Abstract
Genome sequence analysis of Xanthomonas oryzae pv. oryzae KACC10331 provides insight into the X. oryzae gum gene cluster that is composed of 14 open-reading frames (ORFs), designated gumB, -C, -D, -E, -F, -G, -H, -I, -J, -K, -L, -M, XOO3167, and -N. We analyzed the transcriptional linkage of the X. oryzae gum gene cluster by using RT-PCR. Analyses of the gum gene cluster by RT-PCR with the wild-type and mutant strains, which carried a deletion of the promoter-like region upstream of gumB or an insertion of the rrnB transcriptional terminator into the gumF gene, revealed that the ORFs of this gene cluster were transcribed as polycistronic mRNA, from gumB to gumN, and the secondary promoter was located upstream of gumG. Taken together, these results suggest that the genes of this cluster constitute an operon expressed from overlapping transcripts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Xanthomonas oryzae pv. oryzae (X. oryzae) is a Gram-negative bacterium that causes blight on rice and produces copious amounts of exopolysaccharides, mainly xanthan gum, as a virulence factor. X. oryzae also can be used for industrial production of xanthan gum which has a variety of commercial applications as a speciality polymer (Becker et al. 1998). The gum gene cluster of Xanthomonas campestris pv. campestris (X. campestris) is mainly expressed as an operon from gumB to gumM, and is involved in xanthan gum biosynthesis (Katzen et al. 1996, 1998, 1999). Recent studies have also shown that a Tn5 insertion in the gum gene cluster of X. oryzae is associated with a loss of xanthan gum biosynthesis and that the gum gene cluster of X. oryzae is required for xanthan gum biosynthesis in a xanthan gum-deficient mutant (Dharmapuri and Sonti 1999). However, the gum gene cluster of X. oryzae has not been characterized at the molecular level. More recently, the complete genome sequence of X. oryzae KACC10331 was published (Lee et al. 2005), and a cluster of open-reading frames (ORFs) was identified in the X. oryzae genome that consisted of a tandem array of 14 ORFs, gumB, -C, -D, -E, -F, -G, -H, -I, -J, -K, -L, -M, XOO3167, and -N, which was similar to the gum operon of X. campestris. In both species, ORFs of the gum gene cluster were arranged in a similar genome structure.
As a first step toward understanding the molecular function of the gum gene cluster in X. oryzae, we analyzed its transcriptional organization by using RT-PCR and constructing mutants carrying a deletion of the promoter-like region upstream of the gumB or gumF::T rrnB transcriptional block in association with plasmid integration mutagenesis.
Materials and methods
Bacterial strains and growth conditions
Escherichia coli DH5α (Gibco BRL) (F− recA1 endA1 hsdR17[r −k m +k ] supE44 thi-1 gyrA relA1) was used for all recombinant DNA experiments that required a bacterial host. E. coli DH5α cells were grown in Luria–Bertani (LB) broth or on LB-agar plates prepared as described previously (Sambrook et al. 1989). The wild-type Xanthomonas oryzae strain used in this study was KACC 10859. It was grown on SOC agar (Sambrook et al. 1989) and incubated at 28°C for 3–5 days. It was also cultured in NB medium (3 g beef extract and 5 g peptone/l) or XOL medium (Barreras et al. 2004) at 28°C and 200 rpm on a rotary shaker.
Site-specific gene disruption
Site-specific gene disruption was performed by using the nonreplicable integration vector, pK18mobsacB, which enables marker-free deletion of the target gene (Schäfer et al. 1994). A 2,348-bp DNA fragment containing the sequence of ORF gumF was amplified by PCR using X. oryzae KACC10859 genomic DNA as the template and primer pairs gumF1 (5′-CTAGTCTAGAACCGTTACTTCGATAGCGG-3′; XbaI restriction site is underlined) and gumF2 (5′-CTAGTCTAGAGGAAGACCAACGTATGCG-3′; XbaI restriction site is underlined). The resulting 2,348-bp PCR products were digested with XbaI and subcloned into the corresponding sites of pBluescript II KS+ (Stratagene). The resulting plasmid was digested with HincII and ligated with blunt-ended EcoRI-ClaI PCR products of the rrnB terminator amplified by PCR using pTrc99A plasmid DNA (Amann et al. 1988) as the template and primer pairs rrnB1 (5′-ACTCCATCGATGCTGTTTTGGCGGATGAGAG-3′; ClaI restriction site is underlined) and rrnB2 (5′-CCGGAATTCAAAAGGCCATCCGTCAGG-3′; EcoRI restriction site is underlined). The resulting plasmid was digested with XbaI, and the fragment containing the rrnB terminator encompassed by the sequence of ORF gumF was inserted into the corresponding sites of pK18mobsacB to generate the plasmid pSJ861. Plasmid pSJ861 was introduced into X. oryzae KACC 10859 by electroporation as described previously (Sun et al. 2003); integration of the introduced plasmid into the chromosome by a single crossover was monitored on SOC plates containing kanamycin. The single crossover was confirmed by the inability of the cells to grow on SOC plates containing 10 g sucrose/l and by diagnostic PCR (Kim and Cho 2005) using gene-specific primers outside the regions of the targeted gene. For deletion of the targeted gene and vector by a second round of a crossover, the kanamycin-resistant (KmR) and sucrose-sensitive cells were grown overnight in the SOC medium and spread onto SOC plates containing 10 g sucrose/l. Cells growing on the plates were picked and verified for the deletion of the targeted gene and vector by diagnostic PCR using gene-specific primers outside the regions of the targeted gene.
DNA sequences located upstream (−1445 to −826) and downstream (+42 to +1177) from the ORF gumB in the genome of X. oryzae were amplified by PCR using X. oryzae genomic DNA as a template and primer pairs PromUF (5′-GGCCCAAGCTTAGGTGCAGTTGTTCGACC-3′; HindIII restriction site is underlined) and PromUR (5′-CCGGGGTACCGCTCCAGGCTGGGGAATT-3′; KpnI restriction site is underlined), and PromDF (5′-CCGGGGTACCGCCTTGATCTGCTCGATG-3′; KpnI restriction site is underlined) and PromDR (5′-CTAGTCTAGAGTCCAGCAACGTATCGGT-3′; XbaI restriction site is underlined), respectively. The PCR products were digested with XbaI and KpnI, and KpnI and HindIII, respectively, and were ligated into the XbaI and HindIII sites of pK18mobsacB. The resulting plasmid was introduced into X. oryzae KACC 10859 by electroporation; deletion of the putative gumB promoter region in the genome of X. oryzae was monitored and verified as described above.
RT-PCR analysis
For RT-PCR experiments, total RNA was extracted from X. oryzae KACC10859 late-exponentially grown in XOL medium using the hot phenol method, and the extracted RNA was treated with DNase I. The DNase I-treated RNA samples were purified using an RNeasy column (Qiagen). Purified DNase I-treated RNA samples were subsequently incubated with a gene-specific primer for the gumF, ORF XOO3167, or gumN and 200 units of reverse transcriptase at 42°C for 30 min. Reverse-transcribed RNA samples were then added to the gene-specific primers (Table 1) and PCR mix. PCR was routinely carried out as described previously (Kim and Cho 2005).
Determination of xanthan
X. oryzae wild-type and mutant strains were inoculated in XOL medium and incubated for 72 h in a rotary shaker at 200 rpm and 28°C. The amount of xanthan in the supernatant was determined by the colorimetric method for estimation of pentoses and hexoses (DuBois et al. 1956), and was calculated with a standard curve constructed by using a known amount of xanthan (from Fluka Biochemika).
Results and discussion
The gum gene cluster of X. oryzae is transcribed as an operon
The gum gene cluster of X. oryzae spans nearly 16-kbp, from gumB through gumN, and the ORFs of this gene cluster are oriented in the same direction (Lee et al. 2005). RT-PCR was used to analyze the transcriptional linkage in the gum gene cluster; the primer pairs for each neighboring gene lying upstream of the primer used for reverse transcription were applied based on the general premise that sequences of an upstream gene within a given transcript can be readily amplified with those of a downstream gene if these regions form a contiguous transcript. This method was used because Northern blot analysis to determine the size of specific mRNA from the gene cluster might be unsuccessful due to its likely large size. RT-PCR products generated by these primer pairs would, therefore, be contiguous and would be derived from a co-transcribed transcript.
As shown in Table 1, primer pairs for RT-PCR were then chosen to span each intergenic junction. The results from a series of control reactions confirmed the substrate quality and the specificity of the RT-PCR primers. For the negative control, no amplicons were consistently detected when total RNA that was treated with DNase I but was not reverse-transcribed served as the template, ensuring that residual genomic DNA had not contaminated the total RNA preparations. For the positive control, the presence of the expected amplicons when genomic DNA was included in the control samples demonstrated the reliability of the RT-PCR primers. As shown in Fig. 1, RT-PCR products of the predicted length were generated for all primer pairs spanning intergenic junctions from gumB to XOO3167. To determine whether ORF XOO3167 and gumN belong to the same transcriptional unit, RT-PCR experiments were performed as shown in Fig. 2. Pairs of primers specific to each individual gene (19F-14R for XOO3167; 20F-21R for gumN) or to the two adjacent genes (19F-20R for XOO3167-gumN) were chosen. RT-dependent products from each individual ORF were detected, and the expected 874-bp RT-dependent product encompassing both ORFs was obtained. These results suggest that these ORFs (gumB through gumN) are co-transcribed as a single polycistronic transcriptional unit.
Analysis of the gum gene cluster of X. oryzae by RT-PCR. (A) Schematic representation of the X. oryzae gum gene cluster, along with the approximate position and orientation of the gene-specific primers used for generating the cDNA (dotted arrows) and the subsequent PCR amplification step (black arrows). The thick black line represents the DNA sequence, and the overlying white arrows represent the ORFs. (B) RT-PCR products from primers designed to span the intergenic junctions from gumB through ORF XOO3167. The cDNA fractions obtained with the 19R RT primer were used for the subsequent PCR reactions with primer pairs, 2F-2R, 3F-3R, 4F-4R, and 5F-5R (lanes 4, 5, 6, and 7, respectively). The cDNA fractions obtained with the 14R RT primer were used for the subsequent PCR reactions with primer pairs, 6F-6R, 7F-7R, 8F-8R, 9F-9R, 10F-10R, 11F-11R, 12F-12R, and 13F-13R (lanes 8, 9, 10, 11, 12, 13, 14, and 15, respectively). Genome-based PCR and a negative control without reverse transcriptase amplified with the 6F-6R primer pair were included (lane 2 and 3, respectively). DNA size markers in base pairs are indicated (lane 1)
RT-PCR analysis of the 3′ end of the X. oryzae gum operon. (A) Schematic representation of the approximate position and orientation of the gene-specific primers for generating the cDNA (dotted arrow) and the subsequent PCR amplification step (black arrows) in respect to ORF XOO3167 and gumN. The thick black line represents the DNA sequence, and the overlying white arrows represent the ORFs. (B) RT-PCR products from primers designed to span the region of ORF XOO3167 and gumN. The cDNA fractions obtained with the 21R RT primer were used for the subsequent PCR reactions with primer pairs, 19F-14R, 20F-21R, and 19F-20R (lanes 4, 7, and 10, respectively). Genome-based PCR products were obtained with primer pairs, 19F-14R, 20F-21R, and 19F-20R (lanes 2, 5, and 8, respectively). Controls without reverse transcriptase amplified with the primer pairs, 19F-14R, 20F-21R, and 19F-20R, were included (lanes 3, 6, and 9, respectively). DNA size markers in base pairs are indicated (lane 1)
However, no RT-dependent products of the predicted length were obtained when primers extending over 391-bp upstream from the predicted gumB start codon were used (Fig. 3). This indicates that the transcription start is located somewhere in a region between 436- and 391-bp upstream from the predicted gumB start codon, where putative promoter elements were identified. A comparison of the putative promoter region mapped by RT-PCR analysis with the consensus Escherichia coli σ70 promoter elements revealed similarities: 4 of 6 bp in −35 (CTGTCA vs. TTGACA) and 4 of 6 bp in −10 (AATATT vs. TATAAT) E. coli consensus hexamers were conserved in this region. There were also three putative stable RNA secondary structures found at the −416/−425, −438/−464, and −503/−523 regions, respectively. We do not know yet if these regions are involved in any regulatory processes; however, the long leader sequence and the presence of palindromic sequences in the putative promoter region suggest that some type of control may occur at the posttranscriptional level.
RT-PCR analysis of the 5′ end of the X. oryzae gum operon. (A) Schematic representation of the approximate position and orientation of the gene-specific primers for generating the cDNA (dotted arrow) and the subsequent PCR amplification step (black arrows) in respect to the upstream region of gumB. The thick black line represents the DNA sequence, and the overlying white arrows represent the ORFs. The predicted −10 and −35 consensus hexamers are indicated with arrows below the schematic. The number of the first nucleotide of the primer relative to the predicted gumB start codon is indicated by arrows. (B) RT-PCR products from primers designed to span the upstream region of gumB. The cDNA fractions obtained with the 2R RT primer were used for the subsequent PCR reactions with primer pairs, 14F-1R, 15F-1R, 16F-1R, and 17F-1R (lanes 3, 4, 5, and 6, respectively). Genome-based PCR products were obtained with primer pairs, 14F-1R, 15F-1R, 16F-1R, and 17F-1R (lanes 7, 8, 9, and 10, respectively). A control without reverse transcriptase amplified with the primer pair, 14F-1R, was included (lane 2). DNA size markers in base pairs are indicated (lane 1)
The gum gene cluster of X. oryzae constitutes an operon expressed from overlapping transcripts
Further experiments were performed to confirm that gumB through gumN belong to the same transcriptional unit, constituting an operon involved in xanthan biosynthesis. A mutant of X. oryzae KACC10859 was constructed by integrating the rrnB transcriptional terminator into the gumF gene to block transcriptional activity of this operon, as described in Materials and methods. RT-PCR was performed on total RNA isolated from the mutant strain. As expected, RT-PCR products of the predicted length encompassing gumB and gumC, gumC and gumD, gumD and gumE, and gumE and gumF could be obtained. No transcriptional readthrough from the rrnB terminator integration site appeared to occur, since RT-PCR experiments using primer pairs to rrnB terminator and gumG, or gumF and gumG, respectively, yielded no products (Fig. 4B, lane 8 and 9). Surprisingly, RT-PCR products of the predicted length were generated for all primer pairs spanning intergenic junctions from gumG through gumN (Fig. 4B).
Analysis of the gum gene cluster with an insertion of the rrnB terminator into the gumF gene by RT-PCR. (A) Schematic representation of the approximate position and orientation of the gene-specific primers for generating the cDNA (dotted arrows) and the subsequent PCR amplification step (black arrows). The thick black line represents the DNA sequence, and the overlying white arrows represent the ORFs. The rrnB terminator inserted into the gumF gene is indicated with a black triangle above the line. (B) RT-PCR products from primers designed to span the intergenic junctions from gumB through gumN. The cDNA fractions obtained with the 19R RT primer were used for the subsequent PCR reactions with primer pairs, 2F-2R, 3F-3R, 4F-4R, and 5F-5R (lanes 4, 5, 6, and 7, respectively). The cDNA fractions obtained with the 21R RT primer were used for the subsequent PCR reactions with primer pairs, 6F-6R, 18F-6R, 7F-7R, 8F-8R, 9F-9R, 10F-10R, 11F-11R, 12F-12R, 13F-13R, and 19F-20R (lanes 8, 9, 10, 11, 12, 13, 14, 15, 16, and 17, respectively). Genome-based PCR and a negative control without reverse transcriptase amplified with the primer pair, 6F-6R, were included (lane 2 and 3, respectively). DNA size markers in base pairs are indicated (lane 1)
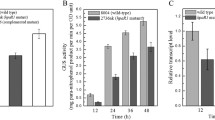
These results indicate that another type of promoter activity exists upstream of gumG. The effect of the integration of rrnB transcriptional terminator into the gumF gene on the transcriptional activity of gum operon was examined in a parallel study. The wild-type and the mutant strain that carried an insertion of rrnB terminator into the gumF gene were grown in XOL medium as described in Materials and methods. The amount of xanthan was quantified from the wild-type (positive control) and the gumF mutant strain. As expected, the transcriptional block of the gumB operon resulting from insertion of the rrnB terminator into the gumF gene had no discernible effect on xanthan accumulation. Analysis of xanthan accumulation from the mutant strain indicated that, in X. oryzae, the absence of the gumF product, an acetyltransferase I homologue catalyzing the acetylation of the mannose residue, would not prevent assembly of xanthan, as seen in X. campestris (Katzen et al. 1998).
If we assume that the synthesis of xanthan in the gumF mutant strain requires the expression of genes downstream of gumF, then insertion of the rrnB terminator into the coding region of gumF would not be polar. We reasoned that this was likely to result from the presence of a secondary promoter internal to the gum gene cluster, particularly one that would lie upstream of gumG, and that it should be possible to test the requirement for secondary promoter in the transcription of genes downstream of gumG by constructing a gumB promoter deletion mutant and examining the consequence of this deletion on transcription of the gum gene cluster. Thus, a promoter deletion mutant carrying an 868-bp internal deletion upstream of gumB was constructed as described in Materials and methods. To confirm whether all downstream genes of gumG could be transcribed from a secondary promoter, total RNA was extracted from the promoter deletion mutant and analyzed by RT-PCR. As shown in Fig. 5, twelve amplifications by RT-PCR covered all of the intergenic junctions of the gum operon, except the junctions between gumB and gumG, which confirms that the secondary promoter is located upstream of gumG.
Analysis of the gum gene cluster with deletion of the putative promoter region upstream of gumB by RT-PCR. (A) Schematic representation of the approximate position and orientation of the gene-specific primers for generating the cDNA (dotted arrows) and the subsequent PCR amplification step (black arrows). The thick black line represents the DNA sequence, and the overlying white arrows represent the ORFs. The putative promoter region upstream of gumB deleted in the X. oryzae wild-type strain was drawn out of the sequence and indicated with a dotted line below the schematic. The predicted −10 and −35 consensus hexamers are also indicated with arrows below the dotted line. The numbers of the nucleotides of the deleted region relative to the predicted gumB start codon are indicated by the dotted line. (B) RT-PCR products from primers designed to span the intergenic junctions from gumB through gumN. The cDNA fractions obtained with the 19R RT primer were used for the subsequent PCR reactions with primer pairs, 2F-2R, 3F-3R, 4F-4R, and 5F-5R (lanes 4, 5, 6, and 7, respectively). The cDNA fractions obtained with the 21R RT primer were used for the subsequent PCR reactions with primer pairs, 6F-6R, 7F-7R, 8F-8R, 9F-9R, 10F-10R, 11F-11R, 12F-12R, 13F-13R, and 19F-20R (lanes 8, 9, 10, 11, 12, 13, 14, 15, and 16, respectively). Genome-based PCR and a negative control without reverse transcriptase amplified with the primer pair, 6F-6R, were included (lane 2 and 3, respectively). DNA size markers in base pairs are indicated (lane 1)
The sequence comprising the intergenic junction of gumF and gumG contains a putative stable RNA secondary structure that differed from the Rho-independent terminator structures, which raised the intriguing speculation that this secondary structure may function as a decay terminator. Many RNA secondary structures serve as barriers to exonucleolytic degradation of the polycistronic mRNA by 3′ to 5′-exonucleases, thereby increasing the stability of upstream mRNA. The functional significance of this transcriptional organization in the gum gene cluster as an overlapping transcriptional unit is not clear, but the overlapping transcriptional arrangement in the gene cluster might protect the downstream genes of gumG from being expressed poorly because of the processing of the primary transcript. Similar stem-loop structures that appear to impart stability to selected regions of polycistronic mRNAs have also been identified in several other bacterial operons (Newbury et al. 1987; Owolabi and Rosen 1990; Schramm et al. 1996; Khun et al. 2000; Maamar et al. 2006).
In conclusion, the results obtained suggest that the X. oryzae gum gene cluster exhibits an operon organization comprised of at least two overlapping transcripts. The predicted stem-loop structure of the intergenic junction of gumF and gumG may have a regulatory role and may partially account for the apparent overlapping transcriptional organization in this gene cluster. In the future, further investigations should be conducted to provide a better understanding of the proposed transcriptional organization of the gum gene cluster and its role in regulating the expression of the gum gene cluster.
References
Amann E, Ochs B, Abel KJ (1988) Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315
Barreras M, Abdian PL, Ielpi L (2004) Funtional characterization of GumK, a membrane-associated β-glucuronosyltransferase from Xanthomonas campestris required for xanthan polysaccharide synthesis. Glycobiology 14:233–241
Becker A, Katzen F, Pühler A, Ielpi L (1998) Xanthan gum biosynthesis and application: a biochemical/genetic perspective. Appl Microbiol Biotechnol 50:145–152
Dharmapuri S, Sonti RV (1999) A transposon insertion in the gumG homologue of Xanthomonas oryzae pv. oryzae causes loss of extracellular polysaccharide production and virulence. FEMS Microbiol Lett 179:53–59
DuBois M, Gilles KA, Hamilton JK, Rebers PA, Smith F (1956) Colorimetric method for determination of sugars and related substances. Anal Chem 28:350–356
Katzen F, Becker A, Ielmini MV, Oddo CG, Ielpi L (1999) New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl Environ Microbiol 65:278–282
Katzen F, Becker A, Zorreguieta A, Pühler A, Ielpi L (1996) Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J Bacteriol 178:4313–4318
Katzen F, Ferreiro DU, Oddo CG, Ielmini V, Becker A, Puhler A, Ielpi L (1998) X. campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol 180:1607–1617
Khun HH, Deved V, Wong H, Lee BC (2000) fbpABC gene cluster in Neisseria meningitides is transcribed as an operon. Infect Immun 68:7166–7171
Kim S-Y, Cho J-Y (2005) A modified PCR-directed gene replacements method using λ-Red recombination functions in Escherichia coli. J Microbiol Biotechnol 15:1346–1352
Lee B-M, Park Y-J, Park D-S, Kang H-W, Kim J-G, Song E-S, Park I-C, Yoon U-H, Hahn J-H, Koo B-S, Lee G-B, Kim H, Park H-S, Yoon K-O, Kim J-H, Jung C-H, Koh N-H, Seo J-S, Go S-J (2005) The genome sequence of Xanthomonas oryzae pathover oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res 33:577–586
Maamar H, Abdou L, Boileau C, Valette O, Tardif C (2006) Transcriptional analysis of the cip-cel gene cluster from Clostridium cellulolyticum. J Bacteriol 188:2614–2624
Newbury SF, Smith NH, Higgins CF (1987) Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131–1143
Owolabi JB, Rosen BP (1990) Differential mRNA stability controls relative gene expression within the plasmid-encoded arsenical resistance operon. J Bacteriol 172:2367–2371
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
Schäfer A, Tauch A, Jäger W, Kalinowski J, Thierbach G, Pühler A (1994) Small mobilizable muti-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69–73
Schramm H-C, Schneppe B, Birkenhäger R, McCarthy JEG (1996) The promoter-proximal, unstable IB region of the atp mRNA of Escherichia coli: an independently degraded region that can act as a destabilizing element. Biochim Biophys Acta 1307:162–170
Sun Q, Wu W, Qian W, Hu J, Fang R, He C (2003) High-quality mutant libraries of Xanthomonas oryzae pv. oryzae and X. campestris pv. campestris generated by an efficient transposon mutagenesis system. FEMS Microbiol Lett 226:145–150
Acknowledgements
This research was supported by the RDA Biogreen 21 Fund (Grant 20050401-034-743-176- 06-00 and 20050401-034-743-006- 06-00).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yoon, KH., Cho, JY. Transcriptional analysis of the gum gene cluster from Xanthomonas oryzae pathovar oryzae . Biotechnol Lett 29, 95–103 (2007). https://doi.org/10.1007/s10529-006-9217-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-006-9217-1