza 2016). Moreover, the use of antibiotics should be limited due to the increasing prevalence of antibiotic-resistant bacteria (Cabello et al. 2013; Santos and Ramos 2018). Consequently, new environmentally-friendly strategies for disease prevention and control are urgently needed. Among, the use of postbiotics is becoming increasingly popular for treatment and/or prevention of diseases (Pérez-Sánchez et al. 2018; Ang et al. 2020). Postbiotics are soluble factors (products or metabolic byproducts) secreted by live bacteria or released after bacterial lysis, which may provide physiological benefit to the host (Aguilar-Toalá et al. 2018). Previous studies, including our own, have demonstrated the probiotic effect of some lactic acid bacteria strains under in vitro and in vivo conditions (Vendrell et al. 2008; Pérez-Sánchez et al. 2011; Zheng et al. 2017). However, the efficiency of a postbiotic obtained as a fermented feed product on the lactococcosis prevention has not been previously evaluated. This postbiotic derives from a fermented feed, which has been previously evaluated in animal husbandry (HEALTSTOCK Project, ID 733627; https://cordis.europa.eu/projects/en).
Given that postbiotics may be potential alternatives to the use of live probiotic microorganisms, it becomes interesting to explore their effects in species of aquaculture interest. The aim of this study was therefore to investigate the effect of a novel postbiotic obtained as a fermented feed product containing lactic acid bacteria on bacterial community structure and composition within the intestinal ecosystem of rainbow trout (Oncorhynchus mykiss), as well as evaluate its capacity to protect rainbow trout from L. garvieae infection.
Materials and methods
Postbiotic and experimental conditions
The postbiotic was obtained as a fermented feed product composed of soy and alfalfa flour. This fermented feed was obtained in two stages. Briefly, a lactic acid bacterium belonging to the genus Lactobacillus, previously isolated from rainbow trout and deposited at the Spanish Type Culture Collection (CECT 9882), was grown in de Man, Rogosa and Sharpe broth (MRS; Oxoid, Basingstoke, UK) overnight at 22 °C. The first stage covered the fermentation of the bacterial pre-culture with other minor components. After the incubation period, bacterial cells were collected and non-bitter beer yeast was added to the raw material and the second fermentation process was then performed, as previously described (Cabello-Olmo et al. 2019). The fermented feed product was micronized to favor the mixture with commercial feed (Inicio Plus 887; BioMar Iberia, S.A., Dueñas, Spain).
A total of 100 pathogen-free rainbow trout weighting 24.6 ± 5.1 g were obtained from a commercial fish farm, which were acclimatized in our experimental fish facility for 2 weeks. The fish were then randomly assigned to three experimental groups and maintained in three tanks. Two groups were fed a commercial feed without any supplement: one group (n = 40) was used as untreated control, whereas the other group (n = 20) was used for the experimental infection as donors. The third group (n = 40) received a diet obtained by adding the postbiotic to the commercial diet at 3.0 mg/g. All fish were fed daily at 1.5% of their biomass.
DNA extraction and sequence analysis
After 30 days of feeding, fish were individually weighed and four fish per treatment were sacrificed to collect the intestinal samples, as previously described (Etyemez and Balcázar 2015). Genomic DNA was extracted using the DNeasy Blood & Tissue kit (QIAGEN; Valencia, CA, USA), and the final concentration and purity were determined using a NanoDrop spectrophotometer (Thermo Scientific; Wilmington, DE, USA). Genomic DNA samples were then submitted to Macrogen Inc. (Seoul, Korea) for high-throughput 16S rRNA gene sequencing on the Illumina MiSeq platform. Analysis of 16S rRNA gene sequences was performed using the MOTHUR software package (Schloss et al. 2009). Briefly, paired-end reads were merged into contigs, screened for quality, aligned to the SILVA reference database, and screened for chimeras. Sequences were then randomly subsampled to contain the same number of sequences (99,729) for further comparisons. Curated sequences were clustered into operational taxonomic units (OTUs) using a 97% similarity cutoff with the average neighbor clustering algorithm. Alpha diversity was calculated using the Shannon diversity index (H’) and the Chao1 richness estimator. Beta diversity was calculated using the Yue & Clayton estimator, which measures the number of shared genera and their relative abundances. The relationship among samples was visualized in a principal coordinate analysis (PCoA) plot based on the Yue & Clayton measure of dissimilarity. Analysis of molecular variance (AMOVA) was used to determine whether the clustering within the ordinations is statistically significant at p < 0.05.
Experimental infection
After 30 days of feeding, fish were challenged with L. garvieae by the cohabitation method. Briefly, L. garvieae strain FLP 33, previously isolated during a natural lactococcosis outbreak in rainbow trout, was grown on tryptic soy agar overnight, resuspended in phosphate-buffered saline (PBS), and adjusted to a concentration of 104 CFU/ml. A volume of 0.1 ml of this suspension was injected intraperitoneally into all fish used for cohabitation (the second group), which were anaesthetized with tricaine methanesulfonate (Tricaine Pharmaq 1000 mg/g) and marked by clipping the adipose fin after injection. Ten infected fish were then transferred into the tanks containing the other two experimental groups. All fish were monitored at least three times daily, and dead fish were immediately removed and examined for external signs of lactococcosis.
Statistical analysis
All statistical analyses were performed using SPSS Statistics v17.0 (SPSS Inc.; Chicago, IL, USA). Differences in the final weight and alpha diversity (number of OTUs, Shannon diversity index and Chao richness estimators) were analyzed using an unpaired two-tailed Student’s t-test. Survival curves were calculated using the Kaplan–Meier method, and significance was determined using the log-rank test. The level of significance was set at p < 0.05.
Results
After 30 days of feeding, no significant difference (p = 0.08) was observed in the final weight between treated (postbiotic) and control groups. Mean final weight of fish was 36.4 ± 7.2 g for treated group and 39.2 ± 6.9 g for control group. Moreover, the number of OTUs observed at a 97% taxonomic cutoff was significantly higher (p < 0.05) in the intestinal samples from fish treated with the postbiotic (414 ± 59), as compared to those samples from untreated fish (334 ± 16). Shannon diversity index and Chao richness estimators were also determined, demonstrating that the intestinal samples from treated fish (1.2 ± 0.2 and 4437.9 ± 241.2, respectively) had significantly higher (p < 0.01) bacterial diversity and richness than those samples from untreated fish (0.9 ± 0.1 and 3788.4 ± 262.5, respectively).
Overall taxonomic characterization of the bacterial community was conducted at the phylum level (Fig. 1). Although Tenericutes and Fusobacteria were found to be the most abundant phyla, there were differences in their abundance between treated and control groups. A higher proportion of sequences affiliated to the phylum Fusobacteria (notably the genus Cetobacterium) was found in the intestinal samples from untreated fish (control), as compared to those treated with the postbiotic. In contrast, a higher proportion of sequences affiliated to the phylum Tenericutes (mainly members belonging to the genus Mycoplasma) was found in fish treated with the postbiotic, as compared to those of the control group. A slight increase in the abundance of Spirochaetes (notably the genus Brevinema) and Bacteroidetes (particularly members belonging to the genera Bacteroides, Dysgonomonas and Flavobacterium) was also observed in fish treated with the postbiotic. At the phylum level, the abundance of sequences affiliated to Proteobacteria, and to a lesser extent, Actinobacteria and Firmicutes was similar between treated and control groups; however, differences were observed at the OTU level (defined at 97% similarity) between the two groups.
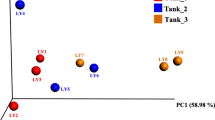
The effect of dietary postbiotic supplementation on bacterial community structure within the intestinal ecosystem was determined using a distance matrix based on the Yue & Clayton measure and visualized using PCoA plots (Fig. 2). The results revealed clear separation between treated and control groups. These observations were further validated using AMOVA tests, as implemented by MOTHUR, which showed that bacterial community structure was significantly different (p < 0.01) between treated and control groups.
After 30 days of feeding, fish were challenged with L. garvieae and Kaplan–Meier analysis revealed a significant difference (p < 0.05) in the cumulative survival between treated and control groups (Fig. 3). Cumulative survivals were 75.0 and 52.5% in treated and control groups, respectively.
Discussion
In the present study, we observed that dietary postbiotic supplementation may modify bacterial community composition and structure within the fish intestinal ecosystem. Previous studies have suggested that the manipulation of the host microbiota may represent a valuable strategy to prevent and/or control pathological and physiological disorders (Pérez et al. 2010). Although there was no significant difference in the final weight between treated and control groups, possibly due to the short period of the study, we observed that dietary postbiotic supplementation conferred significantly improved protection against L. garvieae infection. Postbiotics are nonviable bacterial products or metabolic byproducts from probiotic microorganisms that have biological activity in the host (Patel and Denning 2013). In fact, postbiotics aim to mimic the beneficial effects of probiotics while avoiding the risk of administering live microorganisms. Although the mechanisms underlying their effects seem to be mediated through an interaction between the host and microbial products, there is limited information on their effects on fish intestinal microbiota. In our study, the proportion of sequences affiliated to Tenericutes, and to a lesser extent, Spirochaetes and Bacteroidetes was increased in the intestinal samples of fish treated with the postbiotic as compared to those of untreated fish, whereas the abundance of Fusobacteria was higher in untreated fish. Interestingly, we observed that all sequences affiliated to the phylum Tenericutes were classified as belonging to the genus Mycoplasma, which were dominant in treated fish. A recent study demonstrated that abundance of potential pathogenic Vibrio species appeared to be inversely correlated with the presence of Mycoplasma species within the mid-intestinal microbiota of farmed Chinook salmon (Ciric et al. 2019). Likewise, a recent study demonstrated that Mycoplasma species was the dominant taxon in the gut microbiota of both resistant and susceptible lines of rainbow trout, although it was more abundant in the resistant line (Brown et al. 2019). It is therefore reasonable to assume that disease resistance may be associated with dietary postbiotic supplementation, which increased Mycoplasma species levels within the fish intestinal ecosystem. However, further studies are needed to explore the ability of Mycoplasma species to prevent bacterial infections.
In conclusion, the ability of a novel postbiotic from lactic acid bacteria to modify the intestinal microbiota and confer disease resistance was elucidated in this study. These findings, together with evidence from previous studies in other species (Kareem et al. 2016; Izuddin et al. 2019), suggest that dietary postbiotic supplementation may represent a suitable alternative to the use of probiotics, thereby avoiding potential risks of administering live microorganisms.
Data availability
All data are available on request from the authors.
References
Aguilar-Toalá JE, Garcia-Varela R, Garcia HS, Mata-Haro V, González-Córdova AF, Vallejo-Cordoba B, Hernández-Mendoza A (2018) Postbiotics: an evolving term within the functional foods field. Trends Food Sci Technol 75:105–114
Ang CY, Sano M, Dan S, Leelakriangsak M, Lal TM (2020) Postbiotics applications as infectious disease control agent in aquaculture. Biocontrol Sci 25:1–7
Brown RM, Wiens GD, Salinas I (2019) Analysis of the gut and gill microbiome of resistant and susceptible lines of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol 86:497–506
Cabello FC, Godfrey HP, Tomova A, Ivanova L, Dölz H, Millanao A, Buschmann AH (2013) Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol 15:1917–1942
Cabello-Olmo M, Oneca M, Torre P, Sainz N, Moreno-Aliaga MJ, Guruceaga E, Díaz JV, Encio IJ, Barajas M, Araña M (2019) A fermented food product containing lactic acid bacteria protects ZDF rats from the development of type 2 diabetes. Nutrients 11:2530
Ciric M, Waite D, Draper J, Jones JB (2019) Characterization of mid-intestinal microbiota of farmed Chinook salmon using 16S rRNA gene metabarcoding. Arch Biol Sci 71:577–587
Embregts CW, Forlenza M (2016) Oral vaccination of fish: Lessons from humans and veterinary species. Dev Comp Immunol 64:118–137
Etyemez M, Balcázar JL (2015) Bacterial community structure in the intestinal ecosystem of rainbow trout (Oncorhynchus mykiss) as revealed by pyrosequencing-based analysis of 16S rRNA genes. Res Vet Sci 100:8–11
Gibello A, Galán-Sánchez F, Blanco MM, Rodríguez-Iglesias M, Domínguez L, Fernández-Garayzábal JF (2016) The zoonotic potential of Lactococcus garvieae: an overview on microbiology, epidemiology, virulence factors and relationship with its presence in foods. Res Vet Sci 109:59–70
Izuddin WI, Loh TC, Foo HL, Samsudin AA, Humam AM (2019) Postbiotic L. plantarum RG14 improves ruminal epithelium growth, immune status and upregulates the intestinal barrier function in post-weaning lambs. Sci Rep 9:9938
Kareem KY, Loh TC, Foo HL, Akit H, Samsudin AA (2016) Effects of dietary postbiotic and inulin on growth performance, IGF1 and GHR mRNA expression, faecal microbiota and volatile fatty acids in broilers. BMC Vet Res 12:163
Meyburgh CM, Bragg RR, Boucher CE (2017) Lactococcus garvieae: an emerging bacterial pathogen of fish. Dis Aquat Org 123:67–79
Patel RM, Denning PW (2013) Therapeutic use of prebiotics, probiotics, and postbiotics to prevent necrotizing enterocolitis: what is the current evidence? Clin Perinatol 40:11–25
Pérez T, Balcázar JL, Ruiz-Zarzuela I, Halaihel N, Vendrell D, de Blas I, Múzquiz JL (2010) Host-microbiota interactions within the fish intestinal ecosystem. Mucosal Immunol 3:355–360
Pérez-Sánchez T, Balcázar JL, García Y, Halaihel N, Vendrell D, de Blas I, Merrifield DL, Ruiz-Zarzuela I (2011) Identification and characterization of lactic acid bacteria isolated from rainbow trout, Oncorhynchus mykiss (Walbaum), with inhibitory activity against Lactococcus garvieae. J Fish Dis 34:499–507
Pérez-Sánchez T, Mora-Sánchez B, Balcázar JL (2018) Biological approaches for disease control in aquaculture: advantages, limitations and challenges. Trends Microbiol 26:896–903
Ravelo C, Magariños B, Herrero MC, Costa L, Toranzo AE, Romalde JL (2006) Use of adjuvanted vaccines to lengthen the protection against lactococcosis in rainbow trout (Oncorhynchus mykiss). Aquaculture 251:153–158
Santos L, Ramos F (2018) Antimicrobial resistance in aquaculture: current knowledge and alternatives to tackle the problem. Int J Antimicrob Agents 52:135–143
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Vendrell D, Balcázar JL, Ruiz-Zarzuela I, de Blas I, Gironés O, Múzquiz JL (2006) Lactococcus garvieae in fish: a review. Comp Immunol Microbiol Infect Dis 29:177–198
Vendrell D, Balcázar JL, de Blas I, Ruiz-Zarzuela I, Gironés O, Múzquiz JL (2008) Protection of rainbow trout (Oncorhynchus mykiss) from lactococcosis by probiotic bacteria. Comp Immunol Microbiol Infect Dis 31:337–345
Zheng X, Duan Y, Dong H, Zhang J (2017) Effects of dietary Lactobacillus plantarum in different treatments on growth performance and immune gene expression of white shrimp Litopenaeus vannamei under normal condition and stress of acute low salinity. Fish Shellfish Immunol 62:195–201
Acknowledgements
B. Mora-Sánchez was supported by a fellowship from “Banco Santander – Universidad de Zaragoza”. J.L. Balcázar acknowledges the support from the Economy and Knowledge Department of the Catalan Government through Consolidated Research Group (ICRA-ENV 2017 SGR 1124). This work has been partially supported by the Spanish Ministry of Science and Innovation (AGL2014-54683-R).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Research involving human participants or animals
The study was conducted considering the 3Rs principle (reduction, replacement and refinement). The care and use of fish were performed accordingly with the Spanish Policy for Animal Protection RD53/2013, which meets the European Union Directive 2010/63 on the protection of animals used for experimental and other scientific purposes.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mora-Sánchez, B., Balcázar, J.L. & Pérez-Sánchez, T. Effect of a novel postbiotic containing lactic acid bacteria on the intestinal microbiota and disease resistance of rainbow trout (Oncorhynchus mykiss). Biotechnol Lett 42, 1957–1962 (2020). https://doi.org/10.1007/s10529-020-02919-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-020-02919-9