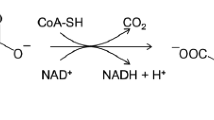
Abstract
Purpose of work
A pair of NAD+- and NADP+-dependent group III-alcohol dehydrogenases was characterized from the enterobacterium, Dickeya zeae, to expand our understanding of the distribution and biochemical properties of this interesting group of enzymes.
Two putative group III-alcohol dehydrogenases (ADHs) were identified in the genome of Dickeya zeae. Amino acid alignments and phylogenetic analysis revealed that Adh3.1 and Adh3.2 are only distantly related (~25 % identity at the protein level). Both proteins were purified to homogeneity after heterologous expression in E. coli. A specific activity of 1.8 U/mg was measured for the NAD+-dependent enzyme Adh3.1 with ethanol used as substrate, while NADPH-dependent Adh3.2 preferred butanal (29.1 U/mg) as substrate. Maximum activity for Adh3.1 was at 50 °C and pH 10 and for Adh3.2 at 70 °C and pH 6. Cell viability assays were used to confirm activity towards butanal and glyoxals. Biochemical characterization and phylogenetic analyses led to the hypothesis that Adh3.1 and Adh3.2 are probably the result of an ancient gene duplication event followed by functional diversification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alcohol dehydrogenases (ADHs) are well-known examples for functional convergent evolution. These enzymes are structurally distinct and show the ability to catalyze similar enzymatic reactions, namely the interconversion of alcohols, aldehydes and ketones. Due to their substrate promiscuity, ADHs are attractive for versatile industrial applications. Beside several NAD(P)+-independent ADHs, three families of non-homologous NAD(P)+-dependent ADHs are currently accepted (Hernandez-Tobias et al. 2011). Insect-type ADHs contain short peptide chains and do not incorporate a metal ion in their catalytic region. Horse liver ADH (HLADH) is the best-studied long-chain zinc-dependent ADH (Hernandez-Tobias et al. 2011; Quaglia et al. 2012). Based on amino acid sequence comparisons of metal-ion containing ADHs from thermophilic and mesophilic microorganisms, it was shown that group III-ADHs have risen evolutionarily independently from a common ancestor and are not related to group I- or group II-ADHs, respectively (Radianingtyas and Wright 2003).
As a result of functional diversification, group III metal-containing ADHs can be further divided into several subgroups with distinct substrate specificities, including glycerol dehydratases, propanediol oxidoreductases, lactaldehyde dehydrogenases and others. Due to their ability to process the valuable 1,3-propanediol (1,3-PD), selected members of two subgroups were investigated in detail. The physiological role of DhaT in Klebsiella pneumoniae is the recycling of NAD+, which is used as co-factor by glycerol dehydrogenase in the oxidative route to produce dihydroxyacetone phosphate, while yqhD from Escherichia coli has been shown to encode a distantly related unspecific NADPH-dependent aldehyde reductase that is important for the detoxification of harmful aldehydes (Perez et al. 2008; Marcal et al. 2009; Lee et al. 2010; Ma et al. 2010). Nevertheless, both enzymes share the ability to convert 3-hydroxypropionaldehyde into 1,3-PD and are therefore of tremendous interest for applications in biotechnology.
Two distantly related group III-ADHs from the phytopathogenic enterobacterium Dickeya zeae were compared in this study. Adh3.1 is a NAD-dependent enzyme preferring short-chain alcohols, while Adh3.2 is a NADPH-dependent aldehyde reductase with activity on butanal and glyoxals. Evolutionary aspects on the distribution in bacteria as well as biochemical properties are reported.
Materials and methods
Strains and culture conditions
Dickeya zeae DSM18068 was obtained from DSMZ (Braunschweig, Germany) and was grown in DSMZ Medium 535. E. coli M15[pREP4] in combination with plasmid pQE-30 (Qiagen) was used as heterologous expression host. E. coli was grown at 37 °C either in Luria-Bertani (LB) medium or in liquid fed-batch fermentation medium according to Horn et al. (1996).
Gene cloning procedures
Genomic DNA isolated from D. zeae DSM18068 was used as template to amplify ADH-encoding genes adh3.1 and adh3.2 by PCR. Primer pairs were as follows: DzDhaT-BamHI-f/DzDhaT-PstI-r (5′-GGATCCAGCAGTGCATTTTACATTCCCGCC and 5′-CTGCAGTTAGAACGCGGCGGCAAAAATTCCG, restriction sites are underlined) and DzYqhD-BamHI-f/DzYqhD-PstI-r (5′-GGATCCCAGAACTTTACGCTTCATACCCCG and 5′CTGCAGTTAGCGGGCGGCTTCGTACACGCG). The following sequence was used: 98 °C for 2 min and 35 cycles of 98 °C for 10 s, 60 °C for 20 s and 72 °C for 30 s, and final elongation at 72 °C for 7 min. After subcloning and sequencing, BamHI/PstI digestion fragments were ligated into linearized vector pQE-30 resulting in plasmids pQE30:adh3.1 and pQE30:adh3.2.
Gene expression and purification of proteins
E. coli M15[pREP4] containing plasmid pQE30:adh3.1 or pQE30:adh3.2 was grown in liquid medium (Horn et al. 1996) in a 1.2 l fed-batch fermentation. Expression of ADH-encoding genes was induced with 2 mM IPTG and cells were harvested by centrifugation after 2 h growth. Cells (5 g wet wt) were resuspended in lysis buffer (50 mM NaH2PO4 buffer containing 300 mM NaCl and 10 mM imidazole, pH 8.0) and disrupted by Spin Aminco French press (Spectronic Instruments). Purification was achieved in a two-step approach with the ÄKTApurifier system (GE Healthcare): (1) Ni-NTA affinity chromatography with imidazole as elution reagent, followed by (2) size exclusion gel filtration using 50 mM NaH2PO4 buffer containing 150 mM NaCl, pH 7.2. Calibration of the size exclusion gel filtration column to determine molecular weights of ADHs was achieved with the following standard proteins according to the information supplied by the manufacturer: ferritin: 440 kDa, aldolase: 158 kDa, conalbumin: 75 kDa and ovalbumin: 44 kDa. Active fractions were pooled, dialyzed against 20 mM Tris buffer (pH 7.0) and stored at 4 °C. SDS-PAGE and Western blotting analysis (His-Tag Monoclonal Antibody, Novagen) were used to visualize the purity of the recombinant ADHs.
Enzyme assays
Enzyme activities were determined by measuring the reduction of NAD(P)+ or the oxidation of NAD(P)H at 340 nm in a spectrophotometer equipped with a Peltier effect-controlled temperature cuvette holder. All experiments were performed in triplicate. Standard assays were performed under optimal pH and temperature conditions (Adh3.1: pH 10.0; 50 °C and Adh3.2: pH 6.0; 70 °C) and substrates were tested at 10 mM, cofactor concentrations were 0.8 mM NAD+/NADH for Adh3.1 and 1 mM NADP/NADPH for Adh3.2. One unit of ADH activity is defined as the amount of enzyme needed to catalyze the reduction or formation of 1 μmol NAD(P)+ per min under standard conditions. In addition, aldehyde reductase activities were indirectly measured in a cell viability assay developed by Lee and Park (2012).
Sequence accession number
The adh3.1 and adh3.2 nucleotide sequences of D. zeae strain DSM18068 were deposited under the accession numbers HF546061 and HF546062 in the EMBL database.
Results
Sequence analysis and phylogenetic relationships of alcohol dehydrogenases
To investigate multiple members of group III-ADHs from a single microorganism, BlastP-analyses were performed using amino acid sequences of characterized alcohol dehydrogenases, 1,3-propanediol oxidoreductases and aldehyde reductases form various bacterial species (Altschul et al. 1990). Two diverse genes encoding putative group III-ADHs were identified within the sequenced genome of D. zeae Ech1591 and were amplified by PCR from genomic DNA of D. zeae DSM18068. The 1,158 bp ORF adh3.1 encodes a protein of 385 amino acids, while the protein Adh3.2 is composed of 387 amino acids and encoded by adh3.2 (1,164 bp). The amino acid identity between both proteins is rather low with 24.9 % in a 349 amino acid overlap. InterProScan assigned both proteins to the group of iron-type ADHs (Quevillon et al. 2005). Phylogenetic analysis also separated both proteins from each other (Fig. 1). Moreover, the unrooted tree clearly opposed group III-ADHs to prototypes of group I or group II, respectively. Adh3.1 belongs to group III-ADHs with sequence identity to proteins involved in lactaldehyde metabolism and production of 1,2-propanediol and 1,3-propanediol. DhaT (49.3 % identity to Adh3.1) from K. pneumoniae and FucO (42.4 % identity to Adh3.1) from E. coli are well-investigated members of this subgroup. However, Adh3.2 belongs to the subgroup of putative butanol dehydrogenases within the group III-ADHs with E. coli YqhD being the characterized prototype. Adh3.2 shares 77.3 % identity with YqhD.
Unrooted phylogenetic tree separating YqhD and DhaT homologues. The phylogenetic relationship is based on an alignment of complete genes from group I-, II- and III-ADHs (Figure S1). Abbreviations and accession numbers are as follows: Clostridium pasteurianum (CpDhaT: AF006034), Dickeya zeae (Adh3.1: HF546061 and Adh3.2: HF546062), Drosophila melanogaster (DmADH: CAA77330.1), Escherichia coli (EcFucO: AAA23825.1 and EcYqhD: NP_417484.1), Granulicatella adiacens (GaYqhD: ZP_05737046.1), Klebsiella pneumoniae (KpDhaT: YP_005956552.1 and KpYqhD: ABR78827.1), Oenococcus oeni (OoAdh3: HE974350), Pectobacterium atrosepticum (Pa12PDO: YP_048854.1 and PaYqhD: HF546063), Zymomonas mobilis (zmADH2: BAF76066.1), and horse liver alcohol dehydrogenase (HLADH: AAB26666). Group I-ADH HLADH and group II-ADH from D. melanogaster were used to illustrate the three groups of ADH
Expression and purification of recombinant Adh3.1 and Adh3.2
Both putative group III-ADHs from D. zeae were cloned into pQE30 expression vector and the genes were expressed in E. coli M15[pREP4]. Proteins were purified in a 2-step purification approach and visualized on SDS-PAGE and by western blotting analyses (Fig. 2). Expression of adh3.1 and adh3.2 gave a single band of ~44–41 kDa, which is in good agreement with the joined molecular masses of proteins (Adh3.1: 43.4 kDa and Adh3.2: 41.3 kDa) and RGS-6xHis-tags (1.1 kDa). Enzymatic activities of 1.8 U/mg (Adh3.1) and 29.1 U/mg (Adh3.2) were obtained from purified enzymes after size exclusion chromatography with ethanol or butanal, respectively (Table 1). The calculated molecular weights from native size exclusion chromatography are 70.5 ± 2.8 kDa (Adh3.1) and 68.4 ± 0.7 kDa (Adh3.2), indicating that both proteins form dimeric structures (Fig. S2).
Expression of D. zeae adh3.1 and adh3.2 genes in E. coli. SDS-PAGE and western blotting analysis (anti-His antibody) of E. coli protein extracts harbouring plasmid pQE30:adh3.1 (a) or pQE30:adh3.2 (b), respectively. Cells were induced with 2 mM IPTG (+IPTG) or not induced (−IPTG) and were grown for 2 h at 37 °C. Total cellular proteins (TCP) were resolved by SDS-PAGE and visualized by Coomassie Blue staining. Cells were disrupted and the insoluble cell debris (Pe Pellet fraction) was separated by centrifugation from the soluble proteins (SN supernatant). His-tagged proteins were purified by affinity chromatography (Ni-NTA) and size exclusion chromatography (SEC)
Enzymatic properties of alcohol dehydrogenases
Enzyme specificities are given in Table 2. Activity measurements revealed that Adh3.1 is active on a broad range of short-chain alcohols with NAD+ being an appropriate co-factor, while Adh3.2 is an NADPH-dependent aldehyde reductase with specific activity towards butanal and 2-oxoaldehydes. Adh3.1 showed optimal activity at 50 °C and pH 10.0, while Adh3.2 is optimally active at 70 °C and pH 6.0 (Fig. 3). To study the kinetic constants of the enzymes with their preferred substrates, non-linear regression analysis of the corresponding Michaelis-Menten curves was performed (Table 3). Sensitivity to butanal and resistance to glyoxal and methylglyoxal of E. coli producing Adh3.1 and Adh3.2 was further tested in plate assays. Expression of adh3.2 completely blocked growth of E. coli on plates containing 20 mM butanal, indicating the production of toxic butanol, whereas expression of adh3.1 allowed the cells to survive (Fig. 4). E. coli expressing either adh3.1 or adh3.2, respectively, exhibited a higher resistance to methylglyoxal (Fig. 5a), and the production of Adh3.2 led to a decomposition of toxic glyoxal (Fig. 5b), which is in good agreement with the enzyme activity towards these compounds (Table 2).
Effect of temperature and pH on ADH activity. a Temperature. Purified Adh3.1 (filled diamonds, black line) was tested at different temperatures under optimal pH conditions with EtOH as substrate. Adh3.2 was tested using butanal (filled squares, dashed line) as substrate at different temperatures. b For the determination of the pH-optimum, Adh3.1 was incubated in citrate-phosphate buffer (pH 4 to 7, filled diamonds, black line), Tris buffer (pH 7–9, filled diamonds, black line), Glycine NaOH buffer (pH 9–10, filled diamonds, black line) and CAPS buffer (pH 10–11, filled diamonds, black line) using ethanol as substrate at 50 °C. The enzyme activity with butanal (filled squares) was measured at 70 °C in citrate-phosphate buffer (pH 4–7, dashed line) and Tris buffer (pH 7–9, dashed line)
Cell viability assay to measure susceptibility to butanal of E. coli expressing adh3.1 or adh3.2, respectively. a Pictures were taken after 24 h of growth. b Pictures were taken after 48 h of growth. Spot assays were performed with E. coli M15[pREP4] either harbouring mock vector pQE-30 (control) or pQE30:adh3.1 or pQE30:adh3.2
Cell viability assay to measure resistance to glyoxals. a LB medium supplemented with different concentrations of methylglyoxal. Pictures were taken after 24 h of growth. b Growth assays were performed on LB medium supplemented with glyoxal. Pictures were taken after 24 h of growth. Spot assays were performed with E. coli M15[pREP4] either harbouring mock vector pQE-30 (control) or pQE30:adh3.1 or pQE30:adh3.2
Discussion
ADH is an excellent example of a versatile enzyme family that has developed diverse functionalities. These proteins execute multiple physiological roles in all organisms by catalyzing the interconversion of alcoholic compounds and aldehydes or ketones. So far, three evolutionary independent groups of NAD(P)-dependent ADHs were described in pro- and eukaryotes and each group contains several subgroups of specialized enzymes probably evolved from ancestral tandem gene duplication and neofunctionalization events. Gene duplication is crucial to functional innovation and has been shown to appear in an unexpectedly high rate in transcription factors, transporters and enzymes. Gene duplication followed by functional innovation has been described for a group-II ADH of Drosophila melanogaster, in which the first of the duplicated genes codes for an enzyme optimized for the reduction of acetaldehyde and the second for the reverse reaction (Conant and Wolfe 2008). Although several members of group III-ADHs have been investigated, little information is available about evolutionary aspects and distribution of genes encoding group III-ADHs. Despite being characterized from E. coli and K. pneumoniae in detail, DhaT- and YqhD-like proteins were mainly used to establish recombinant strains for biotechnological applications including the production of valuable chemical compounds (Xiu and Zeng 2008; Jarboe 2011).
Biochemical and phylogenetic characterization of two distantly related group III-ADHs from the plant-pathogenic enterobacterium D. zeae revealed that these enzymes have undergone an interesting evolution resulting in different substrate specificities and cofactor-dependency. Adh3.1 was a NAD-dependent ADH with preference for ethanol, while Adh3.2 preferred the phosphorylated cofactor and was mostly active on butanal. The ability of Adh3.1 and Adh3.2 to reduce aldehyde compounds was also confirmed by cell viability assays on plates containing butanal, glyoxal or methylglyoxal, respectively. Recently two ADHs from the extreme halophile, Haloferax volcanii, have been compared with regards to their biochemical properties after homologous overexpression in the native host and were shown to accept highly diverse substrates and displayed different stability and activity profiles (Timpson et al. 2012). These isozymes share an identity of 31.5 % at the protein level and belong to group I of ADHs.
The formation of dimers, which has been observed by size exclusion chromatography, is well-known for group III-ADHs and has been shown for several isozymes including E. coli FucO, E. coli YqhD and Zymomonas mobilis zmADH2, respectively (Sulzenbacher et al. 2004; Montella et al. 2005; Moon et al. 2011). So far, structure determination indicated that DhaT-like enzymes contain Fe2+ in their catalytic region, while E. coli YqhD is Zn2+-dependent. Since Adh3.1 and Adh3.2 exhibit all conserved key residues in their primary structures, it can be speculated that these proteins are metallo-enzymes as well (Supplementary Figure 1). Another indication of the presence of a metal-ion in the protein structures is the inactivation during Ni-NTA purification (data not shown). Since imidazole is a chelating agent, elution of metallo-enzymes from Ni-NTA columns might interfere with catalytic activity (Quaglia et al. 2012) However, dialyzing the protein sample against imidazole-free buffer prior to size exclusion chromatography reversed inhibition in case of Adh3.1.
This study expands our understanding of the distribution and biochemical properties of group III-ADHs in enterobacteria. A novel NAD-dependent ADH and a NADPH-dependent aldehyde reductase were characterized from D. zeae. Both enzymes were expressed at a high level in a heterologous host in E. coli and were purified with a simple 2-step purification procedure. Moreover, one enzyme was active on a range of alcohol and aldehyde substrates, while the second enzyme was only active towards aldehydes.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Conant GC, Wolfe KH (2008) Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet 9:938–950
Hernandez-Tobias A, Julian-Sanchez A, Pina E, Riveros-Rosas H (2011) Natural alcohol exposure: is ethanol the main substrate for alcohol dehydrogenases in animals? Chem Biol Interact 191:14–25
Horn U, Strittmatter W, Krebber A, Knupfer U, Kujau M, Wenderoth R, Muller K, Matzku S, Pluckthun A, Riesenberg D (1996) High volumetric yields of functional dimeric miniantibodies in Escherichia coli, using an optimized expression vector and high-cell-density fermentation under non-limited growth conditions. Appl Microbiol Biotechnol 46:524–532
Jarboe LR (2011) YqhD: a broad-substrate range aldehyde reductase with various applications in production of biorenewable fuels and chemicals. Appl Microbiol Biotechnol 89:249–257
Lee C, Park C (2012) Development of a suicidal vector-cloning system based on butanal susceptibility due to an expression of YqhD aldehyde reductase. J Microbiol 50:249–255
Lee C, Kim I, Lee J, Lee KL, Min B, Park C (2010) Transcriptional activation of the aldehyde reductase YqhD by YqhC and its implication in glyoxal metabolism of Escherichia coli K-12. J Bacteriol 192:4205–4214
Ma C, Zhang L, Dai J, Xiu Z (2010) Relaxing the coenzyme specificity of 1,3-propanediol oxidoreductase from Klebsiella pneumoniae by rational design. J Biotechnol 146:173–178
Marcal D, Rego AT, Carrondo MA, Enguita FJ (2009) 1,3-Propanediol dehydrogenase from Klebsiella pneumoniae: decameric quaternary structure and possible subunit cooperativity. J Bacteriol 191:1143–1151
Montella C, Bellsolell L, Perez-Luque R, Badia J, Baldoma L, Coll M, Aguilar J (2005) Crystal structure of an iron-dependent group III dehydrogenase that interconverts L-lactaldehyde and L-1,2-propanediol in Escherichia coli. J Bacteriol 187:4957–4966
Moon JH, Lee HJ, Park SY, Song JM, Park MY, Park HM, Sun J, Park JH, Kim BY, Kim JS (2011) Structures of iron-dependent alcohol dehydrogenase 2 from Zymomonas mobilis ZM4 with and without NAD+ cofactor. J Mol Biol 407:413–424
Perez JM, Arenas FA, Pradenas GA, Sandoval JM, Vasquez CC (2008) Escherichia coli YqhD exhibits aldehyde reductase activity and protects from the harmful effect of lipid peroxidation-derived aldehydes. J Biol Chem 283:7346–7353
Quaglia D, Irwin JA, Paradisi F (2012) Horse liver alcohol dehydrogenase: new perspectives for an old enzyme. Mol Biotechnol 52:244–250
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Radianingtyas H, Wright PC (2003) Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol Rev 27:593–616
Sulzenbacher G, Alvarez K, Van Den Heuvel RH, Versluis C, Spinelli S, Campanacci V, Valencia C, Cambillau C, Eklund H, Tegoni M (2004) Crystal structure of E.coli alcohol dehydrogenase YqhD: evidence of a covalently modified NADP coenzyme. J Mol Biol 342:489–502
Timpson LM, Liliensiek AK, Alsafadi D, Cassidy J, Sharkey MA, Liddell S, Allers T, Paradisi F (2012) A comparison of two novel alcohol dehydrogenase enzymes (ADH1 and ADH2) from the extreme halophile Haloferax volcanii. Appl Microbiol Biotechnol 97(1):195–203
Xiu ZL, Zeng AP (2008) Present state and perspective of downstream processing of biologically produced 1,3-propanediol and 2,3-butanediol. Appl Microbiol Biotechnol 78:917–926
Acknowledgments
The authors thank Henning Piascheck for the help with the fermentation experiments. This work was funded by the Excellence Cluster in the Excellence Initative by the State of Hamburg “Fundamentals of Synthetic Biological Systems (SynBio)”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Elleuche, S., Klippel, B., von der Heyde, A. et al. Comparative analysis of two members of the metal ion-containing group III-alcohol dehydrogenases from Dickeya zeae . Biotechnol Lett 35, 725–733 (2013). https://doi.org/10.1007/s10529-013-1137-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-013-1137-2