Abstract
Objectives
Two genes encoding two acetyl-CoA synthetase (ACS) isoenzymes have been identified in the marine yeast Rhodosporidium diobovatum MCCC 2A00023.
Results
ACS1 encoded a polypeptide with a sequence of 578 amino acid residues, a predicted molecular weight of 63.73 kDa, and pI of 8.14, while the ACS2 encoded a polypeptide containing 676 amino acid residues with a deduced molecular mass of 75.61 kDa and a pI of 5.95. Biological activity of Acs1p and Acs2p was confirmed by heterologous expression in Escherichia coli. A 1.5-kb DNA fragment of the ACS1 gene and a 2.7-kb DNA fragment of the ACS2 gene were deleted using the RNA guide CRISPR-Cas9 system. The strain lacking ACS1 was unable to grow on acetate and ethanol media, while the ACS2 deletant was unable to grow on glucose medium. ACS1-ACS2 double mutants of R. diobovatum were non-viable.
Conclusions
ACS isoenzymes are essential to the yeast metabolism, and other sources of ACSs cannot compensate for the lack of ACSs encoded by the two genes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Acetyl-CoA is a crucial metabolite involved both in the intermediary carbon and energy metabolism and in biosynthetic pathways (Wolfe 2005). Its main function is to transport carbon atoms within the acetyl group to the tricarboxylic acid (TCA) cycle to be oxidized for energy production it is also used for the synthesis of C4 metabolites via the glyoxylate shunt (Chen et al. 2012) and for fatty acid biosynthesis. Notable examples of industrially-interesting products that use acetyl-CoA as a precursor include butanol (Krivoruchko et al. 2013), isoprenoids (Lian et al. 2014), and α-santalene (Chen et al. 2012).
In yeasts, acetyl-CoA can be produced by at least three major pathways. In the first pathway, acetyl-CoA is generated by direct oxidative decarboxylation of pyruvate, catalysed by the pyruvate dehydrogenase (PDH) complex, and it serves as substrate for the TCA cycle (Starai and Escalante-Semerena 2014). In the second, acetyl-CoA is the end-product of fatty acid degradation by the beta-oxidation pathway (Hiltunen et al. 2003). In the third, acetyl-CoA is synthesized via the activation of acetate in an ATP-dependent reaction (De Jong-Gubbels et al. 1997). Acetyl-CoA synthetase (ACS) (EC 6.2.1.1) is the predominant enzyme for the activation of acetate to acetyl-CoA. Two different forms of ACS, Acs1p and Acs2p, are found in yeast, depending on the conditions under which the cells are grown. The two isoenzymes differed with respect to localization, regulation, and kinetic properties, suggesting their different roles in metabolism (van den Berg et al. 1996).
The type II bacterial clustered regularly interspaced short palindromic repeats (CRISPR) and its associated protein (CRISPR-Cas9) systems are robust tools for facilitating genome alteration with high precision (Ran et al. 2013). Here, we identified two ACS genes from the basidiomycetous yeast Rhodosporidium diobovatum, which was isolated from a marine environment, which is characterized by tolerance to low temperature, high hydrostatic pressure, and a high salt concentration (Margesin and Miteva 2011). Compared to enzymes secreted by terrestrial microorganisms, enzymes produced by marine microbes have some unique properties with specialized applications (Brizzio et al. 2007). This has attracted the attention of researchers worldwide.
Materials and methods
Materials and strains
Rhodosporidium diobovatum MCCC 2A00023 was purchased from Marine Culture Collection of China (MCCC). Plasmids pMD19-T, pET-28a+ and pCRCT were obtained from Takara, Novagen and Addgene, respectively. Restriction enzymes, T4 DNA ligase, strains E. coli DH5α and E. coli BL21 (DE3) were obtained from Takara.
Isolation of genomic DNA and RNA
Rhodosporidium diobovatum was grown in YPD medium at 20 °C to extract genomic DNA and total RNA. Total RNA was isolated using TRIzol.
ACS cDNA and DNA cloning
The cDNA was synthesized through reverse transcriptional PCR (RT-PCR). The 5′- and 3′-ends of ACS1 and ACS2 cDNA sequences were obtained by 5′-RACE and 3′-RACE using the BD SMART RACE cDNA amplification kit. The primes used are listed in Supplementary Table 1.
Sequence analysis
Sequences were analyzed using various softwares. The ProtParam tool was used to calculate the amino acid composition, molecular weight, isoelectric point, instability index and aliphatic index. The conserved domain of ACS proteins were analyzed by NCBI Batch Web CD-search tool. The phylogenetic evolutionary analysis were constructed using Molecular Evolutionary Genetics Analysis version 6.06 software.
Expression of recombinant ACSs
The cDNA of ACSs were amplified by PCR and restriction sites XbaI, HindIII, XbaI and NheI were introduced at the 5′-end by primers SP1f (5′-GCTCTAGAGCATGGCCTCCT CCGAG-3′) and SP1r (5′-CCCAAGCTTGGGCTACAGCTTCGAC-3′), SP2f (5′-GCTCTAGAGCATGGCACCGACGTCG-3′) and SP2r (5′-CTAGCTAGCTAGCTAAAGCTTTGCC-3′). The products were inserted into the protein expression vector pET-28a+ (Novagen), and transformed into E. coli BL21 (DE3) competent cells with a Bio-Rad Gene Pulser. The positive recombinant bacteria were cultured in LB medium with 50 μg kananycin ml−1 at 37 °C. The expresssion of recombinant proteins were induced with IPTG for 4 h. Cells were harvested, washed and resuspended in 100 mM potassium phosphate buffer (pH 7.5), 2 mM MgCl2, and 1 mM dithiothreitol. Cells were disrupted by sonication and the cell lysate was centrifuged at 20,000×g for 15 min at 4 °C.
ACS activity assay
Enzyme activity was measured using a coupled assay with malate dehydrogenase and citrate synthase according to Van den Berg et al. (1996). Under assay conditions, malate dehydrogenase (Mdh) and citrate synthase (Cs) activities were in excess, and the rate of NADH formation was limited by ACS activity. The experiments were done in a triplicate. Enzyme activities were calculated assuming an extinction coefficient of NADH of 6.3 ml μM−1 cm−1. One unit (U) was defined as the amount of enzyme catalyzing the formation of 1 μM NADH per min.
Deletion of ACS genes
All mutants were constructed in haploid R. diobovatum (Ura3△) using the CRISPR-Cas9 system. The 20-nt guide RNA sequences, together with the NGG PAM sequence, were searched for on both strands of the ACS genes. These unique sequences were selected to minimize off-target effects. The guide RNA (gRNA) expression constructs, a 114-nt array containing two 20-nt guide RNA sequences and a 226-nt array containing four 20-nt guide RNA sequences, were synthesized and cloned into the pCRCT vector for co-expression with Cas9 (Bao et al. 2015). Plasmids were transformed into R. diobovatum competent cells with a Bio-Rad Gene Pulser. The positive recombinant yeast were cultured in 2 ml synthetic complete (SC) media, and the appropriate mutants were confirmed by PCR.
Results
Identification of ACS genes
Degenerate primers were designed to amplify the core sequences of the encoding gene. The sequence analysis showed that the degenerate PCR products were indeed portions of the ACS genes. The nucleotide sequence obtained by 5′-RACE had the starting codon, ATG, and the one obtained by 3′-RACE had a TGA translation stop codon. Based on these sequences, gene-specific primers were designed for the full-length cDNA and genomic DNA amplification. When the PCR reaction was carried out with the obtained cDNA as the template, the full-length cDNA sequences of ACS1 and ACS2 were obtained severally and were 1737 and 2031 bp long, respectively (Fig. 1a). When the total genomic DNA was used as the template, the genomic DNA of the two ACS isoenzymes were obtained separately and were 2302 and 2922 bp in length, respectively (Fig. 1b). The cDNA sequences and deduced amino acids of Acs1p and Acs2p reported in this paper are available in the GenBank databases under the Accession number KU097683 and KU097684 in the order mentioned. Comparison of ACSs with regard to genomic DNA and cDNA revealed that ACS1 genomic DNA carried seven exons which were separated by six introns (Supplementary Fig. 1a), while ACS2 genomic DNA contained eleven introns (Supplementary Fig. 1b). All of the splice sites of the intron–exon junction were GT-AG, which was the canonical splice site.
Analysis of ACS genes
Both of the corresponding full-length cDNA sequences of the two ACS genes contained a complete ORF. Translation of the ACS1 full length cDNA produced a polypeptide of 578 amino acid residues, with a deduced molecular mass of 63.73 kDa, and a theoretical isoelectric point (pI) of 8.14, while the ACS2 cDNA encoded a polypeptide composed of 676 amino acid residues in length with a calculated molecular weight of 75.61 kDa and a pI of 5.95. The instability index of Acs1p was computed to be 21.28, which indicated that the protein was stable. However, the Acs2p protein, whose instability index was 40.96, was classified as unstable. The aliphatic indices of Acs1p and Acs2p were 77.6 and 87.9, respectively, which demonstrated that both were fat-soluble proteins. The two ACS isoenzymes shared 12.7 % of sequence identity (Fig. 2), which was significantly lower than their similarity in Aspergillus nidulans (62.4 %), Alcaligenes eutrophus (45.4 %), E. coli (48.7 %), Methanothrix soehngenii (44.6 %), Neurospora crassa (59.8 %), Penicillium chrysogenum (61.2 %), and Saccharomyces cerevisiae (61.2 %). Analysis of the existing sequence data from the NCBI database showed that they belong to the AMP-binding family (pfam00501), and it showed strict conservation of the residues in the important sites. The enzymes adopt a conserved structural fold with a large N-terminal and a small C-terminal domain, with the active site lying in the interface of the two domains. Both Acs1p and Acs2p had two transmembrane helices, and none of them had N-terminal sequences that conform to the physicochemical properties of signal peptides or mitochondrial transit peptides. Their C-termini contained the AMP-binding enzyme C-terminal domain. In Acs1p, the 199-210 fragment (VFFTSGTTGAPK) was identified as a putative AMP-binding domain signature. The Ser-Lys-Leu signal in the C-terminus of Acs1p is responsible for directing protein import into peroxisomes. In Acs2p, the 299-310 fragment (ILYSSGTTGKPK) was proposed as a putative AMP-binding domain signature, and this consensus motif (PS00455), [LIVMFY]-{E}-{VES}-[STG]-[STAG]-G-[ST]-[STEI]-[SG]-x-[PASLIVM]-[KR], which is absolutely necessary for the binding of ATP and for adenylate formation, was found in Acs2p. Acs2p harboured a peroxisomal targeting signal, Ala-Lys-Leu, in the C-terminus. A phylogenetic profiling analysis was performed to compare the R. diobovatum ACS protein sequences to other fungi homologs (Fig. 3a, b). The tree indicated that R. diobovatum ACSs were more homologous to the proteins of Basidiomycota than to those of Ascomycota. These were consistent with the currently accepted phylogenies.
Phylogenetic tree of Acs1p (a) and Acs2p (b) revealing evolutionary relationships. Phylogenetic tree was constructed based on the ACS amino acid sequences from different microorganisms using neighbor-joining analysis with 1000 bootstrap replications. The horizontal bar indicates the relative distance in the phylogenetic tree
Functional analysis of R. diobovatum ACSs
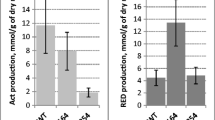
Rhodosporidium diobovatum Acs1p and Acs2p were expressed in E. coli BL21 (DE3). To confirm that the expressed proteins were biologically active, the activity of ACS proteins was determined using the ACS assay (see Methods). Acs1p activity was 0.27 ± 0.06 U mg−1, while Acs2p activity was 0.34 ± 0.1 U mg−1. From these results, we concluded that both the ACS1 and the ACS2 gene cloned in this study encoded functional ACS proteins.
Disruption of ACS1 and ACS2 genes from R. diobovatum chromosomes
The CRISPR-Cas9 system can facilitate precise editing of endogenous genomic loci by simply specifying a 20-nt targeting sequence within its gRNA (Ran et al. 2013). Cas9 promotes genome editing by stimulating a double-strand break (DSB) at a target genomic locus. The gRNA1 and gRNA2 were designed to produce an ACS1 deletant, and a 1.5 kb DNA fragment of the coding sequence for Acs1p was removed (Fig. 4). The gRNA3 and gRNA4 were designed to produce an ACS2 deletant, and a 2.7-kb DNA fragment containing part of the ACS2 gene was deleted (Fig. 4). A plasmid containing four gRNAs was then used to construct the double ACS1 and ACS2 deletion mutants. The ACS1 disruption mutant was unable to grow on acetate or ethanol medium, but its growth on glucose as the sole source of carbon did not differ from that of the wild type. The ACS2 disruption strains grew normally on ethanol or acetate as the sole sources of carbon, but were unable to grow on glucose medium. Disruption of both ACS genes was lethal regardless of carbon source, indicating that ACS isoenzymes are essential for the viability of R. diobovatum.
Agarose-gel electrophoresis of ACS DNA fragments disruption mutant selection of R. diobovatum MCCC 2A00023. M DNA marker (CoWin, China), 1 PCR production of R. diobovatum WT strains, 2 PCR production of R. diobovatum ACS1 disruption mutants, 3 PCR production of R. diobovatum WT strains, 4 PCR production of R. diobovatum ACS2 disruption mutant, 5 PCR production of R. diobovatum double ACS1 and ACS2 disruption mutants
Discussion
Acetyl-CoA acts as a key precursor metabolite in the synthesis of many industrially relevant compounds, such as fatty acids, carotenoids, isoprenoids, vitamins, amino acids, lipids, wax esters, polysaccharides, polyhydroxyalkanoates, sterols, polyketides, acetate esters (such as ethyl acetate and isoamyl acetate), as well as 1-butanol. Our results showed that R. diobovatum had two structural genes, ACS1 and ACS2, each encoding an active ACS isoenzyme that differs from the other in substrate specificity and regulatory characteristics and has a different physiological function. The acetyl-CoA-forming pathways vary with the carbon source, and the need for ACS activity during growth on media with different carbon sources can be met by either of the two ACS isoenzymes (De Jong-Gubbels et al. 1997). Acs1p catalyses the direct formation of acetyl-CoA from acetate, ATP, and CoA (Starai and Escalante-Semerena 2014), and it is essential for the utilization of acetate and ethanol when used as sole carbon sources. When grown on a glucose medium, the ACS2 mutant is not viable. This may be due to glucose catabolite repression and inactivation of ACS1 and its gene product (van den Berg et al. 1996). Other sources of ACSs cannot compensate for the lack of ACSs encoded by the two genes. The explanation may lie in the subcellular compartmentation of the ACS metabolism (De Jong-Gubbels et al. 1997) and appropriate concentrations of ACS must be maintained in the relevant cellular compartment for normal growth and development.
The value of the discovery of two ACS isoenzymes in the marine yeast R. diobovatum is far greater than if it were discovered in terrestrial organisms. Organisms living in marine environments have evolved features to surmount the negative effects of these poor environments. However, it is less clear what specific mechanism regulates the expression of the ACS genes in challenging natural environments. An important issue for future studies is to determine the detailed mechanisms of response to the oligotrophic living environment.
References
Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H (2015) Homology-integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol 4:585–954
Brizzio S, Turchetti B, de García V, Libkind D, Buzzini P, van Broock M (2007) Extracellular enzymatic activities of basidiomycetous yeasts isolated from glacial and subglacial waters of northwest Patagonia (Argentina). Can J Microbiol 53:519–525
Chen Y, Siewers V, Nielsen J (2012) Profiling of cytosolic and peroxisomal acetyl-CoA metabolism in Saccharomyces cerevisiae. PLoS ONE 7:e42475
De Jong-Gubbels P, Van den Berg MA, Steensma HY, van Dijken JP, Pronk JT (1997) The Saccharomyces cerevisiae acetyl-coenzyme A synthetase encoded by the ACS1 gene, but not the ACS2-encoded enzyme, is subject to glucose catabolite inactivation. FEMS Microbiol Lett 153:75–81
Hiltunen JK, Mursula AM, Rottensteiner H, Wierenga RK, Kastaniotis AJ, Gurvitz A (2003) The biochemistry of peroxisomal beta-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 27:35–64
Krivoruchko A, Serrano-Amatriain C, Chen Y, Siewers V, Nielsen J (2013) Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J Ind Microbiol Biotechnol 40:1051–1056
Lian J, Si T, Nair NU, Zhao H (2014) Design and construction of acetyl-CoA overproducing Saccharomyces cerevisiae strains. Metab Eng 24:139–149
Margesin R, Miteva V (2011) Diversity and ecology of psychrophilic microorganisms. Res Microbiol 162:346–361
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Starai VJ, Escalante-Semerena JC (2004) Acetyl-coenzyme A synthetase (AMP forming). Cell Mol Life Sci 61:2020–2030
Strijbis K, Distel B (2010) Intracellular acetyl unit transport in fungal carbon metabolism. Eukaryot Cell 9:1809–1815
van den Berg MA, de Jong-Gubbels P, Kortland CJ, van Dijken JP, Pronk JT, Steensma HY (1996) The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem 271:28953–28959
Wolfe AJ (2005) The acetate switch. Microbiol Mol Biol Rev 69:12–50
Acknowledgments
This work was supported by the Science Research Plan of Hebei Higher Schools (No. Z2010225) and open fund of Key laboratory (No. 3333112).
Supporting information
Supplementary Table 1—Primer sequences used in this study.
Supplementary Fig. 1—(a) Comparison of the nucleotide sequences of R. diobovatum MCCC 2A00023 ACS1 genomic DNA and cDNA. (b) Comparison of the nucleotide sequences of R. diobovatum MCCC 2A00023 ACS2 genomic DNA and cDNA.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Liu, Y., Zhang, M., Wang, T. et al. Two acetyl-CoA synthetase isoenzymes are encoded by distinct genes in marine yeast Rhodosporidium diobovatum . Biotechnol Lett 38, 417–423 (2016). https://doi.org/10.1007/s10529-015-2006-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-015-2006-y