Abstract
A gene responsible for fungicidal activity was identified in the cyanobacterial strain Calothrix elenkinii RPC1, which had shown promise as a biocontrol agent. Functional screening of the genomic library revealed fungicidal (against Pythium aphanidermatum) and endoglucanase activities in two clones. Sequencing revealed an open reading frame of 1,044 bp, encoding 348 amino acid residues with a predicted molecular weight of 38 kDa. Analysis of the deduced amino acid sequence of the putative gene (cael1) showed 99% similarity with the β-1,4-endoglucanase from Anabaena laxa RPAN8 and 97% with the glucanase belonging to the peptidase M20 family of Anabaena variabilis and Nostoc sp. PCC7120, respectively. The putative promoters, ribosomal binding sites and a signal peptide of 22 amino acid residues were identified, revealing the secretory nature of the protein. The phylogenetic tree indicated a close relationship of the gene with Bacillus sp. This study is the first to report on the characterization of an endoglucanase in Calothrix sp.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyanobacteria are known to produce a diverse array of bioactive compounds and enzymes exhibiting antibiotic, algicidal, antifungal, cytotoxic, and immunosuppressive activities (Mundt et al. 2001; Prasanna et al. 2010a, b). In our previous studies, the role of hydrolytic enzymes in fungicidal activity has been demonstrated in several Anabaena strains (Prasanna et al. 2008), and chitosanase/endoglucanase homologs were identified and characterized in Anabaena laxa, A. iyengarii, and A. fertilissima (Prasanna et al. 2010a; Gupta et al. 2010, 2011, 2012). Endoglucanases (1,4-β-d-glucan-4-glucanohydrolases, EC 3.2.1.4) are ubiquitous enzymes, produced by a broad range of organisms, including fungi, bacteria, plants, marine isopods, and insects (Koga et al. 2008; King et al. 2010). Endoglucanases randomly cut the internal bonds within the cellulose polymer. They belong to more than 16 glycoside hydrolase (GH) families, including 5, 6, 7, 8, 9, 12, 16, 17, 44, 45, 48, 51, 55, 61, 74, and 81, on the continually updated carbohydrate active enzymes (CAZy) server (http://www.cazy.org/glycoside-hydrolases.html) (Henrissat 1991; Henrissat and Bairoch 1993, 1996). From a biotechnological viewpoint, endoglucanases have potential applications in the food, animal feed, detergent, and textile industries and in the biological deinking of waste paper (Marques et al. 2003). However, their biocontrol property, especially in cyanobacteria, is far from understood.
In the present study we have characterized an antifungal endoglucanase gene (cael1) from the cyanobacterium Calothrix elenkinii, which had earlier been evaluated for fungicidal activity under laboratory experiments and had shown promise as a biocontrol agent against phytopathogenic fungi challenged vegetable seedlings (Manjunath et al. 2010; Radhakrishnan et al. 2009).
Materials and Methods
Organisms and Growth Conditions
The Calothrix elenkinii (RPC1) strain, available as Calothrix sp. CCC 124 in the Culture Collection of the Centre for Conservation and Utilization of Blue Green Algae, Indian Agricultural Research Institute (IARI), was originally isolated from the rice fields of IARI, New Delhi. Calothrix elenkinii was identified using taxonomic keys provided by Desikachary (1959) and 16S rDNA sequencing (GenBank acc. no. GU292083; data submitted elsewhere). The cyanobacterial strain was purified by standard procedures employing a set of antibiotics (Kaushik 1987), subcultured in nitrogen-free BG11 medium (Stanier et al. 1971), and grown in a light:dark cycle of 16:8 h under white light of 50–55 μE photons m−2 s−1 and temperatures of 28 ± 2°C. Fungicidal activity was evaluated against phytopathogenic fungi (Pythium aphanidermatum ITCC 123) obtained from the Indian Type Culture Collection, Division of Plant Pathology, IARI (Manjunath et al. 2010; Radhakrishnan et al. 2009). The fungal culture was grown at 28°C in a BOD incubator and maintained in potato dextrose agar.
Biochemical Characterization
The cyanobacterial culture was inoculated at a rate of 10% in 250 ml conical flasks, containing 125 ml sterile BG11 medium. The endoglucanase activity was measured spectrophotometrically using carboxymethyl cellulose (CMC) as substrate (Ghosh et al. 1983). Reducing sugars liberated were estimated at 575 nm against the standard curve of glucose. One unit of enzyme represents 1 μmol glucose liberated ml−1 min−1. Activity was recorded in triplicate, and standard error means were calculated.
Construction of Genomic Library and Transformation
A genomic library was constructed in pUC19/SmaI digested vector (Fermentas, USA) using the methodology described earlier (Gupta et al. 2011). The end repaired 2–4 kb DNA fragment was transformed using E. coli JM109 (DE3) competent cells having an isopropyl-β-d-thiogalactopyranoside (IPTG) inducible T7 polymerase gene for expression of the endoglucanase recombinant protein. The transformants were selected on a LB agar plate containing 100 μg/ml ampicillin, 20 mg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and 0.1 M IPTG. All positive white colonies were picked and maintained in Luria–Bertani freeze buffer. Plasmid was isolated from these clones using a high-speed plasmid mini kit (Geneaid PD100, India).
Functional Screening of Genomic Library Clones for Fungicidal Activity
Genomic library clones were analyzed for fungicidal activity using a disc diffusion assay against P. aphanidermatum as a test organism. Sterile discs of 5 mm diameter were soaked in 50 μl of a culture of E. coli clones grown overnight, after overexpression in the 0.4 mM IPTG medium, and placed on the uniform lawn of test organisms, and the zone of inhibition was measured (Prasanna et al. 2008; Gupta et al. 2011). The insert-free vector transformed E. coli was used as a negative control and nystatin (100 U) as a positive control.
Functional Assay for Endoglucanase Activity
Clones were analyzed for endoglucanase activity using 1% CMC as a substrate (Geelen et al. 1995; Teather and Wood 1982). The clones were induced as they were in the assay for fungicidal activity, and a similar negative control was also maintained.
The positive E. coli clones were grown overnight and diluted 1:20 into fresh Luria–Bertani medium until the A 660 reached 0.6. Overexpression of β-1,4-endoglucanase was induced with 0.8 mM IPTG for 4 h. The E. coli cells were harvested by centrifugation, resuspended in lysis buffer (50 mM Tris–HCl, pH 8.0, and 250 mM NaCl) containing 0.1% mercaptoethanol and 2 mM phenylmethylsulfonyl fluoride, and sonicated. Cell debris was removed by centrifugation (10,000×g for 15 min), and the supernatant was used as the enzyme source to evaluate fungicidal and β-1,4-endoglucanase activities using the methodology described above. The insert-free vector transformed E. coli served as a negative control and nystatin as the positive control.
Bioinformatics Tools
BlastN was used to identify nucleotide identity and BlastP to compare amino acid sequences (Altschul et al. 1990). The open reading frame (ORF) was identified using GenDB (Meyer et al. 2003). Nucleotide and amino acid sequences were aligned in the Jalview using Clustal W2 (Larkin et al. 2007). A phylogenetic tree of known endoglucanase sequences from bacteria, fungi, and termites was constructed using the neighbor-joining method as implemented in the program Mega package version 4.1 (Tamura et al. 2007). The stability of the relationships was assessed by boot-strapping (1,000 replicates), and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test was calculated (Felsenstein 1985). All positions containing gaps and missing data were eliminated from the data set (complete deletion option). The conserved domains and motifs were identified using the relevant NCBI search tool. The signal peptide and cleavage site in the translated sequence were predicted, in the form of probability scores, with the hidden Markov model (HMM; Bendtsen et al. 2004), using settings for Gram-negative bacteria and a standard output format in the SignalP 3.0 server. The cleavage site prediction is the probability that a specific position is in the +1 state (i.e., immediately after the cleavage site mature protein starts). The signal peptide prediction is divided into three regions (n, h, and c), used to assign region boundaries to the signal peptides in the data set. For each sequence, SignalP-HMM reports the overall signal peptide probability, equal to the posterior probability of signal peptide (S) for position 1. The bacterial promoter prediction program BPROM (http://linux1.softberry.com/berry.html) was used to identify the position of the promoter sequences (−10 box and −35 box in the input sequence).
Statistical Analyses
To quantify and evaluate the variation, we recorded the antifungal and endoglucanase activities in triplicate for the selected parameters and subjected them to ANOVA (analysis of variance) using SPSS statistical software, as performed earlier (Gupta et al. 2010). Treatment means were compared at 1% level of significance and presented as a mean ± standard deviation of three independent replicates.
Results
Measurement of Endoglucanase Activity
The endoglucanase activity in the culture filtrate of RPC1 (estimated using CMC as substrate) was 0.021 ± 0.004 IU mg−1 protein. The difference in the mean was statistically significant (p < 0.01) in a one-way ANOVA test.
Identification of Fungicidal and Endoglucanase Activities in Genomic Library Clones
Nearly 2,000 clones of 2–4 kb each were generated from the genomic library constructed in pUC19/SmaI. Plate assays for fungicidal and endoglucanase activities were performed to identify the putative gene responsible for both activities (Fig. 1). For fungicidal activity (in terms of zone of inhibition), positive E. coli clones were spotted on the lawn of P. aphanidermatum. Among all the clones, two showed inhibition zones of 12 mm (compared with negative control). Both clones also showed expression on CMC plates in terms of change in color of the streaked region from red to orange, indicating endoglucanase activity. In vitro assays for both fungicidal and endoglucanase activities were performed using cell-free recombinant proteins, which also showed significant activities in both clones (Supplementary Table 1). The insert-free vector transformed E. coli showed no fungicidal/endo-β-1,4-glucanase activity in plate assays or with cell-free recombinant proteins.
Functional screening of genomic library clones of Calothrix elenkinii. (Left) Fungicidal activity of E. coli clones (C1 and C2) against Pythium aphanidermatum. (Right) Endo-β-1,4-glucanase activity of genomic library clones (C1 and C2, E. coli-cael1 clones 1 and 2). N Negative control (insert-free vector transformed E. coli). P Positive control (nystatin)
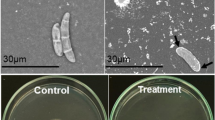
Microscopic analyses of the mycelia revealed the mode of action of both E. coli clones on the fungal filaments (Fig. 2). Curling or disintegration of mycelia of P. aphanidermatum was observed in both clones, whereas only cell wall disintegration was observed in the positive control (nystatin).
Microscopic analysis of fungicidal activity of E. coli-cael1 clone against P. aphanidermatum. a Negative control (insert-free vector transformed E. coli); b positive control (nystatin); c E. coli-cael1 clone 1; d E. coli-cael1 clone 2. Scale bars (lower right of each photo) 50 μm; magnification 2×. Ovals indicate curling or disintegration of the fungal mycelia
Isolation and Sequence Analysis of the Endoglucanase Gene (cael1)
The sequencing results of the positive E. coli clone showed a sequence length of 1,992 bp, including both coding and noncoding regions. An ORF of 1,044 bp was identified, coding for 348 amino acids with a predicted molecular mass of 38 kDa. The BlastN results showed 99% identity with the β-1,4 endoglucanase precursor and glucanase gene from A. laxa and 97% with A. variabilis. The BlastP results comparing amino acid sequences with the NR database (nonredundant protein sequences) showed 98–99% similarity with the glucanases belonging to the peptidase M20 family from A. laxa, A. variabilis, and Nostoc sp. PCC7120. Similarities of 66–75% were observed with the glucanases, peptidases, and cellulases from other cyanobacterial genera (e.g., Lyngbya, Oscillatoria, Arthrospira, Microcystis, Synechococcus) and 36% with the aminopeptidase from Pseudomonas sp. (Table 1). However, BlastP results with the PDB database (protein data bank sequences) revealed significant similarity (24–27%) with proteins coding for glucanase, endoglucanase, and cellulases from Bacillus subtilis, Thermotoga maritima, Streptococcus pneumoniae, and Pyrococcus horikoshii (Table 1). The putative gene coding for an endoglucanase was referred to as cael1. Computational analysis of the nucleotide sequences of cael1 predicted a putative Shine–Dalgarno sequence (positioned at 5 nucleotides before the start codon), a promoter gene containing three signals name transcription start site (TSS), −10 (CGCTATTTT), and −35 (TTCATT) promoter sequences, about 144 nucleotides upstream of the initiation codon (Supplementary Fig. 1). The noncoding sequence of 181 nucleotides upstream and 596 nucleotides downstream of cael1 was also recorded (Fig. 3). The cael1 sequence has been deposited in GenBank (acc. no. GU322875).
Discussion
Cyanobacteria exhibit an array of sophisticated biosynthetic pathways, producing a rich source of novel bioactive products and valuable compounds (Prasanna et al. 2010b). Synthetic fungicidal chemicals have been routinely employed as control measures; their negative implications for the environment and public health, however, have spurred researchers to identify antifungal/fungicidal compounds and enzymes from biological sources. A number of prokaryotic and eukaryotic organisms are known to produce enzymes that degrade cell walls, when chitin or isolated fungal cell wall materials are present in the growth medium. This feature of cyanobacteria has been less explored, and very little published information is available on commercial biocontrol agents developed using cyanobacteria or their metabolites.
Cellulose is one of the cell wall components in Pythium species and other oomycetes. We have previously identified, purified, and characterized two antifungal endoglucanases from the cyanobacterial strain A. laxa (Gupta et al. 2011, 2012). In the current study, we have attempted to identify and characterize similar genes in a promising cyanobacterial strain, C. elenkinii, which has shown promise as a biocontrol agent (Radhakrishnan et al. 2009; Manjunath et al. 2010).
A biochemical assay for CMCase activity revealed the presence of an endoglucanase-like enzyme that may be responsible for the fungicidal activity shown in our earlier studies (Radhakrishnan et al. 2009; Manjunath et al. 2010). In order to identify the gene responsible for the fungicidal activity in this cyanobacterial strain, we constructed a genomic library comprising 2,000 clones, each with a fragment size of 2–4 kb. Functional screening indicated that two clones exhibited fungicidal activity against P. aphanidermatum and showed endo-β-1,4-glucanase activity. The exact mechanism for the expression of E. coli-JM109 (DE3) cells harboring pUC19-cael1 on plate assays without cell lysis is unknown. A possible mechanism may be the secretory nature of the endoglucanase, which was confirmed by the presence of a signal peptide, described earlier for the endoglucanase from A. laxa (Gupta et al. 2011). The plate assay results for both fungicidal and β-1,4-endoglucanase activities were further confirmed by measuring those activities using cell-free recombinant proteins as an enzyme source from both positive clones. This indicates that the endoglucanase responsible for fungicidal activity is functional and is secreted from E. coli cells. Microscopic analyses were undertaken to understand the exact mechanism responsible for fungicidal activity of C. elenkinii. Severe curling or disintegration of the mycelia of fungus P. aphanidermatum was observed, which can be attributed to the activity of the endoglucanase. Similar observations were obtained in the endoglucanase isolated from A. laxa (Gupta et al. 2011). The cael1 encoding endoglucanase in our study can be defined as 1,4-β-d-glucan-4-glucanohydrolase (EC 3.2.1.4), as classified in the BRENDA database and endoglucanases isolated from A. laxa (Gupta et al. 2012).
The deduced amino acid sequence of cael1 for endoglucanase and fungicidal activities showed 98–99% similarities with the glucanases belonging to the peptidase M20 family of Anabaena sp. and Nostoc sp. in the BlastP search (with nonredundant database) (Table 1). The peptidase M20 family is known for consisting of a range of zinc exopeptidases: carboxypeptidases, dipeptidases, and specialized aminopeptidases (Pubmed 7674922), that exhibit hydrolase activity (GO 0016787). A significant similarity of 25–27% was observed with proteins coding for putative glucanases, endoglucanases, and cellulases from Bacillus subtilis, Pyrococcus horikoshii, Streptococcus pneumoniae, and Thermotoga maritima in the BlastP search (PDB database) (Table 1). Incidentally, these enzymes also belong to the peptidase M20 family, and the previously characterized end1 encoding endoglucanase belongs to the same family (Gupta et al. 2011). Thus, our results based on bioinformatics analyses hypothesize that we have sequenced a putative cael1 encoding endoglucanase, belonging to the peptidase M20 family, which is responsible for the observed fungicidal activity.
Genes that encode endoglucanase are known to be regulated by several promoter sequences in bacteria (Han et al. 2003; Endo et al. 2008); however, no similarity with such promoters was observed with the end1 encoding endoglucanase from A. laxa (Gupta et al. 2011). Transcription in cyanobacteria is known to be regulated by the RNA polymerase core enzyme with heterogeneous σ factors (groups 1, 2, and 3) (Imamura and Asayama 2009). Several cyanobacterial promoters have been identified for the expression of the foreign genes such as Anabaena psbAI (Shaw 1975), and Synechococcus psbAII (Haring et al. 1985). Strong cyanobacterial promoters spanning −10 and −35 regions were also identified, showing a very high expression of structural genes such as chloramphenicol acetyl transferase (CAT) compared with other known cyanobacterial promoters (Sode et al. 1996). Therefore, the expression of cael1 in E. coli may be due to the presence of such cyanobacterial promoters. Detailed investigations to identify the functional promoters for this endoglucanase in C. elenkinii are required to gain further insight.
Most of the reported endoglucanases secreted by aerobic microorganisms have a typical structure, with a signal peptide and a mature protein consisting of a catalytic domain linked by a Pro/Thr/Ser-rich linker peptide sequence to a cellulose binding domain (CBD) (Gilkes et al. 1991; Ledger et al. 2006). In our study, the endoglucanase encoded by C. elenkinii lacks a linker and a CBD (Fig. 4; Supplementary Table 2) and therefore is similar to the endoglucanases produced by invertebrates such as termites, phytophagous beetles, crayfish, and mussels (Watanabe and Tokuda 2001; Lo et al. 2003). Further, the bioinformatics analyses predicted a putative ribosome binding site (rbs), which matched with the reported rbs of Zymomonas mobilis ethanologenic genes and A. laxa (Theberge et al. 1992; Rajnish et al. 2008; Gupta et al. 2011).
Earlier reports suggested the secretory/nonsecretory nature of endoglucanases isolated from various bacteria, cyanobacteria, fungi, yeast, and insects, based on the presence of a signal peptide sequence at the amino terminus of the protein sequence (Gao et al. 2004; Wonganu et al. 2008; Gupta et al. 2011). The N-terminal region of the deduced amino acid sequence of RPC1 exhibits a putative peptide signal of 22 residues. The end1 and end2 genes encoding endoglucanases from A. laxa, however, contain a signal peptide of 24 and 20 residues, respectively (Gupta et al. 2011). Unlike RPC1, A. variabilis and Nostoc glucanase sequences available in the database do not show any signal peptides and cleavage sites (Fig. 5; Supplementary Fig. 2). Multiple sequence alignment using the deduced amino acid sequence of endoglucanases from C. elenkinii and Nostoc, A. variabilis, and A. laxa available from NCBI revealed one insertion and a deletion in C. elenkinii. One insertion, a deletion, and a substitution were observed in A. laxa (Fig. 5). Such amino acid exchanges at positions 10 and 21 may be responsible for the secretory nature of the protein and the endoglucanase/fungicidal activity in C. elenkinii seen in the earlier endoglucanases and chitosanases (Gupta et al. 2010, 2011). Further investigation to identify catalytic residues in this strain is necessary to know the exact role of these amino acid residues in fungicidal activity.
Alignment of amino acid sequences of endoglucanase from Calothrix elenkinii (RPC1) and three glucanases available in the NCBI database. Conserved residues are shaded; differences in the sequences are unshaded. N sequence Nostoc (BAB75332), Av Anabaena variabilis (CAA66983), Ce Calothrix elenkinii (ADB64454), Al A. laxa (ADE22238)
The deduced amino acid sequence of cael1, along with other endoglucanase sequences from the bacteria, fungi, and insects available from NCBI, were used to construct a phylogenetic tree to understand the evolutionary relationships. The dendrogram revealed three clusters. Cluster I is divided into two groups, with the endoglucanases of the cyanobacteria (A. laxa, A. variabilis, Nostoc sp. PCC7120) showing close similarity with Pseudomonas syringae pv. tomato str. DC3000, and the endoglucanase from C. elenkinii showing a close relationship with the known Bacillus endo-1,4-β-glucanase (acc. no. Q65JI7, Rey et al. 2004; Veith et al. 2004) and cellulase (acc. no. CAJ70717, Waldeck et al. 2006) belonging to glycoside hydrolase families GH5 and GH2, respectively (Fig. 6; Supplementary Table 2). The endoglucanase from the nematode Meloidogyne incognita was closely related to a Bacillus cellulase in cluster II. Cluster III includes endoglucanases from fungi (Fusarium oxysporum and Staphylotrichum coccosporum). The comparative analysis is depicted in Fig. 4. Further in-depth analyses of structures and proteomes are proposed to reveal the distribution and prevalence of members belonging to this characteristic family of cael1 encoding endoglucanases in cyanobacteria.
Phylogenetic relationships of endoglucanase sequences from cyanobacteria, bacteria, fungi, and insects. Tree constructed using neighbor-joining method (Saitou and Nei 1998) and drawn to scale, with branch lengths in the same units as the evolutionary distances. Bootstrap values indicated above nodes. The evolutionary distances were computed using the method of Tajima and Nei (1984). Scale bar (lower left) 0.2 base substitutions per site. Black triangle sequenced cael1. Brackets on right mark clusters I, II, and III
Although most of the bacterial cellulases are known for their industrial significance, the cellulases produced by Gram-positive endophytic bacteria (Bacillus sp. FP1/2002, Paenibacillus polymyxa DSM36, Bacillus cereus G9667, and Pectobacterium carotovorum LMG2466) have been evaluated for their biocontrol properties (Cho et al. 2007) against several phytopathogenic fungi (Rhizoctonia solani, Fusarium oxysporum, Pythium ultimum, Phytophthora capsici). Such bacteria restrict the growth of phytopathogenic fungi by entering the infected root tissue of the plant, hydrolyzing wall-bound cellulose, or through auxin-induced tumors, water flow, wounds, or at lateral root branching (Al-Mallah et al. 1987). Our earlier study (Manjunath et al. 2010) illustrated the biocontrol potential of this strain against fungi-challenged vegetable seedlings, in pot experiment trials. In the present investigation, the underlying mechanism has been revealed, and studies are under way to overexpress the endoglucanase enzyme and demonstrate its biocontrol potential in the field, either using a purified product or the overexpressing strain.
In conclusion, we have characterized a novel gene (cael1) encoding an endoglucanase belonging to the peptidase M20 super family, which is responsible for the fungicidal activity in C. elenkinii. Apart from this, the identification of promoter region rbs and a signal peptide provide evidence for translation of cael1 encoding endoglucanase in cyanobacterial strains. Thus, our results illustrate the promise of this strain as a potential biocontrol agent against Pythium aphanidermatum. Future research is required to purify and characterize the specific properties of this endoglucanase, to construct knockout mutant forms of this cael1, and to identify the specific promoter sequences and catalytic active site residues.
References
Al-Mallah MK, Davey MR, Cooking EC (1987) Enzymatic treatment of clover root hairs removes a barrier to Rhizobium host specificity. Biotechnology 5:1319–1322
Altschul SF, Gish W, Miller W, Meyers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Bendtsen JD, Neilsen H, Heijne GV, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795
Cho KM, Hong SY, Lee SM, Kim YH, Kahng GG, Lim YP, Kim H, Yun HD (2007) Endophytic bacterial communities in ginseng and their antifungal activity against pathogens. Microbiol Ecol 54:341–351
Desikachary TV (1959) Cyanophyta. ICAR monographs, New Delhi
Endo Y, Yokoyama M, Morimoto M, Shirai K, Chikamatsu G, Kato N, Tsukagoshi N, Kato M, Kobayashi T (2008) Novel promoter sequence required for inductive expression of the Aspergillus nidulans endoglucanase gene eglA. Biosci Biotechnol Biochem 72:312–320
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gao B, Allen R, Davis EL, Baum TJ, Hussey RS (2004) Developmental expression and biochemical properties of a β-1,4-endoglucanase family in the soybean cyst nematode, Heterodera glycines. Mol Plant Pathol 5:93–104
Geelen D, Van Montagu M, Holsters M (1995) Cloning of an Azorhizobium caulinodans endoglucanase gene and analysis of its role in symbiosis. Appl Environ Microbiol 61:3304–3310
Ghosh TK, Bailey HJ, Bisaria VS, Enari TM (1983) Measurement of cellulase activity. Final recommendation, Commission of Biotechnology. Intern Union Pure Appl Chem 59:1–13
Gilkes NR, Henrissat B, Kilburn DG, Miller RC, Warren RAJ (1991) Domains in microbial β-1,4-glycanases: sequence conservation, function, and enzyme families. Microbiol Rev 55:303–315
Gupta V, Prasanna R, Natrajan C, Srivastava AK, Sharma J (2010) Identification, characterization and regulation of a novel antifungal chitosanase (cho) in Anabaena sp. Appl Environ Microbiol 76:2769–2777
Gupta V, Natarajan C, Kumar K, Prasanna R (2011) Identification and characterization of endoglucanases for fungicidal activity in Anabaena laxa. J Appl Phycol 23:73–81
Gupta V, Prasanna R, Chaudhary V, Nain L (2012) Biochemical, structural and functional characterization of two novel antifungal endoglucanases from Anabaena laxa. Biocatal Agri Biotechnol 1:338–347
Han SO, Yukawa H, Inui M, Doi RH (2003) Transcription of Clostridium cellulovorans cellulosomal cellulase and hemicellulase genes. J Bacteriol 185:2520–2527
Haring V, Scholz P, Scherzinger E, Frey J, Derbyshire K, Hatfull G, Willetts NS, Bagdasarian M (1985) Protein RepC is involved in copy number control of the broad host range plasmid RSF1010. Proc Natl Acad Sci USA 82(18):6090–6094
Henrissat B (1991) A classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem J 280:309–316
Henrissat B, Bairoch A (1993) New families in the classification of glycosyl hydrolases based on amino-acid sequence similarities. Biochem J 293:781–788
Henrissat B, Bairoch A (1996) Updating the sequence-based classification of glycosyl hydrolases. Biochem J 316:695–696
Imamura S, Asayama M (2009) Sigma factors for cyanobacterial transcription. Gene Reg Syst Biol 3:65–87
Kaushik BD (1987) Laboratory methods for blue green algae. Associated Publishing, New Delhi
King AJ, Cragg SM, Dymond YLJ, Guille MJ, Bowles DJ, Bruce NC, Graham IA, McQueen-Mason SJ (2010) Molecular insight into lignocellulose digestion by a marine isopod in the absence of gut microbes. Proc Natl Acad Sci USA 107:5345–5350
Koga J, Baba Y, Shimonaka A, Nishimura T, Hanamura S, Kono T (2008) Purification and characterization of a new family 45 endoglucanase, STCE1, from Staphylotrichum coccosporum and its overproduction in Humicola insolens. Appl Environ Microbiol 74:4210–4217
Larkin MA, Blackshields G, Brown NP, Chenna R, Mcgettigan PA, Mcwilliam H, Valentin F, Wallace IM, Wilm A (2007) ClustalW and ClustalX version 2. Bioinformatics 23:2947–2948
Ledger TN, Jaubert S, Bosselut N, Abad P, Rosso MN (2006) Characterization of a new β-1,4-endoglucanase gene from the root-knot nematode Meloidogyne incognita and evolutionary scheme for phytonematode family 5 glycosyl hydrolases. Gene 382:121–128
Lo N, Watanabe H, Sugimura M (2003) Evidence for the presence of a cellulase gene in the last common ancestor of bilaterian animals. Proc Royal Soc Lond B 270(Suppl.):S69–S72
Manjunath M, Prasanna R, Lata, Dureja P, Singh R, Kumar A, Jaggi S, Kaushik BD (2010) Biocontrol potential of cyanobacterial metabolites against damping off disease caused by Pythium aphanidermatum in solanaceous vegetables. Arch Phytopathol Plant Protect 43:666–677
Marques S, Pala H, Alves L, Amaral-Collaco MT, Gama FM, Gino FM (2003) Characterization and application of glucanases secreted by Aspergillus terreus CCM 1498 and Trichoderma viride CCM 184 for enzymatic deinking of mixed office waste paper. J Biotechnol 100:2009–2019
Meyer F, Goesmann A, McHardy AC (2003) GenDB: an open source genome annotation system for prokaryote genomes. Nucleic Acids Res 31:2187–2195
Mundt S, Kreitlow S, Nowotny A, Effmert U (2001) Biochemical and pharmacologiclal investigations of selected cyanobacteria. Int J Hyg Environ Health 203:327–334
Prasanna R, Nain L, Tripathi R, Gupta V, Middha S, Joshi M, Ancha R, Kaushik BD (2008) Evaluation of fungicidal activity of extracellular filtrates of cyanobacteria: possible role of hydrolytic enzymes. J Basic Microbiol 48:186–194
Prasanna R, Gupta V, Natarajan C, Chaudhary V (2010a) Bioprospecting for genes involved in the production of chitosanases and microcystin-like compounds in Anabaena strains. World J Microbiol Biotechnol 26:717–724
Prasanna R, Sood A, Jaiswal P, Nayak S, Gupta V, Chaudhary V, Joshi M, Natarajan C (2010b) Rediscovering cyanobacteria as valuable sources of bioactive compounds. Appl Biochem Microbiol 46:133–147
Radhakrishnan B, Prasanna R, Jaiswal P, Nayak S, Dureja P (2009) Modulation of biocidal activity of Calothrix sp. and Anabaena sp. by environmental factors. Biologia 64:881–889
Rajnish KN, Kishore Choudhary GM, Gunasekaran P (2008) Functional characterization of a putative endo-β-1,4-glucanase gene in the genome of Zymomonas mobilis. Biotechnol Lett 30:1461–1467
Rey MW, Ramaiya P, Nelson BA, Brody-Karpin SD, Zaretskey EJ, Tang M, Lopez de Leon A, Xiang H, Gusti V, Clausen IG, Olsen PB, Rasmussen MD, Andersen JT, Jorgensen PL, Larsen TS, Sorokin A, Bolotin A, Lapidus A, Galleron N, Ehrlich SD, Berka RM (2004) Complete genome sequence of the industrial bacterium Bacillus licheniformis and comparisons with closely related species. Genome Biol 5:R77
Saitou M, Nei M (1998) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shaw WV (1975) Chloramphenicol acetyltransferase from chloramphenicol resistant bacteria. Methods Enzymol 43:737–754
Sode K, Hatano N, Tatara M (1996) Cloning of a marine cyanobacterial promoter for foreign gene expression using a promoter probe vector. Appl Biochem Biotechnol 59:349–360
Stanier RY, Kunisawa R, Mandal M, Cohen-Bazire G (1971) Purification and properties of unicellular blue green algae (order: Chroococcales). Bacteriol Rev 35:171–305
Tajima F, Nei M (1984) Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1:269–285
Tamura K, Dudley J, Nei M, Kumar S (2007) Mega4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Teather RM, Wood PJ (1982) Use of Congo red polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol 43:777–780
Theberge M, Lacaze P, Shareck F, Morosoli R, Kluepfel D (1992) Purification and characterization of an endoglucanase from Streptomyces lividans 66 and DNA sequence of the gene. Appl Environ Microbiol 58:815–820
Veith B, Herzberg C, Steckel S, Feesche J, Maurer KH, Ehrenreich P, Baumer S, Henne A, Liesegang H, Merkl R, Ehrenreich A, Gottschalk G (2004) The complete genome sequence of Bacillus licheniformis DSM13, an organism with great industrial potential. J Mol Microbiol Biotechnol 7:204–211
Waldeck J, Daum G, Bisping B, Meinhardt F (2006) Isolation and molecular characterization of chitinase-deficient Bacillus licheniformis strains capable of deproteinization of shrimp shell waste to obtain highly viscous chitin. Appl Environ Microbiol 72:7879–7885
Watanabe H, Tokuda G (2001) Animal cellulases. Cell Mol Life Sci 58:1167–1178
Wonganu B, Pootanakit K, Boonyapakron K, Champreda V, Tanapongpipat S, Eurwilaichitr L (2008) Cloning, expression and characterization of a thermotolerant endoglucanase from Syncephalastrum racemosum (BCC18080) in Pichia pastoris. Protein Expr Purif 58:78–86
Acknowledgments
The authors are thankful to the Post Graduate School and Director, Indian Agricultural Research Institute, New Delhi, India, for providing a fellowship toward the Ph.D. program of the senior author. The study was also partly funded by the AMAAS Network Project on Microorganisms (Theme: Microbial Genomics) granted by the Indian Council of Agricultural Research, New Delhi. We are extremely grateful to Dr. N. K. Singh, National Research Centre for Plant Biotechnology, IARI, New Delhi, for providing the facilities for nebulization of genomic DNA. We are thankful to the Division of Microbiology, IARI, New Delhi, for providing necessary facilities for undertaking this study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Identification of promoter gene sequences in cael from RPC1 by BPROM software
Supplementary Fig. 2
Identification of signal peptide and cleavage site by hidden Markov model (HMM). (S2A) Eng from RPC1; (S2B) glucanase precursor protein from Anabaena laxa (ADE22238); (S2C) Anabaena variabilis/Nostoc glucanases (CAA66983/BAB75332). Cleavage site assigned by a probability score together with scores for the n-region, h-region, and c-region of the signal peptide
Rights and permissions
About this article
Cite this article
Natarajan, C., Gupta, V., Kumar, K. et al. Molecular Characterization of a Fungicidal Endoglucanase from the Cyanobacterium Calothrix elenkinii . Biochem Genet 51, 766–779 (2013). https://doi.org/10.1007/s10528-013-9605-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10528-013-9605-x