Abstract
Two regulator genes, metK1-sp and afsR-sp, from Streptomyces peucetius ATCC 27952 were heterologously expressed in S. venezuelae ATCC 15439, to produce 14-membered pikromycin antibiotics. The production of pikromycin was increased by 1.6-fold and 2.6-fold by the expression of metK1-sp and afsR-sp, respectively. The overexpression of metK1-sp and afsR-sp in S. venezuelae stimulated the expression of the pathway-specific regulatory genes, pikD and ketosynthase, as demonstrated by RT-PCR. The elevated transcripts of the pikD and ketosynthase genes were consistent with the enhanced production of pikromycin.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pikromycin (Fig. 1), a 14-membered macrolide antibiotic, is structurally related to the semi-synthetic ketolide antibiotics, which are active against multidrug-resistant respiratory pathogens (Agouridas et al. 1998). Due to the wide range of biological activities, the biosynthesis of the pikromycin series of antibiotics has attracted significant interest. Pikromycin was initially isolated from Streptomyces venezuelae ATCC 15439. However, low levels of pikromycin production have led to the development of a strain genetically engineered to produce higher levels of pikromycin. Many of the previous studies on the enhancement of secondary metabolites were focused on either the homologous or heterologous expression of positive regulators. Among the positive regulators, S-adenosylmethionine synthetase (MetK) and a global regulatory gene, afsR, were previously reported to be involved in the enhanced production of various secondary metabolites from different Streptomyces species (Lee et al. 2002; Kim et al. 2003; Okamoto et al. 2003; Wang et al. 2007).
MetK plays an important role in the conversion of ATP and l-methionine to S-adenosyl-l-methionine (SAM), which acts as a major methyl group donor for numerous transmethylation reactions. Furthermore, SAM regulates antibiotic biosynthesis in a manner independent of its role as a methyl donor, in which it acts as a direct intracellular signaling molecule for Streptomyces (Zhao et al. 2006). It also activates the transcriptional activators responsible for the induction of antibiotic synthetic genes, thereby increasing the production of antibiotics (Kim et al. 2003). Recent reports have suggested that SAM induces several ABC transporters in order to modulate secondary metabolism and morphological development in S. coelicolor (Shin et al. 2007).
On the other hand, afsR is a pleiotropic, global regulator that controls the production of secondary metabolites in Streptomyces species. AfsR is widely distributed as the AfsK-AfsR system in Streptomyces and influences the secondary metabolism and morphogenesis of the organism (Horinouchi 2003). As a transcriptional activator, AfsR greatly enhances its DNA-binding activity toward the respective promoter region, thereby enhancing the transcriptional activation for the biosynthesis of secondary metabolites in streptomyces (Tanaka et al. 2007).
On the basis of these findings, we heterologously expressed these two genes, metK1-sp (GenBank accession no. CAJ43278) and afsR-sp (GenBank accession no. AJ786384) from S. peucetius ATCC 27952 (Parajuli et al. 2005), individually in S. venezuelae ATCC 15439 in order to analyze the production of pikromycin. Reverse transcription-polymerase chain reaction (RT-PCR) was used to study the effects of the overexpression of these genes on the regulation of pikromycin biosynthetic gene clusters.
Materials and methods
Bacterial strains and growth conditions
E. coli XL1 Blue MRF′ (Stratagene) was used for DNA amplification. E. coli ET 12567 was used to propagate non-methylated DNA. S. venezuelae ATCC 15439 was the parent strain and was grown in R2YE media for the preparation of protoplasts and isolation of plasmid DNA. Protoplast transformation was carried out according to the standard protocol (Kieser et al. 2000). To isolate pikromycin, wild type and transformant strains of S. venezuelae were cultured SCM liquid medium (15 g soluble starch, 20 g soytone, 1.5 g yeast extract, 0.1 g calcium chloride and 10.5 g MOPS/l distilled water), supplemented with trace amounts of FeSO4 (12.5 mg/l) at 28°C for 60 h (Lee et al. 2006).
To study the growth rate, wild type and transformant strains of S. venezuelae were cultured in 50 ml SCM media at 28°C after 36 h of incubation in R2YE seed media. The cells were collected at intervals of 12 h by centrifuging 50 ml culture broth of each strain at 6000 × g. Cell pellets were washed with distilled water and dried at 72°C in vacuum oven to constant weight. Supernatants obtained after removing cell pellets were used to study the rate of pikromycin production. Values were the average of four determinations. For the isolation of total RNA, S. venezuelae and transformants were grown in APM medium (60 g glucose, 8 g yeast extract, 20 g malt extract, 2 g NaCl, 15 g MOPS and 10 ml trace elements/l distilled water).
Cloning and heterologous expression of metK1-sp and afsR-sp
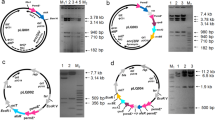
The expression vector, pIBR25, under the control of the ermE* promoter, which leads to the expression of DNA in Streptomyces species, was used for cloning (Basundhara et al. 2004). The metK1-sp gene was amplified by PCR of genomic DNA from S. peucetius, using metK1-F and metK1-R primers. The PCR product (1,209 bp) of the metK1-sp gene was cloned into the BamHI and HindIII sites of pIBR25 to produce the recombinant plasmid pSIBR. Similarly, afsR-sp from S. peucetius was amplified using afsR-pF and afsR-pR primers. The PCR product (2,946 bp) was cloned into the EcoRI and HindIII sites of pIBR25 to produce the recombinant plasmid pGIBR (Parajuli et al. 2005). The primers used in this study are listed in Table 1. Recombinants pSIBR and pGIBR (Fig. 2a and b) were transformed into wild type S. venezuelae (SV) to obtain S. venezuelae/pSIBR (SVSIBR) and S. venezuelae/pGIBR (SVGIBR), respectively. The transformation was accomplished by following the standard protocol (Kieser et al. 2000).
Recombinant Plasmids. (a) pSIBR, metK1-sp from S. peucetius cloned into the pIBR25 vector under the control of the ermE* promoter. (b) pGIBR, afsR-sp from S. peucetius cloned into the pIBR25 vector under the control of the ermE* promoter. (c) Antibacterial activity assay of the isolated pikromycin against B. subtilis. W, isolates from SV; S, SVSIBR; and A, SVGIBR. (d) Comparison of pikromycin production from SV, SVSIBR and SVGIBR. Pikromycin was extracted from each strain, after being grown at 28°C for 60 h in SCM medium, and analyzed by HPLC and LC/MS analysis. The experiment was averaged from 4 different extractions
HPLC analysis of pikromycin production
Each 50 ml culture of SV, SVSIBR and SVGIBR was centrifuged at 6000 × g for 15 min. The supernatants were extracted with 2 vol ethyl acetate. The extract was dried in a rotary evaporator and was reconstituted by 1.5 ml methanol. A 15 μl of the extract was analyzed by HPLC using a reverse-phase C18 column with 80% acetonitrile in 5 mM ammonium acetate buffer containing 0.05% acetic acid, adjusted to pH 8 with NH4OH at 1 ml/min. Detection was at 220 nm. The major peak corresponding to pikromycin was confirmed by LC/MS analysis. The production was averaged from four separate cultivations and extractions. The antibacterial activity of SV, SVSIBR, and SVGIBR was assayed against Bacillus subtilis ATCC 23857 (Lee et al. 2006).
RNA sample preparation and RT-PCR analysis
Each aliquot (5 ml) of the cultures grown for about 48 h was suspended in RNA Protect Bacteria Reagent (Qiagen) for 5 min in order to isolate total RNA. An RNeasy Mini kit (Qiagen) was used for RNA isolation according to the manufacturer’s instructions. Contaminant DNA in the sample was eliminated by using RNase-free DNase (Qiagen) and verified by PCR analysis using the RNA as the template. The total RNA concentration and purity were determined from the 260/280 nm ratio. RT-PCR was performed with a QuantiTech SYBER Green RT-PCR Kit (Qiagen). The primers used for RT-PCR are described in Table 1.
Equal amounts (5.54 μg) of RNA from each sample were used for RT-PCR analysis. The reaction conditions were as follows: first strand cDNA synthesis at 50°C for 30 min; initial denaturation at 95°C for 15 min; and 45 cycles of 1 min at 94°C, 1 min at 63°C and elongation at 72°C for 2 min. The RT-PCR products were electrophoresed on a 1% agarose gel and visualized using ethidium bromide staining. Negative controls were carried out with Taq DNA polymerase without reverse transcripts to confirm that the amplified products were not derived from chromosomal DNA, which could contaminate the RNA preparations. The 16S rRNA gene from S. venezuelae was used as a positive internal control.
Results and discussion
To enhance the production of pikromycin from S. venezuelae wild strain, we chose two genes, namely metK1-sp and afsR-sp, which encode for the MetK protein and global regulatory protein, respectively. Both metK1-sp (1.2 kb) and afsR-sp (2.9 kb) were amplified from S. peucetius using the metK1-F/metK1-R and afsR-F/afsR-R primers, respectively (data not shown). The PCR products of metK1-sp and afsR-sp were separately cloned into pIBR25 to construct recombinant plasmids pSIBR and pGIBR, respectively, as described in the Materials and methods section. The recombinants pSIBR and pGIBR were then transformed into SV to generate SVSIBR and SVGIBR, respectively. Pikromycin produced by strains SV, SVSIBR and SVGIBR is given in Figs. 2d and 3 and confirmed by LC/MS. Production of pikromycin was enhanced by 1.6-fold in SVSIBR and 2.6-fold in SVGIBR in comparison with pikromycin production by SV. Thus, the biological activities of the pikromycin obtained from SV, SVSIBR and SVGIBR were tested against B. subtilis, and the diameters of the inhibition zones in the two mutants showing the least growth were indicative of high levels of pikromycin rather than SV (Fig. 2c). These results revealed that metK1-sp and afsR-sp had a positive influence on the production of pikromycin.
SVSIBR and SVGIBR had higher growth yields than SV itself with. Maximum growth at 48–60 h (Fig. 4a). However, the influence of afsR-sp on the growth rate as well as the production of pikromycin was greater than that induced by metK1-sp (Fig. 4b). Previously, the overexpression of metK stimulated the expression of the pathway-specific regulatory gene actII-ORF4 for the production of actinorhodin in S. coelicolor A3 (2) (Okamoto et al. 2003). Furthermore, AfsR serves as a transcriptional activator of afsS, which activates the pathway-specific genes actII-ORF4, in the actinorhodin biosynthetic gene cluster (Lee et al. 2002). Similarly, strains overexpressing metK1-sp and afsR-sp were most likely to induce the biosynthetic pathway of pikromycin for the enhanced production of pikromycin. The influence of metK1-sp and afsR-sp on the expression levels of two genes, ketosynthase and pikD, involved in the production of pikromycin were assayed by RT-PCR analysis. Ketosynthase is a domain from PikAI that catalyses polyketide chain extension in modular multifunctional polyketide synthases (PKS) (Bisang et al .1999), whereas pikD operates as a pathway-specific activator of the pikromycin biosynthetic gene cluster (Wilson et al . 2001). The RT-PCR results showed that the transcription levels of both the ketosynthase and pikD genes were substantially increased in SVSIBR and SVGIBR when compared to SV (Fig. 5).
Comparison of the growth rate (a) and production rate (b) of SV, SVSIBR and SVGIBR. (a) Cell pellets were collected at 12-h intervals, starting from 12 h to 96 h. (b) Isolation of the compound was carried out at the same intervals and quantification was performed via HPLC analysis. Closed circle, SV; closed square, SVSIBR; and closed triangle, SVGIBR
Quantitative RT-PCR analysis of ketosynthase and pikD which involved in pikromycin biosynthetic pathway using equal amount (5.54 μg) of total RNA isolated from SVSIBR (a) and SVGIBR (b). 16S rRNA gene was used as internal control. M, DNA marker. The relative intensities of the transcriptional bands were measured using a PhosphoImager and were calculated with the Imagequant program. Lane 1, use total RNA from wild type (SV); lane 2, use total RNA from mutants (SVSIBR and SVGIBR)
In S. venezuelae, metK is located downstream of pikD, which might help to provide the methyl group in desosamine synthesis (Xue et al. 1998). Therefore, the increased transcripts of pikD might correspond to the enhanced production of desosamine and pikromycin. Studies on the biosynthesis of desosamine have consistently established DesVI (N,N-dimethyltransferase) as a SAM-dependent methyltransferase in the pikromycin biosynthetic gene cluster (Chen et al. 2002). The increased level of SAM production would apparently aid in the overproduction of desosamine by desVI and thereby enhance the production of pikromycin. In addition, the enhanced expression of metK, which increases the intracellular level of SAM, resulted in the overproduction of actinorhodin from S. lividans (Kim et al. 2003). Although the mechanism underlying the increased production of antibiotics with the increased intracellular level of SAM remains unclear, it has been proposed that SAM activates various transcriptional activators, which are responsible for the induction of antibiotic synthetic genes or serves as a methyl donor directly to the antibiotics.
Similarly, afsR is a global regulator gene that seems to control the secondary metabolism and morphological and physiological differentiation in Streptomyces (Umeyama et al. 2002). Moreover, afsR-sp functioned as a transcriptional activator in order to regulate the production of secondary metabolites in Streptomyces species (Parajuli et al. 2005). Heterologous expression of afsR-sp in SVGIBR enhanced the transcript levels of the ketosynthase and pikD pikromycin biosynthetic genes, resulting in the overproduction of pikromycin. Although the exact mechanism of the increased production of antibiotics should be further elucidated, we found that the metK1-sp and afsR-sp genes influenced the stimulation of the biosynthetic genes to overproduce pikromycin through transcriptional activation in S. venezuelae.
References
Agouridas C, Denis A, Auger JM et al (1998) Synthesis and antibacterial activity of ketolides (6-O-methyl-3-oxoerythromycin derivatives) a new class of antibacterials highly potent against macrolide-resistant and susceptible respiratory pathogens. J Med Chem 41:4080–4100
Basundhara S, Oh TJ, Lamichhane R et al (2004) Neocarzinostatin naphthoate synthase: an unique iterative type I PKS from neocarzinostatin producer Streptomyces carzinostaticus. FEBS Lett 566:201–206
Bisang C, Long PF, Cortés J et al (1999) A chain initiation factor common to both modular and aromatic polyketide synthases. Nature 401:502–505
Chen H, Yamase H, Murakami K et al (2002) Expression, purification, and characterization of two N,N-dimethyltransferases, TylM1 and DesVI, involved in the biosynthesis of mycaminose and desosamine. Biochemistry 41:9165–9183
Horinouchi S (2003) AfsR as an integrator of signals that are sensed by multiple serine/threonine kinases in Streptomyces coelicolor A3(2). J Ind Microbiol Biotechnol 30:462–467
Kieser T, Bibb MJ, Buttner MJ et al (2000) Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kim DJ, Huh JH, Yang YY et al (2003) Accumulation of S-adenosyl-l-methionine enhances production of actinorhodin but inhibits sporulation in Streptomyces lividans TK23. J Bacteriol 185:592–600
Lee SK, Hong JSJ, Choi CY et al (2006) Enhanced production of hydroxylated macrolides from pikromycin pathway of Streptomyces venezuelae. Enzyme Microb Technol 39:778–782
Lee PC, Umeyama T, Horinouchi S (2002) afsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2). Mol Microbiol 43:1413–1430
Okamoto S, Lezhava A, Hosaka T et al (2003) Enhanced expression of S-adenosylmethionine synthetase causes overproduction of actinorhodin in Streptomyces coelicolor A3 (2). J Bacteriol 185:601–609
Parajuli N, Hung TV, Ishida K et al (2005) Identification and characterization of the afsR homologue regulatory gene from Streptomyces peucetius ATCC 27952. Res Microbiol 156:707–712
Shin SK, Park HS, Kwon HJ et al (2007) Genetic characterization of two S-adenosylmethionine-induced ABC transporters reveals their roles in modulations of secondary metabolism and sporulation in Streptomyces coelicolor M145. J Microbiol Biotechnol 17:1818–1825
Tanaka A, Takano Y, Ohnishi Y et al (2007) AfsR recruits RNA polymerase to the afsS promoter: a model for transcriptional activation by SARPs. J Mol Biol 369:322–333
Umeyama T, Lee PC, Horinouchi S (2002) Protein serine/threonine kinases in signal transduction for secondary metabolism and morphogenesis in Streptomyces. Appl Microbiol Biotechnol 59:419–425
Wang Y, Boghigian BA, Pfeifer BA (2007) Improving heterologous polyketide production in Escherichia coli by overexpression of an S-adenosylmethionine synthetase gene. Appl Microbiol Biotechnol 77:367–373
Wilson DJ, Xue Y, Reynolds KA et al (2001) Characterization and analysis of the PikD regulatory factor in the pikromycin biosynthetic pathway of Streptomyces venezuelae. J Bacteriol 183:3468–3475
Xue Y, Zhao L, Liu HW et al (1998) A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: Architecture of metabolic diversity. Biochemistry 95:12111–12116
Zhao XQ, Jin YY, Kwon HJ et al (2006) S-Adenosylmethionine (SAM) regulates antibiotic biosynthesis in Streptomyces spp. in a mode independent of its role as a methyl donor. J Microbiol Biotechnol 16:927–932
Acknowledgments
The work was supported by a medium-term strategic technology development grant from the Republic of Korea to JKS and the authors thank to the Korea Research Foundation (KRF) for the graduate (Ph D) student scholarship.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Maharjan, S., Oh, TJ., Lee, H.C. et al. Heterologous expression of metK1-sp and afsR-sp in Streptomyces venezuelae for the production of pikromycin. Biotechnol Lett 30, 1621–1626 (2008). https://doi.org/10.1007/s10529-008-9735-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9735-0