Abstract
Two biotic stress resistance related genes from the full-length cDNA library of Brassica rapa cv. Osome were identified from EST analysis and determined to be pathogenesis-related (PR) 12 Brassica defensin-like family protein (BrDLFP) and PR-10 Brassica Betv1 allergen family protein (BrBetv1AFP) after sequence analysis and homology study with other stress resistance related same family genes. In the expression analysis, both genes expressed in different organs and during all developmental growth stages in healthy plants. Expression of BrDLFP significantly increased and BrBetv1AFP gradually decreased after infection with Pectobacterium carotovorum subsp. carotovorum in Chinese cabbage. Expression of these two genes significantly changed after cold, salt, drought and ABA stress treatments. These two PR genes may therefore be involved in the plant resistance against biotic and abiotic stresses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Brassicas are an important and diverse group of crops grown worldwide that belong mainly to Brassica rapa, as well as Brassica oleracea and Brassica napus (Cardoza and Stewart 2004). Chinese cabbage (B. rapa) is widely recognized for its economic importance and contribution to human nutrition (Salunkhe and Kadam 1998). Plants undergo continuous exposure to various biotic and abiotic stresses in their natural environment. Biotic stresses include fungi, bacteria, viruses, insects, and abiotic stresses include cold, salt, ABA, drought, high temperature, and heavy metal pollution. Each type of stress functions through different types of molecular mechanisms that affect plants and eventually cause damage. Plants respond to biotic and abiotic stress factors by producing various defense-related proteins, such as pathogenesis-related (PR) proteins, hydroxyproline-rich glycoproteins (HRGPs), and several key enzymes involved in the synthesis of phenylpropanoids, phytoalexins, and lignins (Rajam et al. 2007; Pan et al. 2011; Korkina 2007).
One promising strategy to develop resistance against biotic and abiotic stresses is based on the exploitation of the PRs and other stress responsive genes in transgenic plants. Accumulation of PRs extends into non-inoculated plant parts that, upon challenge, exhibit acquired resistance (Van Loon and Van Kammen 1970; Ryals et al. 1996). PR-proteins are classified into 17 families based on sequence similarity and biochemical functions (Van Loon et al. 2006). It is well known that the birch allergen Betv1 genes belong to the PR-10 family and the defensin genes belong to the PR-12 family (Van Loon and Van Strien 1999). An important common feature of most PRs is their antifungal effect; some PRs also exhibit antibacterial, insecticidal, nematicidal, and anti-viral activity (Van Loon and Van Strien 1999; Van Loon 2001; Selitrennikoff 2001). For instance, the expression of PR-10 protein was significantly higher in Erwinia carotovora resistant chicory inbred lines when compared with susceptible lines, suggesting its involvement in chicory resistance to pathogens attack (Molarda et al. 2004). The product from the defensin gene (BSD1), which belongs to the PR-12 family, shows antifungal activity against several phytopathogenic fungi (Park et al. 2002) and transgenic tobacco plants expressing a defensin gene (Rs-AFP1) showed enhanced resistance to fungal pathogens (Terras et al. 1995).
In this study, we analyzed the sequences of two stress resistance related genes that were identified from the full-length cDNA library of B. rapa cv. Osome and studied their homology with other stress resistance related genes. Expression of these genes was analyzed in different organs and developmental growth stages. In addition, expression was also analyzed after applying abiotic stresses in Chinese cabbage and by microarray data of biotic stress treatments, and their association with biotic and abiotic stress resistance was discussed.
Materials and methods
Plant materials
Chinese cabbage (B. rapa ‘SUN-3061’) plants were grown in the Department of Horticulture, Sunchon National University, Korea. Fresh roots, stems, leaves, and flower buds were harvested, frozen immediately in liquid nitrogen, and stored at −80°C for RNA or DNA isolation.
Stress treatments
Chinese cabbages (B. rapa ‘SUN-3061’) seeds were aseptically grown on MS agar medium in a culture room under a 16 h light photoperiod at 25°C. After 3 weeks of growth, the seedlings were transferred to fresh liquid MSH (half-strength MS medium without sucrose) medium containing 250 mM NaCl and 100 mM abscisic acid (ABA) for 24 h. To induce cold stress, the seedlings were maintained at 4°C for 24 h. Drought treatment was applied by keeping the seedling on the filter paper at 28°C for 24 h. The samples were treated with all the stresses for 0 h (wild type), 30 min, 1, 2, 4, 8, and 24 h and then frozen immediately in liquid nitrogen, and stored at −80°C for RNA or DNA isolation.
RNA extraction
Total RNA was extracted from roots, stems, leaves, flower buds of healthy plants and abiotic stress applied frozen samples using the Rneasy mini kit (Qiagen, USA). RNA was treated with RNase-free DNase (Promega, USA) to remove genomic DNA contaminants.
Sequence analysis of genes
DNA and protein sequences were processed or deduced using Software Editseq (DNAstar Lasergene, USA). ORF was obtained by ORF finder at NCBI (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The primary structure of genes was analyzed using protParam (http://expasy.org/tools/protparam.html) and Augustus (http://augustus.gobics.de/submission). The alignment search was carried out using BLAST from NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). The program used was BLASTp and the database “nr” was selected. Typical domains were analyzed using the web tool from EMBL (http://smart.embl.de/smart/set_mode.cgi?GENOMIC=1). Multiple protein sequences were aligned using PIR (http://pir.georgetown.edu/pirwww/search/multialn.shtml).
Expression analysis
RT-PCR was performed using AMV one step RT-PCR kit (Takara, Japan). The specific primers for BrDLFP, FW-GAGTATCACCCAAGTTTCTGA and RV-TCATTATAGCTTCCTCCTACG, and for BrBetv1AFP, FW-TAGAAAATGGGATTGAGTGG, and RV-AACTGATAGAGATGATCGAACC were used for RT-PCR. Actin primers of Brassica, FW-ATGGTTGGGATGGGTCAAAAA, and RV-TCTTTAATGTCACGGACGATT, were used as control. PCR was performed in 20 μl containing 50 ng cDNA as template in 0.5 ml PCR tubes, 20 pmol of each primer, 150 μM of each dNTP, 1.2 U Taq polymerase, Taq polymerase buffer, and double-distilled H2O. The procedure was pre-denaturing at 94°C for 5 min, followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s and extension at 72°C for 1 min, and was terminated with an additional extension for 5 min at 72°C. The microarray data on B. rapa subsp. pekinensis were obtained from the B. rapa Genomic Project of National Academy of Agricultural Science (Suwon, Korea) (http://brassica-rapa.org/BRGP/index.jsp). Real-time quantitative PCR was performed using 1 μl cDNA in 20 μl using iTaq SYBR Green Super-mix with ROX (California, USA). Specific primers for BrDLFP, FW-GAGTATCACCCAAGTTTCTGA, and RV-CAGCTGAAATCAACAAGACA and for BrBetv1AFP, FW-CTGCTGAGTTTGAGAAGACC and RV-AACTGATAGAGATGATCGAACC were used to perform real-time PCR. The thermal cycler conditions recommended by the manufacturer were used as follows: 10 min at 95°C, followed by 40 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s. The fluorescent product was detected at the last step of each cycle.
Amplification, detection, and data analysis were carried out with a Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science, Australia). Threshold cycle (Ct) represents the number of cycles at which the fluorescence intensity was significantly higher than the background at the initial exponential phase of PCR amplification. Means were separated using Duncan’s multiple range test (P = 0.05) in the SAS (2010) program.
Results and discussion
Identification and sequence analysis
We constructed a full-length cDNA library of B. rapa cv. Osome (Park et al. 2010) and obtained 3,429 ESTs from this library. These ESTs were then annotated and 140 biotic stress related genes were selected (unpublished). From these genes, three chitinase genes have been characterized and their expression after pathogen infection was previously evaluated (Ahmed et al. 2011). In this study, we selected two cDNA clones which are small, cysteine-rich polypeptides and play vital role for stress resistance from these genes and sequenced as full-length cDNA. Several families of small, cysteine-rich basic antimicrobial polypeptides play important roles in protecting plants against invading phytopathogens (Garcia-Olmedo et al. 1998). Two full-length cDNAs encoding biotic stress resistance related genes, designated as Brassica defensin-like family protein (BrDLFP) and Brassica Betv1 allergen family protein (BrBetv1AFP) were characterized. The sequence data for BrDLFP and BrBetv1AFP are deposited in GenBank under accession number of EU186319 and EU186326, respectively.
In the sequence analysis, these genes provided complete coverage of the transcribed region and no introns were identified in the coding region of both genes. The primary structure of these genes was analyzed using bioinformatic tools and the obtained data are presented in the Table 1.
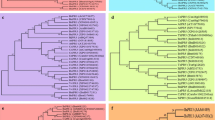
The full-length sequence of the BrDLFP cDNA contained 579 nucleotides and had a putative ORF of 273 bp encoding a defensin protein that contains 90 amino acids, beginning at the initiation codon ATG (position 78) and ending at the stop codon TAA (position 350) of the cDNA. Defensins are small cysteine-rich peptides with a molecular mass between 5 and 10 kDa and some of these peptides have been shown to exhibit potent antifungal activity (Olli and Kirti 2006). BrDLFP has a low molecular weight and contains cysteine residues in identical positions with other published defensin proteins (Fig. 1a). The cysteine-rich motif is believed to be typical for proteins belonging to the plant defensin family.
Alignment of deduced amino acid sequences of a BrDLFP and three other stress responsive defensin genes of A. thaliana (NP_001030645.1 and NP_187224.1) and R. sativus (P30230.4). Cysteine residues in identical positions were indicated by asterisk, b BrBetv1AFP with three other stress responsive genes of A. thaliana (AAM65899.1), A. lyrata subsp. lyrata (XP_002893317.1), and Zea mays (NP_001105279.1) using PIR. Numbers on the right margin indicate the positions of amino acid residues. The identical amino acids are marked in dark background, and more than 50% similarities in light
The full-length sequence of the BrBetv1AFP cDNA contained 636 nucleotides and had a ORF of 468 bp encoding an allergen family protein that contains 155 amino acids, beginning at the initiation codon ATG (position 50) and ending at the stop codon TAA (position 517) of the cDNA. The Betv1 family was named after the major birch pollen allergen and belongs to the PR-10 family (Radauer et al. 2008). In the BrBetv1AFP gene, one Betv1 domain was observed between amino acid residues 2 and 152, which suggest that this protein is a Betv1 family protein and belongs to PR-10 family.
Amino acid sequence comparison
The alignment search was carried out using BLAST from NCBI. The deduced protein sequence of BrDLFP (ABV89608) shared a high homology with the defensin-like protein of Arabidopsis thaliana (NP001030645 and NP187224) (59 and 62% identity, respectively) (Salanoubat et al. 2000). BrBetv1AFP (ABV89615) showed high homology with the pollen allergen-like protein of A. thaliana (AAM65899) (90% identity) (Haas et al. 2002) and Betv1 allergen family protein of Arabidopsis lyrata subsp. lyrata (XP002893317) (91% identity) (unpublished work) (Table 2). Multiple alignment also revealed high similarity among the deduced amino acids of BrDLFP and three other defensin genes (Fig. 1a) and BrBetv1AFP and three other Betv1 allergen family genes (Fig. 1b) indicating their relatively conserved evolutionary relationship at the protein level. Among the three other defensin genes, the Raphanus sativus (P30230) gene conferred enhanced resistance to the foliar pathogen Alternaria longipes in transgenic tobacco (Terras et al. 1995) but the stress resistance related functions of three other species Betv1 allergen family genes have not yet been examined.
Organ-specific and growth-stage expression analysis
Expression analysis was performed using specific primers with equal amounts of cDNA templates prepared from the mRNA of roots, stems, leaves, and flower buds of Chinese cabbage by RT-PCR. BrDLFP showed lower level of expression when compared to the other genes in stems, leaves, and flower buds but no expression was observed in the roots. BrBetv1AFP showed high levels of expression in all the organs, except the root and the highest level of expression was observed in flower buds (Fig. 2a). These organs share characteristics that make them particularly prone to pathogen attack. This include the flower organs are largely devoid of mechanical barriers (e.g., cuticle) to allow reproductive functions such as pollen tube penetration. Thus, adequate protection of these organs against pathogen attack is critical, and the production of an antifungal molecule could be part of a local defense strategy (Hamel and Bellemare 1995). Using microarrays data, expression of these two genes were analyzed at different growth stages and both were found to be expressed at all growth stages from mature seed to 91 days old plants. Very low levels of BrDLFP expression were observed at all growth stages except at the mature seed and 2 days old seedlings stage (CS0D and CS2D). On the other hand, high levels of BrBetvIAFP expression were observed at all growth stages (Fig. 2b). BrDLFP showed the highest level of expression at the mature seed stage (CS0D) and 2 days old seedlings stage (CS2D), which decreased gradually towards the mature stages. Organ-specific expression of this gene was low because the cDNA was extracted from the mature plant samples for RT-PCR. Most of the plants defensin proteins are isolated from seed-derived but the expression of several plants defensin isoforms has been observed in leaves (Kragh et al. 1995; Terras et al. 1995) and flowers (Gu et al. 1992; Moreno et al. 1994). This suggests that these genes are differentially regulated in different tissues or organs, thereby providing more efficient protection against pathogen infection.
Expression analysis of BrDLFP and BrBetv1AFP genes in healthy B. rapa subsp. pekinensis. a RT-PCR expression over different organs, lanes 1–4, PCR products of roots (R), stems (S), leaves (L) and flower buds (Fb). b Microarray expression analysis at different growth stages: CSOD seed, mature; CS2D seedling, 2 days old; CS1W whole plant, 1 week old vegetative stage (7 days old); CS2W whole plant, 2 weeks old vegetative stage (14 days old); CCOD whole plant, 1 day after light-chilled at 4°C (21 days old); CC1D whole plant, 1 day after light-chilled at 4°C (22 days old); CC1W whole plant, 1 week after light-chilled at 4°C (28 days old); CC4W whole plant, 4 weeks after light-chilled at 4°C (56 days old); CC7W whole plant, 7 weeks after light-chilled at 4°C (70 days old); CA1D whole plant, 1 day after greenhouse growth (71 days old); CA2D whole plant, 2 days after greenhouse growth (72 days old); CA1W whole plant, 1 week after greenhouse growth (77 days old); CA2W whole plant, 2 weeks after greenhouse growth (84 days old); CA3W whole plant, 3 weeks after greenhouse growth (91 days old)
Expression analysis after applying stresses
Expression in B. rapa was also analyzed by microarray data after infection with Pectobacterium carotovorum subsp. carotovorum, which is a serious pathogen for Brassica and causes soft rot disease. Both genes responded to infection: BrDLFP was up-regulated at 48 h post-infection (hpi) and BrBetv1AFP was down-regulated from 6 hpi (Fig. 3a, b). BrBetv1AFP was highly responsive to bacterial infection and its expression decreased by about fourfold at 48 hpi relative to the non-infected plants. This gene therefore remains active in the healthy plants and becomes inactive after pathogen infection; this is then an important association in resistance against biotic stresses.
Microarray expression analysis of a BrDLFP and b BrBetv1AFP genes in B. rapa subsp. pekinensis after infection with P. carotovorum subsp. carotovorum. c RT-PCR and real-time quantitative PCR expression of d BrDLFP and e BrBetv1AFP genes after application of cold, salt, drought and ABA stresses. The error bars represent the standard error of the means of three independent replicates. Values denoted by the same letter did not differ significantly at P < 0.05 according to Duncan’s multiple range test
The results of previous studies also support the association of these types of genes with disease resistance. The expression of some defensin genes is developmentally regulated (Epple et al. 1997), whereas expression of other defensin genes is greatly elevated in response to biotic and abiotic external stimuli (Kragh et al. 1995; Broekaert et al. 1997). Defensin genes were induced upon pathogen infection in peas (Chiang and Hadwiger 1991), tobacco (Gu et al. 1992), and Arabidopsis (Epple et al. 1997). Again, the PR-10 class of proteins, first identified as a major pollen allergen (Betv1) from white birch (Breiteneder et al. 1989), are induced by pathogen attack in a wide variety of plant species, including parsley (Somssich et al. 1986), potato (Matton and Brisson 1989), pea (Barratt and Clark 1991), soybean (Crowell et al. 1992), asparagus (Warner et al. 1993), sorghum (Lo et al. 1999) and rice (Midoh and Iwata 1996; McGee et al. 2001). However, there has been no direct evidence of their physiological activity and function in plant-defense mechanisms.
Some genes respond significantly when exposed to various environmental and pathogenic stresses and associated with broad spectrum resistance (Lee et al. 2008). To further explore such resistance, we analyzed the expression of three genes using the same primers after applying abiotic stress treatments including cold, salt, drought, and ABA. Expression of both genes changed compared to control plant after cold, salt, drought, and ABA stress treatments. In case of cold stress, BrDLFP and BrBetvIAFP genes were up-regulated and highest expression was observed after 24 h (Fig. 3c, d, e). In the salt stress, expression of BrDLFP was significantly changed with stress treatment but not directional, and BrBetvIAFP expression was increased from 0 to 1 h and gradually decreased afterwards (Fig. 3c, d, e). After drought stress treatment, BrDLFP and BrBetvIAFP both genes showed the highest expression after 1 h and gradually decreased from 1 to 24 h treatments (Fig. 3c, d, e). Again, expression of BrDLFP and BrBetvIAFP were increased from 0 to 1 h and decreased afterwards while treated with ABA stress (Fig. 3c, d, e). Different types of changes of expression of these genes with abiotic stress treatments indicate their probable association with abiotic stress resistance and we can exploit these genes for overexpression or down-regulation to enhance resistance against abiotic stresses in Brassica. In earlier studies, down-regulation of two genes, homologous to aspartic proteinase 2, and homolog of Jumonji class of transcription factor showed relative drought tolerant phenotypes in Nicotiana benthamiana (Senthil-Kumar et al. 2007). Induction of novel rice Betv1 (PR-10) protein was also observed after salt, drought, and ABA stress treatments (Hashimoto et al. 2004; Moons et al. 1997) but defensin genes had no abiotic stress responding report so far we studied.
In conclusion, BrDLFP and BrBetv1AFP genes in Brassica may be useful sources for engineering transgenic plants to be resistant to biotic and abiotic stresses.
References
Ahmed NU, Park JI, Seo MS, Kumar TS, Lee IH, Park BS, Nou IS (2011) Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica. Mol Biol Rep. doi:10.1007/s11033-011-1139-x
Barratt DHP, Clark JA (1991) Proteins arising during the late stages of embryogenesis in Pisum sativum L. Planta 184:14–23
Breiteneder H, Pettenburger K, Bito A, Valenta R, Kraft D, Rumpold H, Scheiner O, Breitenbach M (1989) The gene coding for the major birch pollen allergen Betv1, is highly homologous to a pea disease resistance response gene. EMBO J 8:1935–1938
Broekaert WF, Cammue BPA, De Bolle MFC, Thevissen K, De Samblanx GW, Osborn RW (1997) Antimicrobial peptides from plants. Crit Rev Plant Sci 16:297–323
Cardoza V, Stewart CN Jr (2004) Brassica biotechnology: progress in cellular and molecular biology in vitro cell. Dev Biol Plants 40:542–551
Chiang CC, Hadwiger LA (1991) The Fusarium solani-induced expression of a pea gene family encoding high cysteine content proteins. Mol Plant Microbe Interact 4:324–331
Crowell DN, John ME, Russell D, Amasino RM (1992) Characterization of a stress-induced, developmentally regulated gene family from soybean. Plant Mol Biol 18:459–466
Epple P, Apel K, Bohlmann H (1997) ESTs reveal a multigene family for plant defensins in Arabidopsis thaliana. FEBS Lett 400:168–172
Garcia-Olmedo F, Molina A, Alamillo JM, Rodriguez-Palenzuela P (1998) Plant defense peptides. Biopolymers 47:479–491
Gu Q, Kawarta EF, Morse M-J, Wu H-M, Cheung AY (1992) A flower-specific cDNA encoding a novel thionin in tobacco. Mol Gen Genet 234:89–96
Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3(6):0029.1–0029.12 (research)
Hamel F, Bellemare G (1995) Characterization of a class I chitinase gene and of wound-inducible, root and flower-specific chitinase expression in Brassica napus. Biochim Biophys Acta 1263(3):212–220
Hashimoto M, Kisseleva L, Sawa S, Furukawa T, Komatsu S, Koshiba T (2004) A novel rice PR-10 protein, RSOsPR10, specifically induced in roots by biotic and abiotic stresses, possibly via the jasmonic acid signaling pathway. Plant Cell Physiol 45(5):550–559
Korkina LG (2007) Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell Mol Biol 53:15–25
Kragh KM, Nielsen JE, Nielsen KK, Dreboldt S, Mikkelsen JD (1995) Characterization and localization of new antifungal cysteine rich proteins from Beta vulgaris. Mol Plant Microbe Interact 8:424–434
Lee SC, Hwang IS, Choi HW, Hwang BK (2008) Involvement of the pepper antimicrobial protein CaAMP1 gene in broad spectrum disease resistance. Plant Physiol 148:1004–1020
Lo SC, Hipskind JD, Nicholson RL (1999) cDNA cloning of a sorghum pathogenesis-related protein (PR-10) and differential expression of defense-related genes following inoculation with Cochliobolus heterostrophus or Colletotrichum sublineolum. Mol Plant Microbe Interact 12:479–489
Matton DP, Brisson N (1989) Cloning, expression, and sequence conservation of pathogenesis-related gene transcripts of potato. Mol Plant Microbe Interact 2:325–331
McGee JD, Hamer JE, Hodges TK (2001) Characterization of a PR-10 pathogenesis-related gene family induced in rice during infection with Magnaporthe grisea. Mol Plant Microbe Interact 14:877–886
Midoh N, Iwata M (1996) Cloning and characterization of a probenazole-inducible gene for an intracellular pathogenesis-related protein in rice. Plant Cell Physiol 37:9–18
Molarda CR, Brugiere N, Moille M, Carrayol E, Limami AM (2004) Molecular characterization of a gene encoding a vegetative storage protein (CiVSP) from Cichorium intybus and its expression in the root and shoot in relation to nitrogen status and pathogen resistance. Physiol Plant 121:568–577
Moons A, Prinsen EIS, Bauw G, Montaguasi MV (1997) Antagonistic effects of abscisic acid and jasmonates on salt stress-inducible transcripts in rice roots. Plant Cell 9:2243–2259
Moreno M, Segura A, Garcia-Olmedo F (1994) Pseudothionin, a potato peptide active against potato pathogens. Eur J Biochem 223:135–139
Olli S, Kirti PB (2006) Cloning, characterization and antifungal activity of defensin Tfgd1 from Trigonella foenumgraecum L. J Biochem Mol Biol 39:278–283
Pan W, Shou J, Zhou X, Zha X, Guo T, Zhu M, Pan J (2011) Al-induced cell wall hydroxyproline-rich glycoprotein accumulation is involved in alleviating Al toxicity in rice. Acta Physiol Plant 33:601–608
Park HC, Kang YH, Chun HJ et al (2002) Characterization of a stamen-specific cDNA encoding a novel plant defensin in Chinese cabbage. Plant Mol Biol 50:59–69
Park JI, Kumar TS, Ahmed NU, Nou IS (2010) Construction of full-length cDNA library and investigation of potential genes in Brassica rapa. J Basic Life Res Sci 10:6–11
Radauer C, Lackner P, Breiteneder H (2008) The Betv1 fold: an ancient, versatile scaffold for binding of large, hydrophobic ligands. BMC Evol Biol 8:286
Rajam MV, Chandola N, Saiprasad GP, Singh D, Kashyap V, Choudhary ML, Sihachakr D (2007) Thaumatin gene confers resistance to fungal pathogens as well as tolerance to abiotic stresses in transgenic tobacco plants. Biol Plant 51(1):135–141
Ryals JA, Neuenschwander UH, Willits MG, Molina A, Steiner HY, Hunt MD (1996) Systemic acquired resistance. Plant Cell 8:1809–1819
Salunkhe DK, Kadam SS (1998) Handbook of vegetable science technology: production, composition, storage, and processing. Marcel Dekker Inc, New York
Selitrennikoff CP (2001) Antifungal proteins. Appl Env Microbiol 67:2883–2894
Senthil-Kumar M, Govind G, Kang L, Mysore KS, Udayakumar M (2007) Functional characterization of Nicotiana benthamiana homologs of peanut water deficit-induced genes by virus-induced gene silencing. Planta 225:523–539
Somssich IE, Schmelzer E, Bollmann J, Hahlbrock K (1986) Rapid activation by fungal elicitor of genes encoding “pathogenesis-related” proteins in cultured parsley cells. Proc Natl Acad Sci USA 83:2427–2430
Terras FRG, Eggermont K, Kovaleva V, Raikhel NV, Osborn RW, Kester A, Rees SB, Torrekens S, Van Leuven F, Vanderleyden J, Cammue BPA, Broekaert WF (1995) Small cysteine-rich antifungal proteins from radish: their role in host defense. Plant Cell 7:573–588
Van Loon LC (2001) The families of pathogenesis-related proteins. Abstracts of the 6th international workshop on PR-proteins, Spa, Belgium, 20–24 May, p 9
Van Loon LC, Van Kammen A (1970) Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. ‘Samsun’ and ‘Samsun NN’ II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40:199–211
Van Loon LC, Van Strien EA (1999) The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiol Mol Plant Pathol 55:85–97
Van Loon LC, Rep M, Pieterse CM (2006) Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol 44:135–162
Warner SAJ, Scott R, Draper J (1993) Isolation of an asparagus intracellular PR gene (AoPR1) wound-responsive promoter by the inverse polymerase chain reaction and its characterization in transgenic tobacco. Plant J 3:191–201
Acknowledgments
This work was supported by the Cabbage Genomics assisted breeding supporting center research program funded by Ministry for Food, Agriculture, Forestry and Fisheries of the Korean Government.
Author information
Authors and Affiliations
Corresponding author
Additional information
Nasar Uddin Ahmed and Jong-In Park equally contributed to this work.
Rights and permissions
About this article
Cite this article
Ahmed, N.U., Park, JI., Jung, HJ. et al. Identification and characterization of stress resistance related genes of Brassica rapa . Biotechnol Lett 34, 979–987 (2012). https://doi.org/10.1007/s10529-012-0860-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-012-0860-4