Abstract
Expression kinetics of six cellulase and four expansin-related genes were studied in the hypercellulolytic Trichoderma reesei CL847 mutant in response to Solka Floc cellulose and soluble inducers. Real-time PCR showed a parallel increase of transcript levels for the cellulase genes cbh1/cel7a, egl1/cel7b, egl4/cel61a, the β-glucosidase genes bgl1/cel3a, bgl2/cel1a, and the swo1 gene, encoding the cell-wall loosening protein swollenin. To evaluate a putative implication of three newly identified expansin/family 45 endoglucanase-like (EEL) proteins in lignocellulose degradation, their expression was also analysed. Only eel2 was found to be transcribed under the present conditions, and showed constitutive expression similar to the endoglucanase encoding cel5b gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The filamentous fungus Trichoderma reesei is an efficient producer of cellulolytic and hemicellulolytic enzymes. It has a great potential for industrial processes, most notably for the production of ethanol from lignocellulose. Mutants with high secretion capacity have been isolated, such as the T. reesei CL847 strain which was selected after several rounds of mutagenesis for its protein secretion capacity of up to 40 g l−1 (Durand et al. 1988).
The cellulolytic system of T. reesei is one of the most extensively studied. Seven cellulase and two β-glucosidase enzymes have been characterized (for a review see Aro et al. 2005), and sequencing revealed the presence of ten cellulase and 16 hemicellulase genes in the T. reesei genome (Martinez et al. 2008). Expression of cellulases is regulated at the transcriptional level, and most cellulase and some hemicellulase genes have been shown to be coordinately induced by cellulose or soluble molecules such as sophorose (Ilmen et al. 1997; Foreman et al. 2003; Margolles-Clark et al. 1997).
In addition to hydrolytic enzymes, other proteins might be involved in lignocellulose degradation, such as the recently characterized swollenin, a protein related to plant expansins which are proteins with a cell wall loosening action (Saloheimo et al. 2002b). Swollenins occur in other fungi, such as Trichoderma asperellum (Brotman et al. 2008), Aspergillus fumigatus, T. virens and T. atroviride. The swollenin cloned from T. pseudokoningii possesses glycohydrolytic activity (Yao et al. 2008).
In T. reesei, three other sequences with similarity to the expansin/family-45 endoglucanase-like domain (PS50842) were found, in addition to the swo1 gene, (Martinez et al. 2008), and were thus called eel1, eel2 and eel3. Unlike swollenin, they neither contain a fungal cellulose-binding domain (CBD) nor the C-terminal CBD of expansins, and their function is still unknown.
To analyse the expression of cellulase genes and genes coding for expansin-related proteins and in the hypercellulolytic T. reesei CL847 strain, expression profiles were established by real-time PCR in response to different inducers. The hypothesis of a possible implication of swollenin and expansin-related genes in lignocellulose degradation is investigated and the results discussed with regard to the promoter structure of the studied genes.
Materials and methods
Strain and media
The Trichoderma reesei CL847 strain, selected for its high level of secreted cellulases (Durand et al. 1988), was used in this study. Cultures were grown at 30°C in shake flasks with minimal medium containing 15 g KH2PO4 l−1, 5 g (NH4)2SO4 l−1, 0.6 g CaCl2 l−1, 0.6 g MgSO4 l−1, trace elements (5 mg FeSO4 l−1, 1.4 mg MnSO4 l−1, 1.4 mg ZnSO4 l−1, 3.7 mg CoCl2 l−1) and a carbon source. After 48 h of preculture with 50 g glucose l−1 as a carbon source, the mycelium was filtered on a sterile gaze, washed three times with minimal medium and resuspended in minimal medium containing 20 g glycerol l−1. Aliquots of 50 ml were introduced in 250 ml Erlenmeyer flasks and induction was started by addition of: 1 mM lactose, 1 mM cellobiose, 1 mM l-sorbose, 1 mM sophorose or 1% (w/v) Solka-Floc, respectively. Samples were taken at the time points indicated, centrifuged and stored at −80°C until use.
RNA preparation
Samples were ground in liquid nitrogen and about 400 mg of mycelium were used for extraction of RNA. Total RNA was extracted using the TRI Reagent Solution (Applied Biosystems) and purified with an additional on-column DNase digestion using the RNeasy Mini Kit (QIAGEN) according to the manufacturer’s instructions. Integrity of RNA preparations was checked with an Agilent 2100 bioanalyzer and quantification was done on a ND-1000 spectrophotometer (Nanodrop, Thermo Scientific, DE, USA). Reverse transcription was performed with 500 ng of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad).
Real-time PCR
For real-time PCR, the iQSYBR Green Supermix (Bio-Rad) was used with 320 nM of forward and reverse primers (see Supplementary Table 1) and 5 μl of 50-fold diluted cDNA in a final volume of 25 μl. Thermocycling was carried out in a MyIQ thermocycler. Every RT-PCR was realised in duplicates on 96 well microplates including negative (water) and positive controls (genomic DNA). and analysed with Bio-Rad IQ5 software. In addition, for all samples negative controls using an RT mix without reverse transcriptase were performed in order to exclude contamination of samples with genomic DNA. Primers were validated by creating standard curves with tenfold serial dilutions of genomic DNA, a primer pair being considered as valid if amplification efficiency ranged between 85 and 115%. Melt curves were realised after each qPCR run, to confirm the specificity of amplification and the absence of primer dimers. The real-time PCR program consisted of an initial denaturation step at 95°C and 40 amplification cycles of 10 s at 95°C and 30 s at 60°C.
In silico sequence analysis
All sequences of expansins and expansin-like modules of T. reesei were identified, analysed and annotated by the Carbohydrate-Active EnZYmes database at AFMB Marseille (CAZy, http://www.cazy.org) (Cantarel et al. 2009). The three newly identified sequences were designated EEL1, EEL2 and EEL3, corresponding to protein IDs 71390, 23115, and 104079, respectively of second annotated version of T. reesei genome (Triree2) in the Joint Genome Institute (http://genome.jgi-psf.org/Trire2/Trire2.home.html). For promoter analysis, 1,500 bp of the 5′ UTR of 4 cellulase genes (cbh1/cel7a, egl1/cel7b, egl4/cel61a, cel5b), 2 β-glucosidases (bgl1/cel3a, bgl2/cel1a) as well as the swo1 and eel1, eel2, eel3 genes were also obtained from the JGI website. Sequencing confirmed that no mutation was present in the studied genes and the genes of transcription factors ACE1, ACE2 and XYR1 in the CL847 strain with respect to the published genome sequence. These promoter regions were analysed with RSA-Tools (http://rsat.ulb.ac.be/rsat/). Structure predictions were realised using the threading server Phyre at http://www.sbg.bio.ic.ac.uk/phyre/.
Results and discussion
The time course of expression of 6 cellulase genes, swollenin and expansin-like genes was followed for 48 h on five substrates, well known for inducing cellulases: lactose, sophorose, cellobiose, Solka Floc and l-sorbose which is the only monosaccharide shown to induce cellulase genes (Nogawa et al. 2001). It is generally considered that cellulase genes are coordinately expressed, but there are differences in time-course of induction and/or extent exist between certain genes in response to different inducers (Foreman et al. 2003). Cellulase genes representative of different profiles were therefore chosen: cbh1/cel7a, egl1/cel7b (stronger induced by sophorose than cellulose), egl4/cel61a (equal induction by cellulose and sophorose in the QM6a strain) and cel5b (no regulation evident). In addition, β-glucosidases bgl1 and bgl2, as well as swo1 were also included. As nothing was known about the expression behaviour of eel genes, their cDNA was amplified from glycerol or cellulose grown cultures in a preliminary experiment. However, only eel2 cDNA could be detected (Fig. 1). Therefore, only this gene was included in the following expression profiling by reverse transcription real-time PCR.
Expression of eel1, eel2 and eel3 genes in the T. reesei CL847 strain. RT-PCR was performed with RNA extracted from T. reesei cultures grown on glycerol (lanes 3, 6 and 9) or glycerol/Solka-Floc (lanes 4, 7 and 10) for 24 h. Amplification of the corresponding genes using genomic DNA was included as a control (lanes 2, 5 and 8) Lane 1: 2Log DNA Ladder
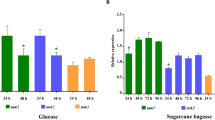
Results show that two expression profiles could be distinguished: 1, Parallel increase of transcript levels of cbh1, egl1, egl4, bgl1, bgl2 and swo1 with a maximum at 4 to 24 h depending on the substrate (Fig. 2a); and 2, no clear induction on any substrate for cel5b and eel2 (Fig. 2b). Considering the first profile type, exposure to sophorose and Solka Floc resulted in the most important increases. cbh1 and egl1 mRNA levels generally showed the highest increases of all tested genes: up to 500-fold on sophorose, between 100 and 200-fold on Solka Floc cellulose and lactose, whereas induction was less important in the presence of l-sorbose and cellobiose. swo1 also showed large increases of its transcript levels on sophorose and Solka Floc (up to 150-fold) but was only a tenth of this on the three other substrates. egl4 and bgl2 mRNA levels were increased to a lesser extent, between about 40 and 10-fold, respectively.
Variations of steady state mRNA levels of cellulase and expansin-like genes in T. reesei CL847 strain from 2 to 48 h of exposure to the indicated substrates. Expression is calculated relative to transcription levels of a glycerol grown culture at the corresponding time point. All values are normalised using the housekeeping gene (β-actin) expression for each time point under the same conditions. Values reported are means of two independent experiments and at least two qPCR experiments for each RNA preparation. a Relative transcript levels of cbh1, egl1, egl4, bgl1, bgl2 and swo1 in response to the 5 substrates. b Relative transcript levels of cel5b and eel2
The strictly correlated expression of cbh1, egl1, egl4 and swo1 genes can be clearly seen on Fig. 3a, and is in contrast to the genes of the second profile which show no induction (Fig. 3b). It is very likely that expression of the genes belonging to the first profile type is induced by the XYR1 transcription factor, as all promoters, except the bgl1 promoter, contain one or more GGCTAA motif(s), the binding site for XYR1 (Rauscher et al. 2006). The bgl1 gene has no GGCTAA sequence, but instead possesses one GGCTAT and two AGGCTA motifs. A GGCTAT sequence was shown to be necessary for full induction of the egl3 by XYR1 (Shida et al. 2008). Thus, it is possible that XYR1 binds to this motif also in bgl1 promoter and maybe others, i.e. those of cbh1, egl1, bgl2, eel2 and swo1 which also contain this sequence.
mRNA increases of selected genes related to their transcript level increases at 2 h after onset of induction. a Crossplot of relative increases of transcript levels of correlated genes against the relative increase of cbh1 mRNA. b Crossplot of relative increases of transcript levels of non-correlated genes against the relative increase of cbh1 mRNA. Values from experiments on all substrates are plotted on (a) and (b). c Relative mRNA level increases on Solka Floc cellulose of cbh1, egl1, egl4 and bgl1. d Relative mRNA level increases of cbh1, egl4, swo and bgl2 at 4 and 10 h on lactose, sorbose and cellobiose
Time course of steady state mRNA levels was also different on the five substrates: on lactose and sophorose, the highest transcript levels were reached later (after 10 h) than in the presence of l-sorbose and cellobiose (4 h). They decreased to control levels at 24 and 10 h, respectively. This fast decrease had also been observed in earlier studies (Mach et al. 1995) and could be due to the low concentrations of each inducer added (1 mM) which was probably rapidly consumed.
Induction kinetics were slower on Solka Floc cellulose, with maximum transcript levels being attained at 10 to 24 h, somewhat later than on soluble substrates. This delay in induction reflects the necessity of formation of a soluble inducer before onset of the signalling cascade and has already been observed (Carle-Urioste et al. 1997). It is also worth noting the earlier decrease of bgl1 transcript levels (at 24 h) on cellulose as compared to the mRNA levels of other genes which were still increased at 48 h compared to their level at 2 h (Fig. 3c). BGL1 participates in the formation of inducer from cellulose (Fowler and Brown 1992) and the present results underline this function.
BGL1 is also necessary for full expression of cellulases on sophorose (Mach et al. 1995). The importance of bgl1 in the presence of this substrate is also corroborated by our results: sophorose induced bgl1 to a higher extent than all other tested substrates (570-fold, Fig. 2a). In this context, an interesting feature is the presence of a CGTAAA motif at −245 bp of the bgl1 promoter which corresponds to the position of the promoter fragment shown to bind proteins of extracts of sophorose grown mycelia (Mach et al. 1995). These motifs are also present on bgl2, egl1 and swo1 promoters and could be involved in binding regulatory proteins induced by sophorose.
Expression kinetics of bgl2 slightly differed from other cellulase genes on lactose, cellobiose and l-sorbose. Its steady-state mRNA levels started to increase earlier but then remained constant for 10 h before decreasing (corresponding to a ratio of mRNA increase of about 1 compared to the value at 2 h in Fig. 3d), in contrast to the swollenin and cellulase genes which continue to increase until 10 h on these substrates. The regulation of this gene was shown to be different from cbh1 or bgl1, since it was not dependent on XYR1 (Stricker et al. 2006). The XYR1 binding sites present in its promoter could also be contacted by ACEII which was shown to bind the same motif in the cbh1 promoter (Aro et al. 2001). An ace2 deletion strain indeed showed a weaker and delayed expression of the bgl2 gene (Saloheimo et al. 2002a).
The cel5b gene, coding for an endoglucanase gene, and the eel2 gene did not show any clear induction on either substrate tested (Figs. 2b and 3b). Considering cel5b, this is in agreement with results from Foreman et al. (2003) who reported a constitutive, but low expression of this cellulase in response to cellulose or sophorose. The expression of eel2 seems also to be constitutive. After 2 h exposure to lactose and cellobiose, eel2 transcript levels were doubled. Unlike cel5b, which contains no XYR1-binding motif, the eel2 promoter contains two perfect match XYR1 binding sites (one on the coding, one on the reverse strand), one GGCTAT and one AGGCTA motif. However, under the tested conditions this gene was clearly not regulated like the major cellulases, suggesting that binding and/or induction by XYR1 might not be functional in this promoter. Alternatively, the repressing protein ACE1 could compete with XYR1 for the same binding sites, as is the case in the promoter of the xyn1 gene coding for xylanase (Rauscher et al. 2006).
Whereas swollenin expression kinetics were shown to follow exactly the behaviour of cellulase genes, suggesting its possible implication in plant cell wall degradation, the expression data for eel genes were shown to be very different: while eel2 seems to be constitutively expressed, eel1 and eel3 were not expressed under the present conditions. In contrast to eel2, their promoters do not contain XYR1-binding motifs which could explain the lack of induction and the missing basal transcription of eel1 and eel3. Indeed, the expression of several cellulase and xylanase genes was shown to be strictly dependent on XYR1 (Stricker et al. 2006). However, there are elements supporting that these genes are not silenced, such as the presence of CCAAT boxes in the promoter sequence. In addition, eel1 and eel3 genes are located in “CAZy clusters”, locations in the genome with enhanced occurrence of CAZy genes, containing a majority of glycoside hydrolases (Martinez et al. 2008). This could suggest a function in polysaccharide degradation.
However, looking at the predicted protein structure of the EEL proteins, they show important similarities with plant expansins, pointing to a different role: the overall sequence identity to expansins is slightly higher (25–30%) than to swollenin (20–22%) and the predicted tertiary structures of EEL proteins have significant similarity (16–17%) to the tertiary structure of Zea mays EXPB1. Another interesting finding is the significant similarity (88% identity) of EEL3 with MRSP1 of Trichoderma virens, a protein with a possible role in the modification of the cell wall structure in germinating conidia (Mukherjee et al. 2006). This suggests that EEL3 could have a similar function in T. reesei.
Conclusion
In the present study, we performed a detailed comparative expression analysis of six genes involved in plant cell wall degradation during T. reesei growth on five carbon sources. The present work also analyses, for the first time, expression of expansin-related genes in parallel to cellulase genes, as a first approach to study a potential implication of the gene products in lignocellulose degradation. Expression studies of EEL proteins during different physiological states of T. reesei, for example in germinating conidia, could help determining their physiological role.
References
Aro N, Saloheimo A, Ilmen M et al (2001) ACEII, a novel transcriptional activator involved in regulation of cellulase and xylanase genes of Trichoderma reesei. J Biol Chem 276:24309–24314
Aro N, Pakula T, Penttila M (2005) Transcriptional regulation of plant cell wall degradation by filamentous fungi. FEMS Microbiol Rev 29:719–739
Brotman Y, Briff E, Viterbo A et al (2008) Role of swollenin, an expansin-like protein from Trichoderma, in plant root colonization. Plant Physiol 147:779–789
Cantarel BL, Coutinho PM, Rancurel C et al (2009) The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238
Carle-Urioste JC, Escobar-Vera J, El-Gogary S et al (1997) Cellulase induction in Trichoderma reesei by cellulose requires its own basal expression. J Biol Chem 272:10169–10174
Durand H, Clanet M, Tiraby G (1988) Genetic improvement of Trichoderma reesei for large scale cellulase production. Enzyme Microb Technol 10:341–346
Foreman PK, Brown D, Dankmeyer L et al (2003) Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J Biol Chem 278:31988–31997
Fowler T, Brown RD Jr (1992) The bgl1 gene encoding extracellular beta-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol Microbiol 6:3225–3235
Ilmen M, Saloheimo A, Onnela ML et al (1997) Regulation of cellulase gene expression in the filamentous fungus Trichoderma reesei. Appl Environ Microbiol 63:1298–1306
Mach RL, Seiboth B, Myasnikov A et al (1995) The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellulose-inducible beta-glucosidase involved in cellulase induction by sophorose. Mol Microbiol 16:687–697
Margolles-Clark E, Ilmen M, Penttila M (1997) Expression patterns of ten hemicellulase genes of the filamentous fungus Trichoderma reesei on various carbon sources. J Biotechnol 57:167–179
Martinez D, Berka RM, Henrissat B et al (2008) Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat Biotechnol 26:553–560
Mukherjee PK, Hadar R, Pardovitz-Kedmi E et al (2006) MRSP1, encoding a novel Trichoderma secreted protein, is negatively regulated by MAPK. Biochem Biophys Res Commun 350:716–722
Nogawa M, Goto M, Okada H et al (2001) l-Sorbose induces cellulase gene transcription in the cellulolytic fungus Trichoderma reesei. Curr Genet 38:329–334
Rauscher R, Wurleitner E, Wacenovsky C et al (2006) Transcriptional regulation of xyn1, encoding xylanase I, in Hypocrea jecorina. Eukaryot Cell 5:447–456
Saloheimo M, Kuja-Panula J, Ylosmaki E et al (2002a) Enzymatic properties and intracellular localization of the novel Trichoderma reesei beta-glucosidase BGLII (cel1A). Appl Environ Microbiol 68:4546–4553
Saloheimo M, Paloheimo M, Hakola S et al (2002b) Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur J Biochem 269:4202–4211
Shida Y, Furukawa T, Ogasawara W et al (2008) Functional analysis of the egl3 upstream region in filamentous fungus Trichoderma reesei. Appl Microbiol Biotechnol 78:515–524
Stricker AR, Grosstessner-Hain K, Wurleitner E et al (2006) Xyr1 (xylanase regulator 1) regulates both the hydrolytic enzyme system and d-xylose metabolism in Hypocrea jecorina. Eukaryot Cell 5:2128–2137
Yao Q, Sun TT, Liu WF et al (2008) Gene cloning and heterologous expression of a novel endoglucanase, swollenin, from Trichoderma pseudokoningii S38. Biosci Biotechnol Biochem 72:2799–2805
Acknowledgements
J. Verbeke was supported by a CIFRE scholarship from the “Association Nationale de Recherche Technique”. The authors thank Antoine Margeot for support in promoter analysis, fruitful discussions and critical remarks, as well as Frédéric Monot for revision of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Verbeke, J., Coutinho, P., Mathis, H. et al. Transcriptional profiling of cellulase and expansin-related genes in a hypercellulolytic Trichoderma reesei . Biotechnol Lett 31, 1399–1405 (2009). https://doi.org/10.1007/s10529-009-0030-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-009-0030-5