Abstract
Aspergillus-derived FAD-dependent glucose dehydrogenases (FADGDHs) were screened from fungal genomic databases, primarily by searching for putative homologues of the Aspergillus niger-derived glucose oxidase (GOD). Focusing on a GOD active-site motif, putative proteins annotated as belonging to the glucose methanol choline (GMC) oxidoreductase family were selected. Phylogenetic analysis of these putative proteins produced a GOD clade, which includes the A. niger and Penicillium amagasakiens GODs, and a second clade made up of putative proteins showing 30–40% homology with GOD. The genes encoding the proteins from the second clade were functionally expressed in Escherichia coli, resulting in dye-mediated glucose dehydrogenase (GDH) activity but not GOD activity. These results suggest that the putative proteins belonging to the second clade are FADGDHs. The 3D structure models of these FADGDHs were compared with the 3D structure of GOD.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the first report of an enzyme sensor, which employed glucose oxidase (GOD) and an O2 electrode for glucose monitoring, thousands of principles and approaches have been reported in the area of biosensors and bioelectronics studies. Among these, electrochemical methods are mainly used due to the simplicity of the detection systems and devices. The majority of biosensors and bioelectronics studies have been devoted to diabetes monitoring. Currently, the most popular principle for the self-monitoring of blood glucose is based on dye-mediated dehydrogenase reactions involving the monitoring of the concentration/quantity of reacted (reduced) artificial electron acceptors on an electrode.
GODs form a category (E.C.1.1.3.4) that is defined as the oxidoreductases capable of oxidizing a glucose its first hydroxyl group, utilizing O2 as the electron acceptor, and harboring FAD as the cofactor. GOD was originally isolated and produced from Aspergillus niger and has been extensively utilized for glucose monitoring. The most distinguishing characteristic of A. niger-derived GOD is its high substrate specificity for glucose. GOD is a representative of the glucose/methanol/choline (GMC) oxidoreductase family, whose members share similar primary structural features: FAD-binding motif(s), FAD-binding domain(s), and catalytic domain(s).
There is currently much attention placed on glucose dehydrogenases (GDHs) harboring FAD (FADGDHs). These GDHs utilize a variety of external electron acceptors but not oxygen; therefore, their systematic name is d-glucose:acceptor 1-oxidoreductase (E.C.1.1.99.10). With the recent and emergent demand for a GDH that is specific for glucose, significant attention has been directed towards fungi-derived FADGDHs, focusing on their narrow substrate specificity, especially their lack of activity toward maltose (FDA Public Health Notification, 2009, http://www.fda.gov/MedicalDevices/Safety/AlertsandNotices/PublicHealthNotifications/ucm176992.htm). The earliest report of the presence of GDH in the fungus Aspergillus sp. dates back to 1937, which reported the presence of an enzyme capable of the oxidation of glucose utilizing certain electron acceptors but not O2 (Ogura and Nagahisa 1937). The GDH from A. oryzae was found to have an FAD cofactor and its enzymatic properties were studied (Bak 1967). Müller (1977) reported that at least three gluconic acid-forming enzymes were identified in cell-free extracts of A. niger and one of them was an GDH. An FADGDH from A. terreus has also been characterized (Tsujimura et al. 2006).
Information describing the fungi-derived FADGDHs and their availability remains limited. In this study, we have carried out the screening of FADGDHs from the fungal genomic database in order to expand our knowledge by discovering a variety of FADGDHs. We focused on amino acid sequences from the A. niger-derived GOD (PDB id: 1CF3) and on the conserved active-site sequence motif Arg-X-Asn-X-His responsible for the recognition of the 1st, 2nd, and 3rd hydroxyl groups of the glucose substrate. The structural genes of some putative FADGDHs were synthesized and recombinantly expressed using Escherichia coli as the host microorganism. The expressed GOD homologues showed dye-mediated glucose specific dehydrogenase activity but not GOD activity. These results help validate our strategy for searching FADGDHs from fungal database. We also discuss the comparison of the 3D structure model of the FADGDHs and 3D structure of GOD.
Materials and methods
Database search and sequence analysis
Protein sequences of putative FADGDHs from Aspergillus sp. were from the NCBI databases and SUPERFAMILY database (Gough et al. 2001) using the GOD of A. niger (PDB id: 1CF3) as reference. The obtained sequences were aligned using ClustalW (Thompson et al. 1994) and verified that the Arg-X-Asn-X-His motif (the highly conserved sequence in fungi-derived glucose oxidoreductases), the Gly-X-Gly-X-X-Gly motif (FAD-binding motif) (Wierenga et al. 1986), and catalytic residues of glucose oxidoreductases were conserved.
Sequence similarities were analyzed using BioEdit v.7.0.9 (Hall 1999) and MEGA4 (Tamura et al. 2007). The phylogenetic tree and evolutionary distances were calculated using the Neighbor-Joining method (Saitou and Nei 1987) and Poisson correction method (Zuckerkandl and Pauling 1965) in MEGA4.
Bacterial strains and plasmids
The putative FADGDH structural genes were synthesized by GenScript USA Inc. with codons optimized for expression in E. coli. The signal sequences were predicted using SignalIP 3.0 (Nielsen et al. 1997; Bendtsen et al. 2004). The region of the synthesized genes encoding the putative mature proteins (minus signal sequences) were amplified by PCR, inserted into the multicloning site of the expression vector pET-30c(+) (Merck KGaA, Germany), and expressed in the host strain E. coli BL21 (DE3).
Preparation of crude cell extract
Recombinant E. coli were cultured in 500 ml conical-shaped flasks containing 150 ml ZYP-5052 medium (0.5% glycerol, 0.05% glucose, 0.2% lactose, 50 mM (NH4)2SO4, 50 mM KH2PO4, 50 mM Na2HPO4, and 1 mM MgSO4) (Studier 2005) in a rotary shaker at 20°C for 24 h. The cells were then harvested, washed twice with 0.85% NaCl, and disrupted by ultrasonication in 20 mM potassium phosphate buffer (pH 6.5). After centrifugation to remove the cell debris and inclusion body (the insoluble fraction), the resulting supernatant was dialyzed against 20 mM potassium phosphate buffer (pH 6.5) and designated as crude extract. The expression of the putative FADGDHs was confirmed by SDS-PAGE analysis of the insoluble fractions and the crude extracts.
Analytical methods
The crude extracts or refolded protein samples were assayed for GDH activity as follows. A 10 μl sample was mixed with 170 μl reaction buffer [20 mM potassium phosphate buffer (pH 6.5), 0.06 mM 2,6-dichlorophenol indophenol (DCIP), and 0.6 mM phenazine methosulfate (PMS)]. The reaction was initiated by injecting 20 μl 400 mM d-glucose and the rate of the reaction was determined by monitoring the decrease in absorbance of DCIP at 600 nm. Samples were also assayed for oxidase activity in 20 mM PPB, pH 6.5, 1.5 mM TODB, 2 U horseradish peroxidase/ml, 1.5 mM 4-aminoantipyrine, and 40 mM glucose at room temperature and measuring the formation of quinoneimine dye at 546 nm. The protein concentrations were measured using the DC Protein Assay Kit (Bio-Rad, CA).
Results
Screening from fungal genomic database
The screening of Aspergillus-derived FADGDHs was primarily carried out by searching for homologues of A. niger-derived GOD (1CF3). From the obtained putative proteins, which belong to the GMC oxidoreductase family, 20 were selected as harboring the Arg-X-Asn-X-His motif, which is the region recognizing the reducing end of glucose that is conserved in GODs. A phylogenic analyses on the putative proteins harboring this motif and 1CF3 clearly show the formation of two clades (Fig. 1). The first clade contains 1CF3 and Penicillium amagasakiens-derived GOD (1GPE) and 12 putative proteins that are assumed to be GODs.
Phylogenetic tree of GODs and putative GDHs. The evolutionary history was inferred using the Neighbor-Joining method (Saitou et al. 1987). The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl et al. 1965) and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset. There were a total of 561 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007). The proteins were annotated as A. niger GOD (1CF3, ADP03053, AAF59929, ACR56326, AAV68194, ABG54443, ABG66642, and CAC12802), A. flavus NRRL3357 putative GMC oxidoreductase (XP_002375824), A. oryzae RIB40 GOD (XP_001727544), A. terreus NIH2624 GOD precursor (XP_001217613, XP_001216461 and XP_001215424), P. amagasakiense GOD (1GPE), A. flavus NRRL3357 putative GOD (XP_002372599), A. oryzae RIB40 GOD (XP_001817515 and XP_001826806), A. niger CBS 513.88 GOD (XP_001391138 and XP_001394544), A. terreus NIH2624 hypothetical protein ATEG_08295 (XP_001216916), A. flavus NRRL3357 putative choline dehydrogenase (XP_002385256), and A. carbonarius ITEM 5010 jgi|Aspca1|33771|fgenesh1_pg.00771_#_19 () (Aspca1 33771)
In order to characterize the putative proteins belonging to the second clade, five were selected for recombinant expression in E. coli. Figure 2 shows an amino acids sequence alignment of these putative proteins with the GODs of A. niger and P. amagasakiens. Among them, XP_002372599 is identical to A. oryzae FADGDH described in WO patent 2004/058958 A1 (Dec. 24, 2003). The sequence of AFL256 has a long deletion in its NCBI entry; however, a complete sequence was reconstructed (Fig. 2) from the genome sequence of A. flavus NRRL3357 by considering the regions annotated as exons, and this sequence was used in this study.
Amino acid sequence alignment of GODs and putative GDHs. The sequences were aligned using ClustalW (Thompson et al. 1994). Gly-X-Gly-X-X-Gly motif and Arg-X-Asn-X-His motif were underlined. Arrows indicate amino acid residues that interact with glucose and which are described in Table 1. Proteins are named as in Fig. 1
Expression of the putative FADGDHs
The constructed genes encoding the putative mature proteins were cloned into pET-30c(+) and expressed in E. coli BL21 (DE3) as described under “Materials and methods” section. All soluble fractions prepared from the recombinant E. coli showed dye-mediated dehydrogenase activities for glucose, but not oxidase activity, indicating that these putative proteins were FADGDHs. However, the GDH expression varied greatly depending on the structural gene. Among the five clones, E. coli expressing AFL 599 showed the highest GDH productivity (13,000 U/l, 52 U/mg with 40 mM glucose), followed by that expressing ANG544 (990 U/l, 1.8 U/mg with 40 mM glucose). The remaining three clones showed much lower activities (ANG138: 93 U/l, 180 mU/mg; AFL256: 200 U/l, 360 mU/mg; AC771: 130 U/l, 330 mU/mg, with 40 mM glucose).
SDS-PAGE analysis of the soluble and insoluble fractions prepared from the recombinant E. coli expression of the putative FADGDHs appeared to reflect the in vitro enzyme activity results (Fig. 3). AFL599 gave the strongest band of about 60 kDa in both the soluble and insoluble fraction. ANG138 gave a very faint band in the soluble fraction and a strong band in the insoluble fraction. No gene products from ANG544, AFL256, and AC771 could be identified by the SDS-PAGE analysis of either the soluble or insoluble fraction. The high productivity of AFL599 may therefore be due to the high expression level and/or high folding efficiency in the E. coli host strain, compared with the other FADGDHs.
SDS-PAGE of crude extracts and insoluble fractions. Molecular weight marker (lane M), crude extracts (odd lanes) and insoluble fractions (even lanes) of AFL599 (lanes 1 and 2), ANG544 (lanes 3 and 4), ANG138 (lanes 5 and 6), AFL256 (lanes 7 and 8), and AC771 (lanes 9 and 10) were separated on a 10% polyacrylamide gel. The arrow indicates the expected molecular weight of the FADGDHs
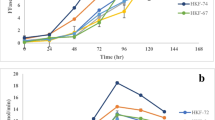
Substrate specificity of the FADGDHs
The substrate specificity of AFL599, ANG138, ANG544, AFL256, and AC771 were investigated using soluble crude extracts (Fig. 4). All gene products showed the highest activity towards glucose and no detectable activity toward galactose, maltose, and cellobiose. Xylose was the second preferred substrate with activities ranging from 9 to 27% of the activity with glucose at 40 mM. The specific activity of ANG138 was too low to accurately measure its activity towards xylose. Low activity towards mannose was also observed with AFL599 (2%) and ANG544 (8%), but could not be detected in the remaining three FADGDHs, which had low specific activity toward glucose.
Discussion
We screened for FADGDHs from fungal genomic database by searching for putative homologues of A. niger-derived GOD that harbor the Arg-X-Asn-X-His motif in their C-terminal catalytic domain. Phylogenetic analysis of the putative homologues produced two distinct clades, a GOD clade and an FADGDH clade, which showed dye-mediated GDH activity but not GOD activity when recombinantly produced in E. coli.
The different structural genes varied greatly in their expression levels and productivity of active enzyme in E. coli. Fungal GODs and FADGDHs are natively produced as extracellular glycosylated proteins (Pazur et al. 1965; Bak 1967). A generally assumed inherent problem with expressing extracellular fungal enzymes in E. coli is the absence of various post-translation modifications that such proteins undergo when natively produced. A major modification that extracellular fungal enzymes undergo is glycosylation, which increases protein solubility and may also play an important role in the natural folding process. The absence of glycosylation in the recombinant GDHs prepared in E. coli might explain the formation of inclusion bodies observed for AFL599 and ANG138.
The absence of their putative N-terminal secretion signal peptide sequences may also have had negative effects on their expression levels. The absence of the secretion process may have resulted in unusually high local concentrations of target proteins, which may have lead to the formation of inclusion bodies. In this study, we attempted to produce FAD-binding oxidoreductases in E. coli. For such proteins to be secreted, they would need to first fold and bind their FAD cofactor in the cytosol, and then be secreted to the periplasm via the twin-arginine translocase (TAT), which secrets fully folded proteins. However, no general strategies have yet been provided for the selection of TAT signal sequences suitable for the expression of fungal FADGDHs in E. coli. As the aim of this study was the screening of FADGDHs from fungal genomic database, the primary focus was on the enzymatic characteristics of the putative enzymes, not their production levels. In the future, the refolding of inclusion bodies may also be considered for the production of high quantities of active enzyme.
Out of the five putative FADGDHs studied, only two could be observed by SDS-PAGE of either the soluble or insoluble fraction (Fig. 3), suggesting that there may also be great variation in their translation rates. Deletion of the fungal secretion signal sequence may have resulted in unfavorable structures at the translational initiating region. The observed differences in the expression level might be due to differences in their N-terminal sequences after deletion of their putative fungal secretion signal sequence.
The 3D structures of AFL599, ANG138, and ANG544 were predicted by homology modeling using the A. niger GOD as template. The FADGDH structures overall showed high similarity with GOD. The amino acid residues at the active the active site show a high degree of conservation between the FADGDHs and GOD, which is not surprising considering their similar substrate specificities. Diversity is observed in the residues predicted to interact with the 6th hydroxyl group of glucose; ANG544 and ANG256 have Ser whereas AFL599, ANG138, and AC771 have Ala. As FADGDH reacts with xylose, which is not a substrate for GOD, the position recognizing 6th hydroxyl group of glucose may have a significant role in recognition of xylose in FADGDH (Fig. 5).
3D structure models of GOD and FADGDHs around active site. The 3D structure models of the putative FADGDHs were constructed by Modeller 9v3 (Šali and Blundell 1993) using the structure of A. niger GOD (PDB id: 1CF3) as template. The residues binding to hydroxyl groups of glucose are colored cyan and those interacting with glucose hydrophobically are colored pink. Images were produced using PyMol 0.99rc6 (DeLano Scientific LLC)
The conservation of active site residues is consistent with the fact that their reductive half reaction, the oxidation of glucose and reduction of FAD, should be similar among these enzymes. Observation of the active site structural models could not explain the differences in their oxidative half reaction, their electron acceptor selectivity, between GODs and FADGDHs. Because the GODs and FADGDHs show relatively low amino acid sequence homology, a more detail investigation may help elucidate the residues and motifs responsible for the differences in oxidative half reaction.
In summary, we have developed a strategy for identifying FADGDHs from fungal genomic data. Further detailed studies may provide characteristic structural motifs of FADGDH that may lead to the discovery and development of FADGDHs ideally suited for self-monitoring of blood glucose applications.
References
Bak TG (1967) Studies on glucose dehydrogenase of Aspergillus oryzae. II. Purification and physical and chemical properties. Biochim Biophys Acta 139(2):277–293
Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Gough J, Karplus K, Hughey R, Chothia C (2001) Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J Mol Biol 313(4):903–919
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Müller HM (1977) Gluconic acid forming enzymes in Aspergillus niger. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg 132(1):14–24
Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 10:1–6
Ogura Y, Nagahisa M (1937) Untersuchungen über die Atmung und die Dehydrasesysteme von Aspergillus oryzae. Bot Mag Tokyo 51:597–612
Pazur JH, Kleppe K, Cepure A (1965) A glycoprotein structure for glucose oxidase from Aspergillus niger. Arch Biochem Biophys 111:351–357
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Studier FW (2005) Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif 41:207–234
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tsujimura S, Kojima S, Kano K, Ikeda T, Sato M, Sanada H, Omura H (2006) Novel FAD-dependent glucose dehydrogenase for a dioxygen-insensitive glucose biosensor. Biosci Biotechnol Biochem 70(3):654–659
Wierenga RK, Terpstra P, Hol WGJ (1986) Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol 187:101–107
Wohlfahrt G, Witt S, Hendle J, Schomburg D, Kalisza HM, Hecht HJ (1999) 1.8 and 1.9 Å Resolution structures of the Penicillium amagasakiense and Aspergillus niger glucose oxidases as a basis for modelling substrate complexes. Acta Crystallogr D55:969–977
Wohlfahrt G, Trivić S, Zeremski J, Peričin D, Leskovac V (2004) The chemical mechanism of action of glucose oxidase from Aspergillus niger. Mol Cell Biochem 260:69–83
Zuckerkandl E, Pauling L (1965) Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ (eds) Evolving genes and proteins. Academic Press, New York, pp 97–166
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Mori, K., Nakajima, M., Kojima, K. et al. Screening of Aspergillus-derived FAD-glucose dehydrogenases from fungal genome database. Biotechnol Lett 33, 2255–2263 (2011). https://doi.org/10.1007/s10529-011-0694-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-011-0694-5