Abstract
A novel transcription factor, TcAP2, was isolated from Taxus cuspidata by yeast one-hybrid strategy. This factor interacts with jasmonate- and elicitor-responsive element. Analysis of the deduced TcAP2 amino acid sequence revealed that TcAP2 contained a conserved AP2/ethylene-responsive element binding protein domain that consisted of 268 amino acids in a potential nuclear localization sequence. The factor of TcAP2 had a high homology, in its AP2 domain, to other AP2 family members. Based on phylogenetic analysis, it was different from other five DRE-binding proteins in their evolutionary relationship. The transcription of TcAP2 gene in yew accumulated primarily in young organs, such as young stems. Quantitative real-time RT-PCR analysis indicated that TcAP2 gene was inducible to express by treatments with methyl jasmonate plus salicylic acid, high salinity, and cold. This gene showed no response to either abscisic acid or drought treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methyl jasmonate (MeJA) and salicylic acid (SA) function as induction signals during phytoalexin synthesis in plant cells (Yazaki et al. 1997; Sun et al. 2001). Jasmonate (JA) and SA are regarded as stress hormones since they are able to induce biosynthesis of some secondary metabolites that are usually synthesized when plants respond to environmental stresses. Octadecanoid-responsive Catharanthus AP2 protein (ORCA3) has been isolated from Catharanthus roseus with a DNA activation tagging (van der Fits and Memelink 2000). The gene for ORCA3 is a JA-responsive AP2-domain transcription factor and also the first transcription factor in terpene biosynthesis pathway. Over-expression of ORCA3 enhanced expression of several genes that are involved in terpene indole alkaloids biosynthesis, such as genes DXS, AS, TDC and STR (van der Fits and Memelink 2000). Regulation of biosynthetic genes by ORCA3 affected metabolite synthesis when plants respond to stresses (Aerts et al. 1994; Gantet et al. 1998; Vázquez-Flota and De Luca 1998; van der Fits and Memelink 2000; Memelink et al. 2001). ORCA3 binds specifically, via a JA- and elicitor-responsive element (JERE), to the promoter of the STR gene (van der Fits and Memelink 2000; Memelink et al. 2001). MeJA, JA precursor and intermediates of JA biosynthetic pathway could induce ORCA3 gene expression. The AP2/ERF-domain transcription factor ORCA3 has been proven to be an important regulator for both primary and secondary metabolism in Catharanthus roseus (van der Fits and Memelink 2000; Memelink et al. 2001). Functional characterizations in yeast and plant cells have revealed an N-terminal acidic activation domain and a serine-rich C-terminal domain in ORCA3 protein.
Biosynthetic gene regulation by JA-responsive AP2 transcription factors may cause changes in secondary metabolism when plants respond to stresses. Plants may regulate both primary and secondary metabolism via a single transcription factor. Since JA is able to induce many secondary metabolites, identification of the AP2-domain protein as a common regulator for several genes may uncover a control mechanism of metabolic pathways that are stress-responsive (Vom Endt et al. 2003).
Taxol is an effective and essential integrant in anti-cancer medicines. In previous studies, Taxol concentrations were increased in Taxus suspension cultures by adding of MeJA to culture media (Sun et al. 2001; Moon et al. 1998). During Taxol biosynthesis, transcriptional expression of key enzyme genes was up-regulated by MeJA (Walker and Croteau 2000; Walker et al. 2002; Laskaris et al. 1999). Currently, no transcription factor, which is involved in taxol biosynthesis, has been isolated from Taxus. In order to understand more about taxol biosynthesis, in this study, we isolated a novel cDNA encoding a JERE-binding factor, TcAP2, from T. cuspidata cDNA library by using the yeast one-hybrid method. This factor is a gene encoding a nuclear protein and inducible by MeJA plus SA.
Materials and methods
Plant materials, growth conditions and stress treatments
Three-year-old seedlings of T. cuspidata were purchased from Weihe Forestry Bureau, Heilongjiang Province, P. R. China. Suspension cell cultures were initiated and maintained with B5 medium (Gamborg et al. 1968) plus 2, 4-dichlorophenoxyacetic acid (1 mg/l), naphthalenic acid (1 mg/l), and 6-benzylaminopurine (0.2 mg/l). MeJA (100 μM, Sigma) was filter-sterilized before adding into the autoclaved medium. Flasks containing cell cultures were placed on a shaker at 100 rpm, in the dark at 15 ± 2°C.
For chemical treatments, the roots were immerged into sterile water, NaCl (250 mM), MeJA (100 μM) plus SA (100 μM), or ABA (100 μM), respectively during 24 h. Low temperature treatment was performed by placing seedlings in the refrigerator at 4 ± 1°C for 12 h. For drought treatment, the seedlings were desiccated by stopping watering. Seedlings were cultured at 15 ± 2°C, in a dark period of 24 h if not specifically mentioned. After treatment, leaves and roots, were removed from seedlings and quickly immersed into liquid N2 and stored at −75°C for subsequent RNA extraction.
Strains and plasmid
Saccharomyces cerevisiae strain Y187 (MATα, ura3-52, his3-200, ade2-101, trp1-901, leu2-3, 112, gal4Δ, met−, gal80Δ, URA3 : : GAL1UAS-GAL1TATA-lacZ, MEL1) (BD & Clontech, Franklin Lakes, NJ, USA) was used for transformation and screening. To construct the bait plasmid, oligonucleotides containing three tandem copies of wild-type JERE were synthesized by PCR using two primers (JERE cis Z: 5′2032-CCGAATTCCTCTTAGACCGCCTTCTTTGAAAGCTCTTAGACCGCCTTCTTTGAAAGCTCTTAGACCGCCTTCTTTGAAAG-3′; JERE cis F: 5′-CCGAGCTCCTTTCAAAGAAGGCGGTC-3′). The fragment was digested with restriction enzymes, and inserted into pHIS2 at the EcoRI and SacI sites. As the one-hybrid reporter vector, the pHIS2 contained HIS3 nutritional reporter gene and had a multiple cloning site (MCS) on the upstream of the HIS3 reporter gene. It can be inserted into a cis-acting DNA target sequence to construct a bait plasmid and to be linked to the minimal promoter of the HIS3 locus (PminHIS3). The cDNA library of T. cuspidata was constructed into a pGADT7-Rec2 vector, which was a LEU-marked homologous recombination-mediated expression plasmid that harbored a galactose-inducible (GAL4) domain under the control of yeast alcohol dehydrogenase (ADH1) promoter.
Screening of T. cuspidata cDNA library and 5′ cDNA fragment cloning
The plasmid pHIS-JERE was transformed into yeast Y187 by following lithium acetate protocol. After yeast one-hybrid screening, the pGADT7-Rec2 vector was transferred into yeast cells that were able to grow on the Leu-deficient medium. These yeast cells were than plated on Leu-, His- and Trp-deficient selective media. After a 4 to 6-day culture at 30°C, about 3.5 × 106 yeast transformants were overlaid on the medium. Two oligonucleotide primers of 5′-TTCCACCCAAGCAGTGGTATCAACGCAGAGTGG-3′, derived from the DNA sequence flanking the GAL4 activating domain, and 5′-GTATCGATGCCCACCCTCTAGAGGCCGAGGCGGCCGACA-3′, derived from the ADH1 terminator region in pGADT7-Rec2 vector, were used to amplify the T. cuspidata cDNA inserts in pGADT7-Rec2. The 3′cDNA fragments were obtained from yeast one-hybrid screening as described previously. Through analysis with BLAST (Basic Local Alignment Search Tool), only one transformant had high homology to 3′ fragment of a putative Arabidopsis thaliana.AP2 transcription factor gene (GenBank accession no. NM_102039). According to that, we used 5′ smart race strategy and two primers (first:5′-CCTGGAGGAACACTACTGGTAACAGCC-3′;second:5′-GAAGAAGTATATTGCCCATTGGCTGCG-3′) to clone the 5′ cDNA fragment of target gene under the instruction of SMART RACE cDNA Amplification kit manual (BD & Clontech, Franklin Lakes, NJ, USA). The PCR conditions were: 25 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 3 min.
DNA sequence analysis
The nucleotide and deduced amino-acid sequences were compared with those in GenBank databases by BLAST program. The alignments were produced by Vector NTI Suit 8.0 program. Associated information was analyzed by BioEdit and Clustal W. Phylogenetic tree analysis was aligned with the program CLUSTAL X (Thompson et al. 1994). All the alignment gaps were eliminated.
Subcellular localization
TcAP2 protein was fused to the N-terminal of a green fluorescent protein (GFP) by constructing a expression plasmid p35S::TcAP2-GFP. For this purpose, the termination codon of TcAP2 gene was removed by PCR using a pair of specific oligonucleotide primers. The resulting fragment was fused to the coding region of GFP in the binary vector pCAM35S-GFP. One inner onion peel (2 × 2 cm2) was placed on Murashige and Skoog medium (Murashige and Skoog 1962) containing 30 g glucose/l as described by Lin et al. (2007). The plasmids containing the fusion gene and control GFP were transformed into onion epidermal cells under vacuum by particle bombardment at a helium pressure of 200 psi. After bombardment, the transformed onion peel was incubated at 24°C in darkness for 16–18 h. The GFP fluorescence of TcAP2-GFP fusion protein and control GFP in the transformed onion cells was checked under a microscope with visible and UV lights (Lin et al. 2007).
Quantitative real-time RT-PCR
The primers of (1) qRT-PCR forward primer, 5′-CAGCAGGCAAGATAACCCAGA-3′, and (2) qRT-PCR reverse primer, 5′-CCTCATTCTCACACCCTTGTATTG-3′ were used for quantitative real-time RT-PCR. Quantitative Realtime-PCR was performed under the conditions: denatured at 94°C for 2 min, followed by 40 cycles of 15 s at 94°C, 1 min at 60°C. The real-time PCR products were detected by SYBR Green I method. DNA intensity ratio of the TcAP2 gene was analyzed on icycler Real-time qPCR Detecting System (BIORAD, Hercules, California, USA) to evaluate the expression patterns of TcAP2 gene.
Results
Isolation of cDNAs encoding JERE-binding proteins
In order to isolate the genes encoding JERE (CTCTTAGACCGCCTTCTTTGAAAG) binding proteins from T. cuspidata cDNA library, the yeast one-hybrid system was employed. This system includes two plasmids including the T. cuspidata cDNA library cloned in a fusion of pGADT7-Rec2 yeast expression vector with the GAL4 activation domain and pHIS-JERE fused to HIS3 selection marker as the bait. These two plasmids were transformed into the same strain Y187. 3.5 × 106 transformants of pHis-JERE were screened. Seventy-two positive transformants showed high growth on Leu-, Trp- and His- deficient medium with 130 mM 3-AT. On analysis with BLAST, only one insert cDNA may be derived from the AP2-type gene, via 5′ SMART RACE strategy. The full-length cDNA contained a 804 bp open reading frame of the JERE-binding cDNA (Fig. 1a), which encodes a 268-amino-acid protein with a calculated molecular mass of 23.8 kDa and a pI of 5.81 (Fig. 1b). After analysis with BLAST, the gene was named TcAP2 and accessed in GenBank (accession no. EU549860).
Nucleotide and deduced amino acid sequence of TcAP2 gene and schematic representation of TcAP2 protein. a Nucleotide and deduced amino acid sequence of TcAP2 (NM_102039). The AP2 domain is indicated by a thin line below the sequence. The putative acidic domain in C-terminal regions is underlined by wave-line. A broken line represents a putative nuclear localization signal located adjacent to N-terminal region. Thick underline indicates putative oligomerization site. b Schematic representation of the TcAP2 protein. The box represents the encoding open reading frame, starting from the first ATG codon and the line indicates putative untranslated region. Numbers below the boxes refer to positions of the last nucleotides. Black box represents the conservative AP2 domain. Numbers below it indicate the first and the last positions of amino acid residues, respectively. Dark-grey box with black triangle presents a putative nuclear localization signal. Hatched-box with black represents acidic domains and circles showed rich-acidic amino acids sites. The hatched-box with “+” indicates putative oligomerization site
Sequence and structural analyses of TcAP2
The nucleotide sequence analysis revealed that TcAP2 contains one conserved AP2-domain, some key sites to bind to the DRE element (Figs. 1 and 2) and a potential nuclear localization sequence in its N-terminal (Fig. 1). The C-terminal region of this protein might act as a transcriptional activation domain (Fig. 1). The serine (Ser)- and threonine (Thr)-rich region in the C-terminal is the phosphorylation site (Fig. 1a), which is mainly involved in the phosphorylation modification process to regulate transcription factor’s localization in the nucleus. The putative amino acid sequence of the TcAP2 protein was compared with AP2/EREBP proteins by Vector NTI suite 8.0 and CLUSTAL X biosoftwares (Fig. 2) showing a very high homology to Arabidopsis thaliana At1g21910 and Physcomitrella patens PpAP2 in the AP2 domain (Fig. 2a). It implies that their similar function is only present in the DNA binding ability. The nuclear localization region sequence and transcriptional activation domain showed a large difference between gymnosperms and monocots (Fig. 2a). The LWSF motif in the C-terminal (Fig. 1a) and DRE-binding site (Fig. 2a) suggest that TcAP2 belongs to the DREB-subfamily. Based on phylogenetic tree analysis with ten proteins, TcAP2 was different from Arabidopsis CBF subgroup and rice OsDREB1A-D (Fig. 2a). Therefore, TcAP2 might have a different function than other CBF family members.
Alignment and phylogenic analysis. a Alignment and phylogenic analysis of TcAP2 and other DRE binding proteins. The amino acid sequences shown are: TcAP2 EU549860, Arabidopsis AtAP2 (accession no. NM_102039), PpAP2 (accession no. XP_001763400), CBF3 (accession no. AB007787), CBF1 (accession no. AB007788), CBF2 (accession no. AB013817), OsDREB1A (accession no. AF300970), OsDREB1B (accession no. AF300972), OsDREB1C (accession no. AP001168), OsDREB1D (accession no. AF243384.1). The conserved AP2/EREBP domains are broken lined. Identical amino acid residues and conservative amino acid residues in the alignment are marked in black and gray background, respectively. A thin broken line and thick broken line show the location of the YRG and RAYD elements in the AP2 domain. The three anti parallel β-sheets and one α-helix were marked. The black filled circles showed two amino acids (Val105 and Ala110) in β-sheet for binding DRE element and one amino acid (Ala128) in α-helix for binding GCC Box. b Phylogenetic analysis of the deduced amino acid sequence of the TcAP2 gene and other CBF proteins from Arabidopsis and rice. The bootstrapped tree was produced by CLUSTAL X
Organ-specific expression and subcellular localization
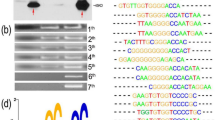
Total RNA was extracted from various organs for analysis of organ-specific expression. TcAP2 gene expression appeared to be constitutive like OsDREB1 (Dubouzet et al. 2003). The transcripts were detected at a higher level in young stems than that in roots, leafs and old stems (Fig. 3a).
Expression and subcellular localization of TcAP2. a Expression of TcAP2 in different organs of T. cuspidata. Each lane was loaded with 40 μg realtime RT-PCR product of total RNA extracted from the roots (lane 1), leaves (lane 2), young stems (lane 3) and old stems (lane 4). b Subcellular localization of TcAP2 protein. The plasmids expressing the fusion gene and GFP control were transformed into onion epidermal cells by particle bombardment. The fluorescence images of GFP control (the upper row) and the fusion protein (the lower row) under white light (the most left column, a and d) and UV light (the right two columns, b, c, e, and f) are shown (each of the bars = 1 μm)
Based on sequence analysis, a putative NLS is located in the N-terminal region of TcAP2 protein. In order to identify its subcellular localization, a plasmid p35S::TcAP2-GFP was constructed, and the fusion gene expression was promoted by a 35S promoter of cauliflower mosaic virus (CMV). The plasmid DNA for the fusion gene and GFP control was introduced into onion epidermal cells. From fluorescence observations, it is clear that TcAP2 is able to direct the fusion protein to localize at the nucleus of the transformed cells, whereas the GFP protein distributed nonspecifically in the cells transformed with vector plasmid DNA (Fig. 3b). These results indicated that the TcAP2 protein is located in the plant nucleus.
Expression patterns of TcAP2 responding to abiotic stresses
By quantitative real-time PCR analysis, TcAP2 transcripts quickly accumulated after MeJA plus SA treatment. The accumulation peaked 24 h after the treatment. In 8 h following high salinity treatment, the expression of TcAP2 accumulated and then decreased gradually when compared with the control treated with sterile water (Fig. 4). It appears that TcAP2 is a main switch in regulating stress-inducible target genes.
QRT-PCR analysis of TcAP2 transcripts. Expression profiles of the TcAP2 gene in response to various abiotic stresses and hormones. Seedlings were treated from 0 to 24 h with NaCl, cold (4°C), drought, MeJA plus SA, or ABA, respectively. Each lane was loaded with 8 μl QRT-PCR product obtained from each treatment
Discussion
A novel cDNA encoding JERE-binding protein, TcAP2, is isolated by the yeast one-hybrid strategy. Currently, the yeast one-hybrid system is regarded as an effective approach to isolate DNA-binding transcription factor genes and to investigate interactions between cis-elements and transcription factors (Liao and Fang 2000). This method is commonly applied to angiosperm species. However, the isolation of a transcription factor gene by the yeast one-hybrid system application was only achieved for a gymnosperm, Catharanthus roseus (van der Fits and Memelink 2000). This is possibly due to more difficulties in application of this method to gymnosperm species. The successful isolation of a transcription factor gene with the yeast one-hybrid system from T. cuspidata could contribute to develop a genetic manipulation approach of the taxol biosynthetic pathway. Since TcAP2 is inducible with MeJA, TcAP2 could function as a regulator in MeJA-responsive gene expression process.
Multiple alignment of TcAP2 with other AP2/EREBP transcription factors from various species indicates high conservation in the DNA-binding domain. The consensus AP2 domain forms a three-stranded anti-parallel β-sheet and an α-helix that bind to base pairs in the DNA major groove, and the binding site V14 in β-sheet of YRG element and A37 in α-helix of RAYD element also reflects AP2 domain’s conservative binding ability to DRE element and GCC Box. The conservation of the AP2/EREBP domain indicates the importance of this domain in the AP2-type genes for the TF function. Similar to other AP2 proteins, TcAP2 contains an alkaline amino acid-rich region that is presumed to be a nuclear localization signal (NLS) at the upstream of the DNA binding domain. This fact suggests that TcAP2 might be transported into the nucleus through the nuclear pore complex as implied by subcellular localization in this research.
The process leading to stress-inducible gene expression includes the complicated sequences of events, which usually go through elicitors, signal amplification and integration, and signal response by the activation of specific genes. Most of the events are regulated at the transcriptional level (Latini et al. 2007). The dehydration responsive element (DRE, 5′-TACCGACAT-3′) binding transcription factor (DREB) is a major subfamily in the AP2/EREBP family, which plays a key role in resistance of plants to various environmental stimuli, such as cold, high salinity and drought. As the expression profile of rice OsDREB1A (Dubouzet et al. 2003), TcAP2 was not inducible with drought or ABA, which suggests that its signal transduction pathway in Taxus may be ABA-independent. Since ABA is commonly produced under drought conditions for generating drought tolerance (Seki et al. 2007), the drought stress non-sensitive genes may also have no response to ABA treatment. On the other hand, TcAP2 expression was induced by low temperature and high salinity in this study. The expression pattern of RCBF2 was different from those of the CBF genes in Arabidopsis. It is also similar to OsDREB1 subfamily in rice, which was induced within 5 h after salt and cold treatment. These facts suggest that TcAP2 could be classified into DREB1 subfamily that is involved in cold stress (Liu et al. 1998; Pradeep et al. 2006). Furthermore, RD29A/LTI78/COR78, KIN1, COR6.6/KIN2 and COR47/RD17 genes do not require ABA for their expression under drought, salt and cold stress conditions (Shinozaki and Yamaguchi-Shinozaki 2000). The responsiveness of TcAP2 to high salinity and cold indicates that TcAP2 plays an important role in resistance to abiotic stresses. Apparently, TcAP2 is best inducible with MeJA plus SA if compared to other stress treatments. This protein is definitely a MeJA -responsive transcription factor and its nuclear localization has been validated by this study.
References
Aerts RJ, Gisi D, De Carolis E (1994) Methyl jasmonate vapor increases the developmentally controlled synthesis of alkaloids. Plant J 5:635–643
Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM (2003) A role for the GCC-Box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol 132:1020–1032
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:151–158
Gantet P, Imbault N, Thiersault M, Doireau P (1998) Necessity of a functional octadecanoic pathway for indole alkaloid synthesis by Catharanthus roseus cell suspensions cultured in an auxin-starved medium. Plant Cell Physiol 39:220–225
Laskaris G, Bounkhay M, Theodoridis G (1999) Induction of geranylgeranyl diphosphate synthase activity and taxane accumulation in Taxus baccata cell cultures after elicitation by methyl jasmonate. Plant Sci 147:1–8
Latini A, Rasi C, Sperandei M, Cantale C, Iannetta M, Dettori M, Ammar K, Galeffi P (2007) Identification of a DREB-related gene in Triticum durum and its expression under water stress conditions. Ann Appl Biol 150:187–195
Liao M, Fang F (2000) Yeast one-hybrid system—one effective method studying DNA-protein interaction. Acta Acad Med Sin 22:388–391
Lin RM, Zhao WS, Meng XB, Peng YL (2007) Molecular cloning and characterization of a rice gene encoding AP2/EREBP-type transcription factor and its expression in response to infection with blast fungus and abiotic stresses. Physiol Mol Plant Pathol 70:60–68
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Liu JG, Zhang Z, Qin QL, Peng RH, Xiong AS, Chen JM, Xu F, Zhu H, Yao QH (2007) Isolated and characterization of a cDNA encoding ethylene-responsive element binding protein (EREBP)/AP2-type protein, RCBF2, in Oryza sativa L. Biotechnol Lett 29:165–173
Memelink J, Verpoorte R, Kijne JW (2001) ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci 65:212–219
Menke FL, Champion A, Kijne JW, Memelink J (1999) A novel jasmonate- and elicitor-responsive element in the periwinkle secondary metabolite biosynthetic gene STR interacts with a jasmonate- and elicitor-inducible AP2-domain transcription factor, ORCA2. EMBO J 18:4455–4463
Moon WJ, Yoo BS, Kim DI, Byun SY (1998) Elicitation kinetics of taxane production in suspension cultures of Taxus baccata Pendula. Biotechnol Tech 12:79–81
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15:473–497
Pradeep K, Agarwal PA, Reddy MK, Sopory SK (2006) Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 25:1263–1274
Seki M, Umezawa T, Urano K, Shinozaki K (2007) Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol 10(3):296–302
Shinozaki K, Yamaguchi-Shinozaki K (2000) Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr Opin Plant Biol 3:217–223
Sun XE, Mei XG, Gong W, Zhang ZN (2001) Effects of the combination of methyl jasmonate with salicylic acid or fungal elicitor on the cell suspension of Taxu chinensis. Biotechnol 11:10–13
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
Vázquez-Flota FA, De Luca V (1998) Jasmonate modulates development- and light-regulated alkaloid biosynthesis in Catharanthus roseus. Phytochemistry 49:395–402
Vom Endt D, Kijne JW, Memelink J (2003) Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry 61:107–114
Walker K, Croteau R (2000) Molecular cloning of a 10-deacetylbaccatin III-10-O-acetyl transferase cDNA from Taxus and functional expression in Escherichia coli. Proc Natl Acad Sci USA 97:583–587
Walker K, Long R, Croteau R (2002) The final acylation step in Taxol biosynthesis: Cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc Natl Acad Sci USA 99:9166–9171
Yazaki K, Takeda K, Tabata M (1997) Effects of methyl jasmonate on shikonin and dihydroechinofuran production in Lithospermum cell cultures. Plant Cell Physiol 38:776–782
Acknowledgements
This work was supported by “973” Program (2007CB108805) and Shanghai Science and Technology Committee (08391911800).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Dai, Y., Qin, Q., Dai, D. et al. Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata . Biotechnol Lett 31, 1801–1809 (2009). https://doi.org/10.1007/s10529-009-0068-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-009-0068-4