Abstract
Objectives
To explore the role of thioesterases in Rhodococcus opacus PD630 by endogenously overexpression in this bacteria for increased lipid production.
Results
Overexpression of four thioesterases from R. opacus PD630 in E. coli led to a 2- to 8-fold increase in C16:1 and C18:1 fatty acids while, when overexpressed in R. opacus PD630, only two recombinants had significant effect on the quantities and compositions of total fatty acid. The contents of total fatty acids (FAs) in two recombinants, pJTE2 (OPAG_00508 thioesterase) and pJTE4 (WP_012687673.1 thioesterase), were 400–460 mg/g (CDW) which is 1.5 times of wild-type strain PD630 (300-350 mg/g CDW), and 20–30 % (w/w) more than that of the control strain PDpJAM2 (330-370 mg/g CDW). The contents of 17:1 and 18:1 fatty acids increased by about 27 and 35 %, respectively, in pJTE2 and by 35 and 20 %, respectively, in pJTE4 compared with the control strain.
Conclusions
The engineered strains showed improved production of lipid (as total fatty acids), and could also tailor the composition of the fatty acid profile when cultured in mineral salts medium using glucose as sole carbon source.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Microbial lipids are increasingly being considered as sources for biodiesel or high-valued fatty acids (FAs). Major efforts have been focused on microbial production of lipids by using metabolic and genetic engineering tools. Rhodococcus opacus PD630, as a model oleaginous prokaryote, can accumulate large amount of neutral lipids up to 25 % (w/w) of its cell dry weight when cultivated in the mineral salt medium using glucose as the sole carbon source (Alvarez et al. 2008). Studies on lipid metabolism in R. opacus PD630 have focused on the synthesis of triacylglycerols (TAG). For example, R. opacus PD630 overexpressing diacylglycerol acyltransferase (DGAT) or phosphatidic acid phosphatase (PAP) had an increase of 10–15 % (w/w) in TAG content (Hernandez et al. 2013, 2015). However, the regulation of genes involved in fatty acid (FA) biosynthesis in PD630 has not been reported.
Thioesterases (TE) hydrolyze the thioester bond between a carbonyl group and a sulfur atom of acyl-ACP and play an essential role in FA biosynthesis. Based on their substrate specificities, TEs are classified into EC 3.1.2.1 to EC 3.1.2.27 as well as EC 3.1.2.- for unclassified TEs. EC 3.1.2.14 enzymes occur in both eukaryotes and bacteria and show fatty acyl–acyl carrier protein (ACP) hydrolase activity. In bacteria, the FA biosynthesis pathway occurs in the cytoplasm and is composed of a repeated cycle of reactions involving the sequential condensation of two-carbon units onto a growing fatty acyl-ACP derivatives. The successive elongation of the acyl-ACP chain can be terminated by acyl-ACP TEs (E.C. 3.1.2.14) that hydrolyze the thioester bond of acyl-ACP to release the FAs. The FAs are then converted to fatty acyl-CoAs that are further converted to TAGs and other lipids for storage within the cell.
The production and composition of FAs are determined by acyl-ACP TEs. Many acyl-ACP TEs from plants have been heterologous-expressed in Escherichia coli. For example, acyl-ACP TEs genes from Cuphea were overexpressed in E. coli to produce high content of specific chain length FAs (Joseph et al. 2013). Expression of acyl-ACP TE cDNA from Umbellularia californica in E. coli resulted in a very high accumulation of 12:0 which is a minor FA in the wild type strain (Fan et al. 2013). In addition, the overexpression of two plant TEs from Ricinus communis and Jatropha curcas in E. coli increased total FAs content from 0.2 g/l to 2 g/l (Zhang 2011). Although acyl-ACP TEs from plants have been extensively reported, little attention has been paid to bacterial TEs.
Like plant acyl-ACP TEs, bacterial acyl-ACP TEs can also increase the content of total FA and have broad substrate specificities for FAs with various chain lengths (Lee 2013). A codon-optimized acyl-ACP TE of Streptococcus pyogenes overexpressed in E. coli yielded 339 mg total FA/l, which was 1.5-fold higher than that of the control strain (Lee 2013). Overexpression of Acinetobacter baylyi TE (AcTesA) achieved 500 mg FA/l in shake-flask culture, in contrast to only 20.5 mg/l obtained in wild-type E. coli (Zheng 2012). Bacterial acyl-ACP TE may provide superior FA production compared to plant acyl-ACP TE, given that bacterial proteins can be more optimally expressed in bacterial cell than plant proteins (Jing et al. 2011). Thus, in this study, the genes of one putative acyl-ACP TE (OPAG_00508) and three E.C 3.1.2.14 enzymes (EHI47208.1, WP_005241865.1 and WP_012687673.1) from R. opacus PD630, which have not been studied before, were chosen and homologously overexpressed in PD630. The substrate-specificity of these proteins and their influence on FAs synthesis in R. opacus PD630 were studied and are reported here.
Materials and methods
Strains and medium
The sources of all bacteria are listed in Table 1. E. coli Top10 was used for plasmid copy and storage. Rhodococcus. opacus PD630 and E. coli BL21(DE3) transformed with recombinant plasmids were used as host strain for protein expression and the production of lipids. Lysogeny broth (LB) was used as seed medium to culture R. opacus strains and E. coli strains. For the production of FAs, R. opacus strains were cultivated in the mineral salts medium (MSM) as described by Schlegel et al. (1961). Glucose was used in MSM as the sole carbon source at 1 % (w/v). For nitrogen-limiting conditions, NH4Cl was decreased to 0.1 g/l (MSM0.1/1.0). E. coli strains were cultivated in M9 to produce FAs. SOC medium (20 g tryptone/l, 5 g yeast extract/l, 0.5 g NaCl/l, 2.5 mM KCl, 10 mM MgCl2, 20 mM glucose) was used for recovery of competent cells after electroporation.
Plasmid construction and transformation
Plasmids and primers used in this study are listed in Supplementary Table 1. All DNA manipulations were performed according to standard procedures. Chromosomal DNA of R. opacus PD630 was isolated and used as template for PCR. The DNA fragments of four putative TEs from R. opacus PD630 were amplified without its native start codon and stop codon using specific primers (Supplementary Table 1), and inserted into BamHI/XbaI sites of pJAM2 to generate pJAM2/TEs (pJAM2/TE1, pJAM2/TE2, pJAM2/TE3 and pJAM2/TE4) which contains an inducible acetamidase (ace) promoter and six histidine codons downstream of the XbaI site (Triccas et al. 1998). The TE gene in the constructed fusion lacked its own stop codon but contained the stop codon after the His6-tag linker sequence of pJAM2. Therefore, the amino acids SRHHHHHH occurred in the C-terminal region of the protein (Hanisch et al. 2006). To obtain pCold1/TEs, the DNA fragments of four putative TEs were inserted into BamHI/HindIII sites of pCold1.
Plasmid pJAM2 and its derivatives were introduced into R. opacus via electroporation (Shao et al. 1995; Kalscheuer et al. 1999) with minor modifications. To obtain electrocompetent cells of R. opacus, 2 ml LB pre-culture grown at 28 °C for 24 h were inoculated into 50 ml LB supplemented with 8.5 g glycine/l and 10 g sucrose/l in a 250 ml flask. Before electroporation, 200 μl competent cells were mixed with DNA and pre-incubated at 40 °C for 5 min followed by holding for 10 min on ice and then electroporated. Electroporation was performed in an electrocuvette (Bio-Rad) with gaps of 2 mm and the following settings: 2.5 kV/cm, 600 Ω and 25 μF. Time constants of 6 ms were used. Pulsed cells were immediately diluted with 800 μl SOC medium and regenerated at 28 °C for 3–4 h before they were plated on solid SOC agar plate containing kanamycin (50 μg/ml). Transformants could be scored after 3–4 days. Plasmid pCold1 and its derivatives were introduced into E. coli BL21 (DE3) by heat shock at 42 °C for 90 s. Transformants were grown overnight on LB agar plates containing ampicillin.
Cultivation conditions in shake-flasks
For preparation of the inoculum, R. opacus PD630 recombinant strains were cultured aerobically in liquid LB containing kanamycin (50 μg/ml) for 24 h at 28 °C with shaking at 200 rpm. These cultures were inoculated at 1 % (v/v) into 1 l shake-flasks with liquid MSM0.1/1.0 containing corresponding antibiotics and an appropriate concentration of acetamide (0.5 % w/v) for the induction of the acetamidase (ace) promoter of pJAM2 and its derivatives (Triccas et al. 1998). Cells were harvested and washed twice with sterile saline (0.85 % w/v NaCl) before analysis. E. coli BL21(DE3) recombinant strains were grown in LB medium containing ampicillin (100 μg/ml) for 12 h at 37 °C with shaking at 200 rpm. This was used at 1 % (v/v) to inoculate 1 l shake-flasks with liquid M9 medium. When the OD600 reached 0.5–0.6, IPTG was added at 0.1 mM to induce the expression of the pCold1. Cells were grown for additional 24 h at 16 °C and were harvested by centrifugation.
Preparation of crude cell extracts and soluble fractions, SDS-PAGE, and western blot analysis
Cells of recombinant strains were harvested by centrifugation (4000×g) for 15 min at 4 °C, washed twice with sterile saline (0.85 % w/v NaCl), and suspended in 500 μl 50 mM sodium phosphate buffer (pH 7.4). All procedures were carried out in ice to avoid changes in lipids. Subsequently, cells were lysed by ultrasonication on the ice. In order to obtain the whole cell extracts, the lysates obtained were centrifuged as described above to remove residual cells. The cytoplasmic components were obtained by centrifuging the whole cell extracts at 100,000×g at 4 °C for 30 min. Protein concentrations were determined by the Bradford method using bovine serum albumin as standard. All crude cell extracts and soluble fractions were added with loading buffer. Proteins were denatured by holding at 100 °C for 5 min and separating using 12 % (w/v) SDS-PAGE. Proteins were stained with Coomassie Brilliant Blue R-250. All western blotting was performed as described by Burnette (1981). LifeTein anti-His monoclonal antibodies (LifeTein, Chian) were used as the primary antibody at a dilution of 1:2000–4000. Goat anti-mouse horseradish peroxidase (HRP)-affinipure antibodies (LifeTein, Chian) were used as secondary antibody at a dilution of 1:10,000.
Fatty acid (FA) extraction and analytical techniques
Total FAs were extracted as described previously (Sakuradani et al. 2003): lyophilized cells were mixed with 2 ml 6 M HCl and held at 80 °C for 3 h. Tridecanoic acid (13:0) was added into the acidified samples as internal standard. Three ml chloroform/methanol (2:1, v/v) was added to extract lipid. The mixture was vortexed for 15 min and then centrifuged at 3000×g for 5 min. The chloroform phase was evaporated under N2. Samples were methylated using 1 ml 10 % (w/w) HCl/methanol at 60 °C for 3 h. The fatty acid methyl esters (FAMEs) were quantified by GC equipped with a flame ionization detector (FID). The separation of FAMEs was performed using a PB-wax column (30 m × 0.32 mm; 0.25 μm film thickness). The oven was initially held at 120 °C for 3 min, and raised with a gradient of 5 °C/min to 190 °C, then raised with a gradient of 4 °C/min to 220 °C for 2 min. N2 was used as the carrier gas. The injector and detector were held at 240 °C and 260 °C, respectively.
Results and discussion
Selection of four TEs
Based on the classification of the thioesterases from R. opacus PD630, as given on ThYme database (http://www.enzyme.cbirc.iastate.edu), the three thioesterases—EHI47208.1 (TE1), WP_005241865.1 (TE3) and WP_012687673.1 (TE4) were selected, and they were all belong to EC 3.1.2.14 family. Bacterial thioeserases in this family were acyl-ACP thioesterase(Cantu 2010. The one putative acyl-ACP TE OPAG_00508 (TE2) was not included in ThYme database, but selected through a multiple amino acid sequence alignment, which was built by aligning the sequnces of OPAG_00508 and 12 publically-available acyl-ACP TEs from other Rhodococcus species. The OPAG_00508 amino acid sequence shared similarity with its orthologs from Rhodococcus sp. JVH1 (98 % identity), Rhodococcus sp. AD45 (64 %), R. erythropolis SK121 (65 %), R. equi ATCC 33707 (56 %) and R. rhodochrous ATCC 21198 (53 %). In addition, we identified the conserved domain of these TEs—see Fig. 1. This result demonstrated that the OPAG_00508 from R. opacus PD630 probably belongs to acyl-ACP TE group.
Expression of the four TEs in E. coli BL21(DE3)
Escherichia coli BL21(DE3) was used for the expression of four TEs (TE1, TE2, TE3 and TE4) from R. opacus PD630 to identity enzyme activities. To obtain functionally active TEs, we used pCold1 as expression vector that can express proteins at low temperatures to improve the solubility of TEs in E. coli BL21(DE3). The recombinant E. coli BL21(DE3) strains were cultured at 16 °C for 24 h in induced medium, with the OD600 of all strains reaching approx 5 to 6 to make sure the consistency of growth status.
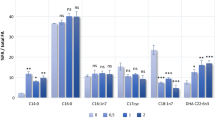
Total FA composition analysis (Fig. 2b) showed that the recombinant E. coli BL21(DE3) strains expressing TE1, TE2, TE3 and TE4 alone had considerable increases in contents of 16:1 and 18:1 by 2- to 3-fold and 4- to 8-fold, respectively, and led to a 25–50 % decrease in the content of 16:0 as compared to strain BL1. The results demonstrated that the four TE genes of R. opacus PD630 expressed in E. coli encode functional thioesterases with primary activity on 16:1 and 18:1. The TEs are therefore similar to plant FatA thioesterases that are preferentially specific for unsaturated acyl-ACPs with greatest specificity for 18:1-ACP (Serrano-Vega et al. 2005). This result suggested that TE can indirectly alter FA composition by removing feedback inhibition of acyl-ACPs (Jiang 1994). Interestingly, the unsaturated FAs—16:1, 17:1 and 18:1 occupied a large proportion of FA composition in R. opacus PD630.
a SDS-PAGE analysis of crude cell extracts and soluble fractions from the recombinant E. coli strains. Lane M protein molecular weight marker; lanes 1, 3, 5, 7, 9 whole cell proteins of BL1, BLTE1, BLTE2, BLTE3 and BLTE4; lanes 2, 4, 6, 8, 10: soluble fractions of recombinants. b The relative proportion of fatty acids (16:0, 16:1, 18:1) in recombinants
Overexpression of TEs in R. opacus PD630 strains
Recombinant R. opacus PD630 strains harbouring four reconstructed plasmids were named, respectively, pJTE1, pJTE2, pJTE3 and pJTE4 and that harbouring pJAM2 was named PDpJAM2 as the control strain (Table 1). To examine the expression of TE1, TE2, TE3 and TE4 in the recombinants, whole cell extracts and cytosol components were analyzed by SDS-PAGE and western blotting. The vector pJAM2 has been confirmed to replicate stably in R. opacus (Hetzler et al. 2013). SDS-PAGE electropheretograms of whole cell extracts and cytosol components of four recombinants are shown in Fig. 3: there were no obvious additional, visible protein bands at the expected molecular mass in the recombinants in comparison with PDpJAM2. This may due to the low plasmid copy numbers of pJAM2 which was calculated as 6 ± 1 copies per chromosome in R. opacus PD630 grown in MSM0.1/1.0. The expression of pJAM2, controlled by ace promoter from Mycobacterium smegmatis, was significantly lower in R. opacus than in M. smegmatis. However, the results of western blot analysis showed a single protein band on each track from the recombinant strains except for PDpJAM2 (Fig. 3). The EHI47208.1 protein molecular mass was the smallest one at ~25 kDa; OPAG_00508 was at ~30 kDa; the other two proteins molecular mass were similar at ~29 kDa.
SDS-PAGE and western blot analysis of crude cell extracts and soluble fractions from the recombinant R. opacus PD630 strains. Lane M, protein molecular weight marker; lane 1 whole cell extracts of the control strain; lanes 2–5 whole cell proteins of pJTE1, pJTE2, pJTE3 and pJTE4, respectively; lanes 6–9 cytosol components of four recombinants
Thus the TEs were indeed expressed as soluble intracellular proteins. The soluble products of the expressed proteins suggest that they are in an active form. This may be partly due to their low expression in the recombinant strains and also that bacterial proteins can be more optimally expressed in their own cells than in an exogenous host. Furthermore, the synthesis of FAs also occurs in the cytoplasm where the TEs can play an effective role.
Effects of acetamide concentration on cell growth and total FA accumulation
Acetamide as inducer was added into the medium to promote the expression of pJAM2 plasmid. To study the effects of acetamide concentration on the total FA accumulation, strain pJTE2 was cultured in 50 ml MSM0.1/1.0 containing acetamide from 0.25 to 1.5 % (w/v) and incubated at 28 °C, with shaking at 200 rpm for 72 h. The optimum concentration of acetamide was 0.5 % (w/v) and the content of total FA with this concentration was 45.6 % (w/w) of CDW which was 24 % higher than that in uninduced culture (Fig. 4a). The increased content of total FAs in pJTE2 cells grown with acetamide is more likely to be associated with a more active acyl-ACP TE.
a The content of total fatty acid produced by recombinant pJTE2 and the corresponding OD660 values (y = 0.5122 × g/l, y: cell dry weight, x: optical density) with various concentrations of acetamide. b Growth and c total fatty acids contents of four recombinants and the control strain. Values were mean of two independent experiments. Error bars represent the standard error of the mean
Oleaginous microorganisms cultivated in the nitrogen-limited medium accumulate more lipids than in nitrogen-rich medium, when their growth is restricted (Ratledge and Wynn 2002). The maximum cell dry weight of pJTE2 reached to 2.56 g/l when growing in MSM0.1/1.0 without acetamide, which was similar to wild-type strain PD630. However, when growing the bacteria in MSM0.1/1.0 with addition of 0.25 or 0.5 % acetamide, the cell dry weight reached to 6.81 and 6.15 g/l respectively. This result demonstrated that the addition of acetamide has a positive influence on the growth of the strains possibly by increasing the amount of available N to the cells.
Effect of TEs overexpression on the cell growth and FA accumulation in R. opacus PD630
To determine whether the overexpression of the four TEs could boost the total FA synthesis and affect the growth status of cells, we determined the total FA yield and composition as well as the cell dry weight of the control strain and recombinant strains. The engineered R. opacus PD630 strains were cultivated in MSM0.1/1.0 and induced with acetamide (0.5 %, w/v). Figure 4b shows that the growth rate of PDpJAM2 was similar to that of pJTE1, pJTE2, pJE3 and pJTE4 until the late growth phase (~72 h). This was expected as cells would not have had to divert too many resources to produce the low-expressed TEs. However, after the main growth phase, the growth rate and the final OD660 of strains pJTE2 and pJTE3 were a little higher than that of PDpJAM2. These results suggest that the overexpression of each of the four TEs did not have an inhibitory effect on the growth of engineered strains.
TEs have substrate specificities and can hydrolyze the thioester bond of oleoyl-ACP, which then regulates the FA composition of strain. Thus, the intracellular activity of TE to some degree can be confirmed by FA analysis. As shown in Fig. 5, decreases in amounts of 16:0 in pJTE2 and pJTE4 strains were more significant than that of PDpJAM2 over time and, at the same time, the contents of 17:1 and 18:1 increased by 27 and 35 %, respectively, in pJTE2, by 35 and 20 %, respectively, in pJTE4 during stationary phase. These changes were greater than seen with PDpJAM2 and wild-type R. opacus PD630 (Holder et al. 2011; Alvarez et al. 2000). This is because cells start to accumulate lipids in the stationary phase and TEs then become more effective as there is now more substrate available in cells. However, overexpression of TE2 and TE4 alone did not lead to significant change of FA composition in PD630 compared to that in E. coli. Thus, we speculate that, besides the four TEs, other genes or factors might also be involved in controlling the FA composition in PD630. Overall, the changes in FA composition suggested that TE2 has a preference for 18:1 and that TE4 is more active with 17:1.
The lipid content, expressed as total FAs, of engineered strains pJTE2 and pJTE4 reached, respectively, 41 and 38 % (w/w) of CDW at 48 h, and 46 and 44 % (w/w) of CDW, respectively, at 72 h. As shown in Fig. 4c, quantitative analyses revealed an increase of the lipids, as total FAs, in the recombinant strains pJTE2 and pJTE4 by 15–30 % and 10–25 %, respectively, in comparison to PDpJAM2 after 48 h of cultivation, while pJTE1 and pJTE3 showed no obvious changes.
In order to decouple the impact of the acetamide addition, the wild type strain was cultivated in the absence and presence of 0.5 % acetamide. We found that the wild type strain grown in the absence of acetamide can accumulate more lipids than that in the presence of acetamide, while the latter led to 2–3 times as much biomass (measured as the OD660 value) as the former (Table 2). It is suggested that the wild type strain probably uses acetamide for growth and thus the cells cannot accumulate as much lipid in this nitrogen-richer medium. In the presence of acetamide, the pJTE2 and pJTE4 had 1.5 times as much total FAs as the wild type strain. However, the PDpJAM2 also accumulated more lipids than the wild type strain. Thus, we speculate that the PDpJAM2 needs to use some acetamide for inducing the expression of the vector, and the addition of acetamide decreased lipid accumulation in wild type strain more than that in PDpJAM2.
In PD630 cells, FAs are mainly produced through FA synthase system I, and they are probably key intermediates for the biosynthesis of many of the lipids such as TAGs, phospholipids and wax esters (Holder et al. 2011). For example, the fatty acyl-CoA products originating from FAs are attached to glycerol thereby producing the lipids in PD630 (Holder et al. 2011), and a fatty acid transport protein in Rhodococcus strain RHA1 was involved in the provision of FAs to TAG metabolism (Villalba and Alvarez 2014). In addition, the accumulated acyl-ACPs would inhibit upstream enzymes in FA synthesis (Jiang 1994). Therefore, we can utilize the critical function of acyl-TEs in the alleviation of feedback inhibition of acyl-ACPs, during the FA synthesis process, to improve the pool of FAs, which may contribute to the accumulation of lipids in PD630.
Conclusion
The four TEs from R. opacus PD630 when heterologously overexpressed in E. coli BL21(DE3) led to considerable changes in FA composition. When the four TEs were homologously overexpressed in R. opacus PD630, the putative acyl-ACP TE2 (OPAG_00508) and TE4 (WP_012687673.1) led to significant increases in lipid accumulation and evident changes in FA composition. Thus, the two putative acyl-ACP TEs contribute to the production of total FAs and also have a specific influence on the FA composition in R. opacus PD630. However, the TE1 (EHI47208.1) and TE3 (WP_005241865.1) did not led to obvious changes in lipid content and FAs composition.
References
Alvarez HM, Kalscheuer R, Steinbuchel A (2000) Accumulation and mobilization of storage lipids by Rhodococcus opacus PD630 and Rhodococcus ruber NCIMB 40126. Appl Microbiol Biotechnol 54:218–223
Alvarez AF, Alvarez HM, Kalscheuer R, Waeltermann M, Steinbuechel A (2008) Cloning and characterization of a gene involved in triacylglycerol biosynthesis and identification of additional homologous genes in the oleaginous bacterium Rhodococcus opacus PD630. Microbiology 154:2327–2335
Burnette WN (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anim Biochem 112:195–203
Cantu CD, Chen Y, Reilly JP (2010) Thioesterases: a new perspective based on their primary and tertiary structures. Prot Sci 19:1281–1295
Fan L, Liu J, Nie K, Liu L, Wang F, Tan T, Deng L (2013) Synthesis of medium chain length fatty acid ethyl esters in engineered Escherichia coli using endogenously produced medium chain fatty acids. Enz Microb Technol 53:128–133
Hanisch J, Waltermann M, Robenek H, Steinbüchel A (2006) The Ralstonia eutropha H16 phasin PhaP1 is targeted to intracellular triacylglycerol inclusions in Rhodococcus opacus PD630 and Mycobacterium smegmatis mc2155, and provides an anchor to target other proteins. Microbiology 152:3271–3280
Hernandez MA, Arabolaza A, Rodriguez E, Gramajo H, Alvarez HM (2013) The atf2 gene is involved in triacylglycerol biosynthesis and accumulation in the oleaginous Rhodococcus opacus PD630. Appl Microbiol Biotechnol 97:2119–2130
Hernandez MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM (2015) Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol 99:2191–2207
Hetzler S, Broeker D, Steinbüchel A (2013) Saccharification of cellulose by recombinant Rhodococcus opacus PD630 strains. Appl Environ Microbiol 79:5159–5166
Holder JW, Ulrich JC, Debono AC et al (2011) Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet 7(9):e1002219
Jiang P, CronanJr JE (1994) Inhibition of fatty acid synthesis in Escherichia coli in the absence of phospholipid synthesis and release of inhibition by thioesterase action. Bacteriol 176:2814–2821
Jing F, David CC, Jarmila T, Jay PC, Basil JN, Marna DY, Peter JR (2011) Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem 12:44–59
Kalscheuer R, Arenskötter M, Steinbüchel A (1999) Establishment of a gene transfer system for Rhodococcus opacus PD630 based on electroporation and its application for recombinant biosynthesis of poly-3-hydroxyalkanoic acids. Appl Microbiol Biotechnol 52:508–515
Lee S, Park S, Lee J (2013) Improvement of fatty acid production in Escherichia coli using codon-optimized Streptococcus pyogenes acyl-ACP thioesterase. Bioprocess Biosyst Eng 36(10):1519–1525
Ratledge C, Wynn JP (2002) The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol 51:1–51
Sakuradani E, Shimizu S (2003) Gene cloning and functional analysis of a second delta 6-fatty acid desaturase from an arachidonic acid-producing Mortierella fungus. Biosci Biotechnol Biochem 67(4):704–711
Schlegel HG, Kaltwasser H, Gottschalk G (1961) Ein Submersverfahren zur Kultur wasserstoffoxydierender Bakterien: wachstumsphysiologische Untersuchungen. Arch Mikrobiol 38:209–222
Serrano-Vega MJ, Garces R, Martinez-Force E (2005) Cloning, characterization and structural model of a FatA-type thioesterase from sunflower seeds. Planta 221:868–880
Shao Z, Dick WA, Behki RM (1995) An improved Escherichia coli-Rhodococcus shuttle vector and plasmid transformation in Rhodococcus spp. using electroporation. Lett Appl Microbiol 21:261–266
Torella JP, Ford TJ, Kim SN, Chen AM, Way JC, Silver PA (2013) Tailored fatty acid synthesis via dynamic control of fatty acid elongation. Proc Natl Acad Sci 110:11290–11295
Triccas JA, Parish T, Britton WJ, Giquel B (1998) An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett 167:151–156
Villalba MS, Alvarez HM (2014) Identification of a novel ATP-binding cassette transporter involved in long-chain fatty acid import and its role in triacylglycerol accumulation in Rhodococcus jostii RHA1. Microbiology 160:1523–1532
Zhang X, Li A, Agrawal A, San KY (2011) Efficient free acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab Eng 13:713–722
Zheng Y, Li L, Liu Q et al (2012) Boosting the fatty acid synthesis of Escherichia coli by expression of a cytosolic Acinetobacter baylyi thioesterase. Biotechnol Biofuel 5:76–88
Acknowledgments
This study was funded by the National Natural Science Foundation of China (31271812), the National High Technology Research and Development Program of China (2012AA022105C), and the Strategic Merieux Research Grant.
Supporting Information
Supplementary Table 1—List of plasmids and primers used.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical statement
This article does not contain any studies with human participants or animals performed by any of the authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, L., Zhao, L., Zan, X. et al. Boosting fatty acid synthesis in Rhodococcus opacus PD630 by overexpression of autologous thioesterases. Biotechnol Lett 38, 999–1008 (2016). https://doi.org/10.1007/s10529-016-2072-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-016-2072-9