Abstract
A phytoene synthase gene SePSY was isolated from euhalophyte Salicornia europaea L. The 1655 bp full-length SePSY has an open reading frame of 1257 bp and encodes a 419-amino acid protein. The overexpression of SePSY enhanced the growth of transgenic Arabidopsis. When the plants were exposed to 100 mM NaCl, the photosynthesis rate and photosystem II activity (Fv/Fm) increased from 92% to 132% and from 9.3% to 16.6% in the transgenic lines than in the wild-type, respectively. The transgenics displayed higher activities of SOD and POD and lower contents of H2O2 and MDA than the WT. In conclusion, the transgenic lines showed higher tolerance to salt stress than WT plants by increased photosynthesis efficiency and antioxidative capacity. This is the first report about improving the salt tolerance by genetic manipulation of carotenoid biosynthesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High salinity is the most severe environmental abiotic stress. Salinity not only causes physiological drought and ion toxicity, but also results in a reduction in photosynthesis and the production of reactive oxygen species (ROS) (Zhu 2002). Furthermore, ROS is an inevitable byproduct in electron transport chain during oxygenic photosynthesis in plant (Møller 2001).
Carotenoids are the most diverse and wide spread group of pigments found in nature (Bartley and Scolnik 1994). ROS can be effectively eliminated by carotenoids, In addition, carotenoids are indispensable in light collection and energy transmission during photosynthesis (Tao et al. 2007) suggesting carotenoids play a vital role in plant tolerance to salinity.
So far, the plant carotenoid biosynthetic pathway has been well documented (Cunningham and Gantt 1998; Römer and Fraser 2005). Genetic manipulation of carotenoid biosynthesis has been applied to develop plant species with increased carotenoid content in many crops (Sandmann 2001). However, there has been no report about improving the salt tolerance by genetic manipulation of carotenoid biosynthesis.
Phytoene synthase (PSY) may be a rate-limiting enzyme in carotenoid biosynthesis, which catalyzes the conversion of two molecules of geranylgeranyl pyrophosphate into phytoene(Cunningham and Gantt 1998). Gene encoding phytoene synthase has been identified from numerous higher plants (Sandmann 2001). A recent report has shown that the transcript level of PSY3 in maize was regulated in response to abiotic stresses (Li et al. 2007), suggesting that PSY is involved in plant tolerance to abiotic stress.
Salicornia europaea L. is a salt-accumulating halophyte which can survive under high salinity. It not only exempted from the damaged caused by ROS, but also showed high photosynthetic efficiency under 200 mM NaCl (our unpublished data), suggesting there is a special mechanism in S. europaea to deal with the ROS caused by high salinity, which might be related, at least partially, to its hither carotenoids. In this study, the putative phytoene synthase gene was cloned from the halophyte S. europaea and overexpressed in Arabidopsis with the aim to understand the relationship between carotenoid synthesis pathway and salt tolerance.
Materials and methods
Isolation and sequence analysis of SePSY
Total RNA was extracted from young shoots of S. europaea using Trizol reagent (Gibco, BRL, USA). The first strand cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen, USA). Single strand-cDNA was subjected to PCR amplification with the following degenerated primers: 5′-GAAGCTTATGAT(C/A)G(T/A)TGTGG-3′ and 5′-CA(A/T/C)ACCGGCCATCT(A/G)CTAGC-3′. The rapid amplification of cDNA end (RACE) approach was used to isolate the 5′- and 3′-ends of SePSY gene using the kit (Invitrogen, USA) according to the manufacturer’s instructions. A putative full-length PSY cDNA was obtained by PCR using a specific pair of primer at the 5′-end, 5′-CAGATAGTGGAAGGGTTTGG-3′ and 3′-end, 5′-GGCGGAAAAAGAAAATGTTGC-3′. A 1655 bp product was cloned into pGEM-T-Easy vector (Promega, USA) and sequenced. Multiple sequence alignment was performed by the ClustalW (1.82) software. Phylogenetic analysis and transmembrane prediction were conducted using DNAman software. Chloroplastic transit peptide was predicted on the website: http://www.cbs.dtu.dk/services/ChloroP/.
Plasmid construction and plant transformation
The complete coding region of SePSY amplified by PCR with forward primer (5′-TTGGATCCATGCCTCTTGCTTTGCTATG-3′ BamHI site underlined) and reverse primer (5′-TTGGATCCCTACACCTTTGACATGCTAG-3′, BamHI site underlined) was inserted into the plant expression vector SN1301 at BamHI sites. The resultant plasmid containing cauliflower mosaic virus (CaMV) 35S promoter was introduced into Arabidopsis thaliana (ecotype: Columbia) by Agrobacerium tumefaciens (strain LBA4404)-mediated transformation using the floral-dip method (Clough and Bent 1998). The seeds collected were screened in Murashige and Skoog (MS) medium supplemented with 20 mg hygromycin l−1, and seedlings were identified by PCR. RNA was extracted from the T3 generation seedling, and Northern blot was carried out following the method by Sambrook et al. (1989).
Measurements of photosynthesis parameters and antioxidative enzymes
Homozygous T3 plants were grown in plastic pots containing a commercial soil mix and grown in a growth chamber at 23 ± 1°C under a 12 h photoperiod. Three-week seedlings of WT and three transgenic lines were treated with 100 mM NaCl. About 3 days after salt treatment was applied, seedlings were subjected to measurements of photosynthesis parameters, content of H2O2 and malondialdehyde (MDA), and activity of peroxidase (POD) and superoxide dismutase (SOD).
The net photosynthesis rate (P N ) was determined with an LI6400 portable apparatus (LI-COR, USA) in leaves 4 and 5 from the apex from at least 8 plants for each treatment. The photosystem II (PS II) activity was determined with a portable modulated fluorometer (FMS2, Hansatech, England), and expressed as Fv/Fm to assess damage to leaf photosynthetic apparatus. The measurements of H2O2 and MDA contents, the extraction of antioxidative enzymes, and activity assay of SOD and POD were based on the method by Parida et al. (2004).
Statistical analyses
Statistical differences between WT and transgenic lines were assessed based on the analysis of variance ANOVA using SPSS (Chicago, IL, USA). Differences were considered significant at a probability level of P ⩽ 0.05.
Results
Isolation and sequence analysis of SePSY
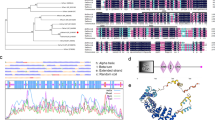
A 716 bp cDNA fragment encoding PSY was amplified by PCR reaction using degenerate primers from S. europaea. The full-length cDNA sequence was 1655 bp in length, obtained from 3′-RACE and 5′-RACE. It contained an open reading frame of 1257 bp, and encoded a polypeptide of 419 amino acids with a predicted molecular mass of 47.2 kDa (GenBank accession number No. AY789515). A putative chloroplastic transit sequence was predicted to be the N-terminal residues 1–65 through the Chlorop 1.1 program based on the website: http://www.cbs.dtu.dk/services/ChloroP/ (Fig. 1a). The transmembrane domains of SePSY were analyzed with DNAman program, which located in 1–19 and 242–264 amino acids, respectively (Fig. 1b).
(a) Deduced amino acid sequence of novel phytoene synthase cDNA (SePSY) from S. europaea (GeneBank accession number: AY789515). The putative signal of chloroplastic transit peptide is underlined. (b) Transmembrane domains of SePSY located in 1–19 and 242–264 amino acid. (c) The phylogenetic tree of the S. europaea PSY to other various specious PSY. Phylogenetic analysis is based on the deduced amino acid sequences of PSY from various species. The tree was generated by DNAman software. The cDNA sequences used for amino acid translation and GenBank accession numbers are: Zea mays (U32636), Cucumis melo (Z37543), Helianthus annuus (AJ304825), Tagetes erecta (AY099482), Lycopersicon esculentum (M84744), Arabidopsis thaliana (L25812), Citrus unshiu (AF220218), Salicornia europaea (AF789515), Daucus carota (AB032797) and Agrobacterium tumefaciens str. C58 (NP354560)
The deduced amino acid of SePSY was compared with those of other known PSY proteins. SePSY has a high degree of similarity with various plant species (69.9–76.6%). By contrast, a low identity (19%) was observed with Agrobacterium tumefaciens. The phylogenetic analysis demonstrated similar patterns (Fig. 1c).
Overexpression of SePSY conferred salt tolerance in Arabidopsis
To validate the function of the SePSY gene during salt stress, we transformed Arabidopsis with the SePSY gene under the control of a CaMV 35S constitutive promoter. The transformed plants were verified by PCR (Fig. 2a). The positive transgenic lines were subjected to RNA gel blot analysis, and the results indicated that SePSY was expressed in transgenic Arabidopsis (Fig. 2b).
Three-week-old seedlings of wide type (WT) and transgenic T3 Arabidopsis lines were subjected to 100 mM NaCl treatment. The results demonstrated that no significant difference in phenotype was observed between WT and any of the transgenic plants under non-stress condition. However, 3 days after salt treatment, the WT displayed serious chlorosis and wilting while all three transgenic lines showed normal phenotypes under 100 mM NaCl (Fig. 2c), suggesting that overexpression of SePSY in Arabidopsis conferred salt tolerance to the transgenic plants to some extent.
Due to the role of carotenoids as accessory pigment in photosynthesis and photo-protectors preventing photo-oxidative damage, the effects of transgenic lines on photosynthesis parameters and activity of some antioxidative enzymes under salt stress were further characterized.
Transgenic plants exhibit greater photosynthesis rate and PSII activity
The photosynthesis rate and PSII activity (Fv/Fm ratio) were determined in WT and lines overexpressing SePSY gene under both 1/4 Hoagland and 100 mM NaCl. NaCl treatment decreased significantly the photosynthesis rate in both WT and transgenic lines. However, there was a marked increase in the photosynthesis rate in transgenic lines regardless of whether the transgenic lines were subjected to salt stress. The photosynthesis rate increased from 69.2% to 121% for 1/4 Hoagland treatment and from 92.4% to 131.7% for 100 mM NaCl treatment. Salt stress induced a reduction in the PSII activity in WT and transgenic plants. However, PSII activity increased from 9.3% to 16.6% in the transgenic lines than in the wild-type under 100 mM NaCl. (Table 1).
Transgenic plants have lower contents of H2O2 and MDA
To assess the role of overexpression of SePSY on oxidative stress, the content of H2O2 and malondialdehyde (MDA) were monitored in both WT and transgenic plants. Salt stress induced significant increase in H2O2 content in both wild type and transgenic plants. However, the transgenic lines maintained significantly lower H2O2 than WT under both control as well as salt treatment (Fig. 3). There was no significant difference in MDA content between wild type and transgenic lines under 1/4 Hoagland. However, salt stress significantly increased MDA content for WT, while MDA content in transgenic lines remained unchanged by salt treatment, resulting in 1.2–1.6 times MDA content in wild type than in the transgenic lines under 100 mM NaCl (Fig. 3).
Overexpression of SePSY enhanced the POD, SOD activity
There was no significant difference in the activity of POD between WT and transgenic lines under 1/4 Hoagland. However, significant increase of POD activity was observed in both wild type and transgenic lines in response to salt stress. In addition, overexpression of SePSY showed significant increase in the POD activity in transgenic lines as compared with WT under salt stress (Fig. 4).
The activity of SOD in wild type was significantly lower than that in transgenic lines under 1/4 Hoagland. However, there was no significant difference between wild-type and transgenic lines under salt stress because the salt stress induced a significant increase in SOD activity for WT but not for transgenic plants (Fig. 4).
Discussion
Carotenoids play a crucial role in eliminating the ROS which is produced during salt stress and normal oxygenic photosynthesis. Previous work about genetic manipulation of carotenoid biosynthesis was done with the aim to provide sources for the isolation of desired carotenoids or to generate food plants with increased carotenoid content including rice, tomato, canola and potato (Cunningham and Gantt 1998; Sandmann 2001; Römer and Frased 2005). Overexpression of β-carotene hydroxylase enhances stress tolerance caused by increased sunlight in Arabidopsis (Davison et al. 2002). However, there have been few reports on transformation of carotenoids biosynthesis genes to improve the plant salt tolerance. In this study, a phytoene synthase gene SePSY was isolated from halophyte S. europaea, and successfully overexpressed in Arabidopsis. The results demonstrated that the transgenics suffered less than the WT under salt stress, which might be resulted from an enhanced activity of antioxidative enzymes and photosynthesis rate.
The carotenoid pigments are synthesized in the plasmids of plants. In chloroplasts they accumulate primarily in the photosynthetic membranes in association with the light-harvesting and reaction center complexes (Cunningham and Gantt 1998). Our results indicate that there are two transmembrane domain located in 1–19 and 242–264 amino acids in SePSY, respectively (Fig. 1b), which demonstrate that SePSY is a membrane-bound protein. To exhibit its role, the protein products have to be imported into organelles. A putative transit sequence for plastid targeting in SePSY (Fig. 1a) was predicted to be the N-terminal residues 1–65; this result is line with previous results in daffodil (Schledz et al. 1996), Arabidopsis (Scolnik and Bartley 1994), and sunflower (Salvini et al. 2005).
H2O2 is produced in response to many physiological stimuli such as UV stress, high salinity in plant cells, which can cause oxidative stress and subsequently protein inactivation (Izumi and Schroeder 2004). To control the level of ROS and protect cells under stress conditions, plant tissues contain several enzymes scavenging ROS, such as POD, SOD, CAT (Blokhina et al. 2003). In this study, the lines overexpressing SePSY accumulated less H2O2 than WT plants under salt stress (Fig. 3), suggested that overexpression of SePSY induced the activity of SOD, and POD in the transgenics (Fig. 4). Meanwhile, the increased carotenoid can function directly as antioxidants by reacting with ROS (Krinsky 1989). Similar results were obtained through overexpression of SOD, glutathione peroxidases (GPX), and aldehyde dehydrogenase (Bartels and Sunkar 2005; Kotchoni et al. 2006). Increased capability of antioxidation inhibited effectively the lipid peroxidation in the transgenic lines, as measured by a delay in MDA formation.
Reductions in plant growth due to salt stress are often associated with decreases in photosynthetic activities which cause the inevitable oxidative to PSII. Overexpression of SePSY enhanced significantly the photosynthesis rate and PSII activity (Fv/Fm) as compared with WT (Table 1), which suggested the role of SePSY in plant salt tolerance. In addition to the enhanced activity of antioxidative enzymes, the increased carotenoids content play important role in improving the photosynthetic efficiency. Recent studies have reported the reversible enzymatic interconversion between the carotenoids violaxanthin and zeaxanthin (the xanthophylls cycle) regulates the induction of photoprotective non-photochemical quenching (NPQ) in the thylakoid membranes of plants (Demming-Adams 1990). The dissipation of excess energy absorbed by light-harvesting antenna was transferred from chlorophyll a to a low-lying carotenoid excited state, identified as one of the two luteins (lutin1) in LHCII by a twist in the configuration of the LHCII-bound carotenoid neoxanthin (Ruban et al. 2007). Two carotenes are bound to the D2 protein and this position was discussed in relation to their ability to act as quenchers of singlet oxygen (Telfer 2005). The further work needs to be done to identify the exact mechanism of improved the photosynthetic efficiency in the transgenic lines of SePSY.
In conclusion, for the first time in this field, the phytoene gene was successfully isolated from halophyte S. europaea and overexpressed in Arabidopsis. The improved salt tolerance of the transgenics suggested that carotenoid could be used as a potential target for manipulating plants salt stress. Further detailed studies are required to establish the metabolism of involvement of carotenoid in plant adaption to stress response.
References
Bartels D, Sunkar R (2005) Drought and salt tolerance in plants. Critical Rev Plant Sci 24:23–58
Bartley GE, Scolnik PA (1994) Molecular biology of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 45:287–301
Blokhina O, Virolainen E, Fagerstedt KV (2003) Antioxidants, oxidative damage and oxygen deprivation stress: a review. Ann Botany 91:179–194
Clough SJ, Bent AF (1998) Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Cunningham JFX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol 49:557–583
Davison PA, Hunter CN, Horton P (2002) Overexpression of β-carotene hydroxylase enhances stress tolerance in Arabidopsis. Nature 418:203–206
Demming-Adams B (1990) Carotenoids and photoprotection in plants: a role of the xanthophylls zeaxanthin. Biochem Biophys Acta 1020:1–24
Izumi CM, Schroeder JI (2004) Reactive oxygen species activation of plant Ca2+ channels: a signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol 135:702–708
Kotchoni SO, Kuhns C, Ditzer A, Kirch H, Bartels D (2006) Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ 29:1033–1048
Krinsky NI (1989) Antioxidant functions of carotenoids. Free Radic Biol Med 7:617–635
Li FQ, Vallabhaneni R, Wurtzel ET (2007) PSY3, a new member of the phytoene synthase gene family conserved in the Poaceae and regulator of abiotic-stress-induced root carotenogenesis. Plant Physiol 10.1104/pp.107.111120
Møller IM (2001) Plant mitochondria and oxidative stress: electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu Rev Plant Physiol Plant Mol Biol 52:561–591
Parida AK, Das AB, Mohanty P (2004) Defense potentials to NaCl in a mangrove, Bruguiera parviflora: differential changes of isoforms of some antioxidative enzymes. J Plant Physiol 161:531–542
Römer S, Fraser PD (2005) Recent advances in carotenoid biosynthesis regulation and manipulation. Planta 221:305–308
Ruban AV, Berera R, Ilioaia C, van Stokkum IHM, Kennis JTM, Pascal AA, van Amerongen H, Robert B, Horton P, van Grondelle R (2007) Identification of a mechanism of photoprotective energy dissipation in higher plants. Nature 450:575–579
Salvini M, Bernini A, Fambrini M, Pugliesi C (2005) cDNA cloning and expression of the phytoene synthase gene in sunflower. J Plant Physiol 162:479–484
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, Cold Spring Harbor
Sandmann G (2001) Genetic manipulation of carotenoid biosynthesis: strategies, problems and achievements. Trends Plant Sci 6:13–17
Schledz M, Al-Babili S, Lintig J, Haubruck H, Rabbani S, Kleinig H, Beyer P (1996) Phytoene synthase from Narcissus pseudonarcissus: functional expression, galactolipid requirement, topological distribution in chromoplasts and induction during flowering. Plant J 10:781–792
Scolnik PA, Bartley GE (1994) Nucleotide sequence of an Arabidopsis cDNA for phytoene synthase. Plant Physiol 104:1471–1472
Tao NG, Hu ZY, Liu Q, Xu J, Cheng YJ, Guo LL, Guo WW, Deng XX (2007) Expression of phytene synthase gene (Psy) is enhanced during fruit ripening of Cara Cara navel orange (Citrus sinensis Osbeck). Plant Cell Rep 26:837–843
Telfer A (2005) Too much light? How β-carotene protects the photosystem II reaction centre. Photochem Photobiol Sci 4:950–956
Ye XD, Al-Babili S, Kloti A, Zhang J, Lucca P (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273
Acknowledgements
This work was financially supported by the National High Technology and Research Development Program of China (“863” project) (Grant No.2003AA627010 and No.2007AA091705).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Han, H., Li, Y. & Zhou, S. Overexpression of phytoene synthase gene from Salicornia europaea alters response to reactive oxygen species under salt stress in transgenic Arabidopsis. Biotechnol Lett 30, 1501–1507 (2008). https://doi.org/10.1007/s10529-008-9705-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-008-9705-6