Abstract
Indirect genetic effects from relatives may result in misleading quantifications of heritability, but can also be of interest in their own right. In this paper we propose Trio-GCTA, a model for separating direct and indirect genetic effects when genome-wide single nucleotide polymorphism data have been collected from parent-offspring trios. The model is applicable to phenotypes obtained from any of the family members. We discuss appropriate parameter interpretations and apply the method to three exemplar phenotypes: offspring birth weight, maternal relationship satisfaction, and paternal body-mass index, using real data from the Norwegian Mother, Father and Child Cohort Study (MoBa).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Most human traits exhibit some degree of heritability (Polderman et al. 2015). Some phenotypes are characteristics not only of individuals, but also depend on the influence of other individuals. While direct genetic effects refer to how the phenotype of an individual depends on their own genotype, indirect genetic effects refer to how it depends on the genotypes of others (McAdam et al. 2014). In this paper we describe a model for separating direct genetic effects from the indirect genetic effects of family members when genome-wide single nucleotide polymorphism (SNP) data have been collected from parent-offspring trios.
As parents transmit half their complement chromosomes to their children, the genomes of parents and offspring are correlated. Because the same genetic variants can have both direct and indirect effects, failing to account for the indirect genetic effects of relatives when attempting to measure heritability can result in misleading quantifications of the importance of direct genetic effects (Eaves et al. 2014; Young et al. 2019).
Indirect genetic effects can also be of interest in their own right. With respect to the focal individual (i.e., the individual whose phenotype is the focus of study), indirect genetic effects are part of the environment and may be of great interest when trying to understand causes of individual differences. In this paper we are concerned with indirect genetic effects underlying intra-familial dynamics. This can include instances where heritable characteristics of parents affect offspring development. For example, maternal influence on offspring health through the intrauterine environment (Evans et al. 2019), or where parents affect offspring development by providing an advantageous rearing environment. It also includes instances where heritable characteristics of the offspring evoke responses in their parents. For example, when child behavior influences the mental well-being of their parents.
The quantitative genetics literature distinguishes between two approaches to modelling indirect genetic effects. Trait-based models specify indirect genetic effects on the phenotype of the focal individual mediated by the phenotypes of other individuals. Variance-partitioning models avoid specification of the phenotypes that underlie the indirect genetic effects, instead quantifying the total contributions from these effects while being agnostic as to the underlying mechanisms (Bijma 2014).
The emergence of large-scale genotype data in population-based cohorts has provided new opportunities for developing methods to separate direct and indirect genetic effects. This was leveraged by Eaves et al. (2014) who proposed a variance-partitioning method for separating indirect maternal genetic effects from direct genetic effects with respect to an offspring phenotype, relying on genome-wide SNP data from mother-offspring pairs. In the current manuscript we extend the work of Eaves et al. (2014) to separate direct and indirect genetics effects within parent-offspring trios. We discuss alternative interpretations of variance components depending on the role of the focal individual, useful restricted model specifications and apply the method to three etiologically diverse exemplar phenotypes (offspring birth weight, maternal partner relationship satisfaction and paternal body mass index) using real data from the Norwegian Mother, Father and Child Cohort Study (Magnus et al. 2016).
Model formulation
Yang et al. (2010) introduced a method for quantifying additive genetic variance contributions from all measured SNPs using a linear mixed effects model. Extensions of this methodology include formulations for quantifying dominance genetic effects (Zhu et al. 2015), gene–environment interactions (Yang et al. 2013), parent-of-origin effects (Laurin et al. 2018), maternal effects (Eaves et al. 2014) and avoiding bias from environmental effects (Young et al. 2018). The current approach (Trio-GCTA) uses parent-offspring trios to quantify the importance of direct and indirect genetic effects within the nuclear family. We refer to the individual whose phenotype is under study as the focal individual, noting that the method is applicable regardless of who is the ’owner’ of the phenotype.
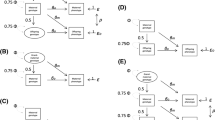
In order to formulate a model for direct and indirect genetic effects, we assume that phenotypic measures have been obtained from a focal individual in \(K\) parent-offspring trios, and that genotypes for the same \(M\) SNPs are available for all individuals. We represent the three \(K \times M\) matrices of maternal, paternal and offspring standardized genotype dosages (Zhu et al. 2015) by \(\mathbf {Z}_m\), \(\mathbf {Z}_p\) and \(\mathbf {Z}_o\), respectively, arranged so that row \(k\) corresponds to the same parent-offspring trio. A linear model for the phenotypes can then be formulated as
where \(\varvec{y}\) is a \(K \times 1\) vector of continuous phenotypes, \(\mathbf {X}\) is a \(K \times P\) matrix of measured covariates with \(P \times 1\) vector of coefficients \(\varvec{\beta }\), \(\varvec{u}_m\), \(\varvec{u}_p\) and \(\varvec{u}_o\) are \(M \times 1\) random vectors of additive genetic effects associated with the maternal, paternal and offspring standardized genotype dosages, respectively, and \(\varvec{e}\) is a \(K \times 1\) vector of residual effects.
The genetic and residual effects are assumed to follow a multivariate normal distribution, where the different types of genetic effects may be dependent but individual SNP effects are independent. The residual effects are assumed to be independent of the genetic effects and across individuals
Although independence is assumed for the effect size of individual SNPs, this formulation makes no assumption about the structure of linkage disequilibrium (Yang et al. 2016). Because the effect sizes are assumed identically distributed, the standardization we use for genotypes does however imply that the unstandardized SNPs have effect sizes that decrease with increasing allele frequency (Yang et al. 2017). The expected covariance structure of the phenotype across all individuals is given by:
\(\sigma ^2_m\), \(\sigma ^2_p\) and \(\sigma ^2_o\) are the variances of the maternal, paternal and offspring genetic effects, respectively, \(\sigma _{om}\) is the covariance between the offspring and maternal genetic effects, \(\sigma _{op}\) is the covariance between the offspring and paternal genetic effects, \(\sigma _{pm}\) is the covariance between the paternal and maternal genetic effects and \(\sigma ^2_e\) is the residual variance. When mating is random, the covariance between the maternal and paternal effects are not expected to contribute to the variance of the phenotype and the total variance decomposition is therefore
Depending on the role of the focal individual, the model parameters have different interpretations. If it is an aspect of the offspring phenotype that is under study, \(\sigma ^2_m\) and \(\sigma ^2_p\) corresponds to variance attributable to indirect genetic maternal and paternal effects, respectively, whereas \(\sigma ^2_o\) is the variance due to direct genetic effects. The components \(\sigma _{om}\) and \(\sigma _{op}\) are the covariances between the direct offspring genetic effect and the indirect maternal and paternal genetic effects, respectively. These parameters quantify the extent to which the same variants contribute to direct and indirect genetic effects. With respect to the offspring, the maternal and paternal genetic effects form part of the environment so these covariance terms may therefore also be interpreted as measuring variability due to gene–environment correlations. The component \(\sigma _{pm}\) is the covariance between the indirect maternal and paternal effects and is a measure of the extent to which the same variants contribute to indirect genetic effects. Sex-dependent expression of genetic effects has been studied with respect to a variety of phenotypes using family designs (Neale and Cardon 2013). A weak correlation between maternal and paternal effects would indicate a qualitative sex difference, wherein mothers and fathers influence their offspring through different heritable traits (alternatively it could be that ostensibly the same trait is under the influence of different genetic factors when expressed in mothers and fathers). A correlation of unity but different magnitude between the maternal and paternal effect would indicate a quantitative sex difference, wherein mothers and fathers influence the offspring by the same heritable traits, but to a quantitatively different extent. Sex-dependent expression of parental effects can therefore potentially reveal insights into differences in maternal and paternal effects on the offspring. \(\sigma ^2_e\) is the residual variance of the phenotype.
If it is an aspect of a maternal phenotype that is under study, \(\sigma ^2_m\) is the variance due to direct genetic effects, whereas \(\sigma ^2_p\) and \(\sigma ^2_o\) measure variability due to indirect genetic effects. The paternal and offspring genetic effects are environmental from the perspective of the mother. Although the underlying mechanisms may be distinct, a maternal phenotype may depend on interactions with both their partner and offspring. \(\sigma _{pm}\) and \(\sigma _{om}\) are the covariance between the direct maternal genetic effect, and the indirect paternal and offspring genetic effects, respectively. If the same genetic variants contribute to direct and indirect genetic effects, these covariance terms are expected to differ from zero. Assuming that mating is random, a genetic correlation between the direct maternal and indirect paternal effect is not expected to affect the phenotypic variance, because maternal and paternal genotypes are independent. However, as the offspring and maternal genotypes are correlated, a genetic correlation between the direct maternal and indirect offspring effect implies a gene–environment correlation that will either increase or decrease the phenotypic variance depending on the sign of \(\sigma _{om}\). \(\sigma _{op}\) is the covariance between the indirect paternal and offspring effects and is a measure of the extent to which the same additive genetic effects contribute to the indirect genetic effects. \(\sigma ^2_e\) is the residual variance of the phenotype. These interpretations are conversely the same if it is a paternal phenotype that is under study. Table 1 summarizes how interpretation of parameters change depending on the role of the focal individual.
Special cases
Several other models of potential interest can be obtained as special cases of the general model described above. Young et al. (2018) introduced relatedness disequilibrium regression (RDR) as a method to avoid environmental bias in heritability estimates by modelling parental genetic nurturing effects in addition to direct genetic effects. The RDR model can be specified by setting \(v_g =\sigma ^2_o\), \(v_{e \sim g}={\sigma ^2_m}/{2}={\sigma ^2_p}/{2}={\sigma _{pm}}/{2}\) and \(c_{g,e}={\sigma _{om}}/{2}={\sigma _{op}}/{2}\), where \(v_g\) is the variance due to direct genetic effects, \(v_{e \sim g}\) is the variance due to parental genetic effects and \(c_{g,e}\) is the covariance between the direct and the parental genetic effects. Therefore, the RDR model can also be seen as assuming the maternal and paternal genetic effects are the same and of equal magnitude. If maternal or paternal effects are not of specific interest on their own, this will likely be a more effective way of accounting for indirect parental effects, as only four variance parameters are required compared to seven under the general model.
Eaves et al. (2014) proposed a method (M-GCTA) for jointly estimating the variance explained by direct genetic effects, indirect maternal genetic effects and their covariance with respect to an offspring phenotype. The M-GCTA model can be obtained with the constraints \(\sigma ^2_p = \sigma _{op} = \sigma _{pm} = 0\). For many research questions, especially those related to pre- and peri-natal phenotypes, this may be a sufficient model.
Under the original GCTA model (Yang et al. 2010) all genetic effects are attributed to the focal individual and can be obtained by omitting all indirect genetic effects from the model.
In the applications below we explore further interpretations of the model parameters when the focal individual has different roles. In the supplementary material we provide a simulation study demonstrating that parameters can be recovered when a trait is generated as a function of correlated direct and indirect genetic effects.
Applications
We applied the Trio-GCTA method to a set of phenotypes measured in parent-offspring trios participating in the Norwegian Mother, Father and Child Cohort Study (MoBa, Magnus et al. 2016). MoBa is a population-based pregnancy cohort study conducted by the Norwegian Institute of Public Health. Participants were recruited from all over Norway from 1999-2008. The women consented to participation in 41% of the pregnancies. The cohort comprises 114,500 children, 95,200 mothers and 75,200 fathers. The current study is based on version 11 of the quality-assured data files. Information was also obtained via a linkage to The Medical Birth Registry (MBR), a national health registry containing information about all births in Norway.
Blood samples were obtained from both parents during pregnancy and from mothers and children (umbilical cord) at birth. The project Better Health by Harvesting Biobanks (HARVEST) sampled 11,000 parent-offspring trios for genotyping from MoBa’s biobank at random. Genotyping was performed using llumina HumanCoreExome-12 v.1.1 and HumanCoreExome-24 v.1.0 arrays. The pre-imputation quality control and imputation procedure is described in Helgeland et al. (2019). Post-imputation, we removed individuals with more than 10% missing genotypes and SNPs with imputation info score less than 0.9 or minor allele frequency less than 0.05. This procedure left 8157 complete triads and four and a half million SNPs eligible for analysis.
Closely related individuals can disproportionally influence genetic variance estimates and introduce confounding from environmental effects not specified in the model (Yang et al. 2017). We used a threshold of 0.10 for the largest allowed genetic correlation between any two individuals (ignoring parent-offspring pairs), reasoning that this will exclude most relations likely to share environments without substantially reducing the sample size. For pairs of individuals exceeding the threshold, we removed one individual at random. This procedure left 7612 complete trios.
Out of the retained trios, 7605 had response data on birth weight, 6702 on relationship satisfaction and 7290 on body mass index. Due to attrition, more responses are missing from later waves of data collection. We refer to Magnus et al. (2016) for a description of attrition from the MoBa study.
Example 1: birth weight (offspring phenotype)
Both offspring and maternal genes are likely to be involved in determining birth weight as the intrauterine environment is provided by the mother. Both traditional family (Lunde et al. 2007; Magnus 1984) and molecular genetic designs (Warrington et al. 2019) have previously indicated substantial portions of variance in birth weight determined by both direct offspring and indirect maternal genetic effects. We applied to current method to birth weight measures in order to obtain a comparison to previous findings. This method further allows the correlation between maternal and offspring genetic effects to be estimated.
Example 2: relationship satisfaction (maternal phenotype)
Maternal reports of relationship satisfaction between mothers and fathers have been found to decrease on average following the birth of a child (Dyrdal et al. 2011). A possible explanation for this decrease is that relationship satisfactions to some degree depend on aspects of the infant phenotype. We therefore investigated whether maternal reports of relationship satisfaction six months after birth are influenced by offspring genotype. Measures of relationship satisfaction were obtained by summation of the ten items comprising the Relationship Satisfaction scale (Røysamb et al. 2014).
Example 3: body mass index (paternal phenotype)
Body mass index (BMI) in adulthood has both genetic and environmental components of causation. Yang et al. (2015) found that 27% of variability in BMI could be accounted for by direct genetic effects based on a detailed analysis of genome-wide SNP data. We analyzed paternal BMI obtained from maternal ratings of their partner’s weight and height. If any maternal biases are inherent in these ratings, including an indirect maternal genetic effect may allow us to still obtain valid estimates of the contributions from direct genetic effects.
A box–cox transformation and a scaling to zero mean and unit variance was applied to all phenotype measures. Because of the expected mean difference in birth weight between boys and girls, we included gender as a covariate. All models were fit using the OpenMx package (Neale et al. 2016) in R (R Core Team 2019).
Results from applying the full model to the three phenotypes are presented in table 2. The strongest genetic influences on birth weight were due to direct offspring effects, accounting for 10.6% of the variation. Indirect maternal effects accounted for another 7.5%, whereas there was no indication of indirect paternal effects. A positive covariance between direct offspring and indirect maternal genetic effects accounted for 2.4% of the variance, corresponding to a correlation estimated as \(\sigma _{om} / (\sigma _o\sigma _m)=0.27\).
For maternal relationship satisfaction, direct maternal genetic effects accounted for 10.3% of the variance. An almost equally large fraction of 10.2% was attributable to indirect offspring genetic effects, while indirect paternal genetic effects accounted for 6.4%. The correlation between direct maternal and indirect offspring genetic effects was estimated as \(-0.52\), the correlation between direct maternal and indirect paternal genetic effects as 0.92 and the correlation between indirect paternal and offspring genetic effects as \(-0.15\).
Genetic influences on paternal BMI were mainly attributable to direct paternal effects, accounting for 30.4% of the total variance.
For all three phenotypes, direct effects accounted for the largest fraction of genetic influences. These results are consistent with the general findings from twin studies, pointing to direct additive genetic effects as the major systematic source of variation for most traits (Polderman et al. 2015; McAdams et al. 2014).
In the analysis of birth weight, we considered the offspring as the focal individual. Our analysis indicated contributions from both offspring and maternal genetic effects, and a larger fraction from direct than from indirect maternal effects. Estimates from biometric analysis of pedigrees have attributed 30–50% of the variability in birth weight to direct genetic effects and around 20% to indirect maternal genetic effects (Magnus 1984; Lunde et al. 2007). Two other studies, relying on similar methodology as in our application, have estimated direct offspring genetic effects to account for nearly 30% of the variation and indirect maternal genetic effects to account for nearly 10% (Warrington et al. 2019; Qiao et al. 2020). Similar to our estimate, both studies also reported a positive correlation between direct and indirect effects, suggesting that partially the same genes may be involved in these effects. The relative importance of direct versus indirect maternal genetic effects estimated in our analysis are thus consistent with prior findings. The absolute magnitudes of our estimates are however generally smaller. Compared to findings from pedigree designs, this is expected based on the different assumptions underlying these methodologies (see Yang et al. (2017) and Young (2019) for discussions). It is more difficult to reason about discrepancies between other studies using similar approaches until large enough samples are available to obtain estimates with satisfactory precision.
In the second exemplar analysis, the mother was the focal individual reporting on her satisfaction with the relationship to her partner. We estimated that a fraction of the trait variance could be ascribed to all family members, with strongest contributions from direct maternal and indirect offspring genetic effects. The strong positive correlation between maternal and paternal effects may suggest that the same genes contribute to the maternal and paternal effects, whereas the negative correlation between maternal and offspring genetic effects may indicate that genes have opposing effect when expressed in mothers and offspring. A prior twin study estimated that around half of the variability in relationship satisfaction could be ascribed to direct genetic effects (South et al. 2016), but we are unaware of other attempts to quantify the importance of genetic effects expressed in other family members. These initial findings may motivate further studies into how relationship satisfaction may depend on characteristics of partners and children.
The last application concerned BMI where fathers were the focal individual in the analysis. Because weight and height values were provided from their partner, we considered the possibility that a component of the BMI value could be attributed to maternal genetic effects. This was not indicated in the analysis, and we estimated that approximately 30% of the variability was due to direct genetic effects. This is close to the estimates from Yang et al. (2015) of 27% and Young et al. (2018) of 34% which relied on genome-wide SNP data. Results from twin and family designs are typically larger, with estimates ranging from 40 to 90% and 24 to 81%, respectively (Maes et al. 1997; Elks et al. 2012).
Considering the relatively large uncertainty associated with the parameter estimates, the results from the applications should be interpreted with caution. We emphasize that our analyses are not intended as a comprehensive study of the causes of variation for the phenotypes we examined, but rather are meant to illustrate how the proposed model can be used to investigate a diverse range of research questions. Considerably larger sample sizes may be necessary to justify reliable inferences about the model parameters (Visscher et al. 2014; Yang et al. 2017). For a more detailed analysis it would likely be preferable to fit alternative nested models as described above and compare whether simpler models are equally supported by the data. We did not pursue this approach here because with the current sample size it is unlikely that we could detect relevant aspects of alternative model specifications. However, sufficiently large samples are increasingly available.
Discussion
We proposed a new method, Trio-GCTA, for resolving direct and indirect genetic effects within parent-offspring trios when genome-wide SNP data is available. The model formulation is invariant to which of the family members is the focal individual in the analysis; only the interpretation of parameters (in terms of direct and indirect genetic effects) changes in different cases. We illustrated this by applying the method to three exemplar phenotypes using real data on offspring, maternal and paternal phenotypes. Results from the applications highlighted the potential of the method for clarifying intra-familial dynamics.
An advantage of the proposed method is the ability to gain insights into the dynamics of intra-familial processes without requiring specification of the specific traits that mediate the indirect genetic effects. Variance-partitioning of direct and indirect genetic effects may therefore serve as a useful first step, potentially motivating more detailed studies of specific processes. Trait-based models (Bijma 2014), including explicit formulations of the hypothesized mediating variables may potentially provide better understanding of such mechanisms. However, in addition to the computational challenges, such specifications would also contradict one of the initial motivations for the GCTA model which avoid bias from common environmental effects by relying on measures obtained from unrelated individuals (Yang et al. 2011).
Several other methodological approaches outside those we have already discussed have been developed to address questions related to indirect genetic effects from relatives. Various kinships have been used to specify variance partitioning (York et al. 2009, 2013) and trait-based models (Maes et al. 1997), and have a long history in quantitative genetics (Lynch and Walsh 1998). The polygenic score approach taken in Bates et al. (2018) and Kong et al. (2018) is related to our method, estimating the contributions of indirect genetic effects associated with specific parental traits.
There are several issues related to estimating genetic variance parameters from genome-wide SNP data. Yang et al. (2017) emphasized that genetic variance parameters based on measured (or imputed) genome-wide SNPs differ from population parameters because they are dependent on the specific set of SNPs included in the analysis. They addressed several issues relating to estimating genetic variance parameters from genome-wide SNP data, and these considerations apply also to the method proposed in the current paper. There are likely further challenges that are specifically related to the use of parent-offspring trios and the method we have proposed here. First, the full model has seven variance parameters, which will likely require large sample sizes in order to obtain reliable estimates. Second, we have assumed that mating is random, and it is currently unclear how assortative mating could affect inferences under different models of intra-familial interactions. Third, although the distinction between direct and indirect genetic effects of parents and offspring may be an adequate description of many phenotypes, other relatives such as siblings may also play important roles in determining individual differences. Fourth, we have assumed that direct and indirect genetics effects combine additively in influencing the phenotype. Both dominance and epistatic effects within individuals, but also interactions between direct and indirect genetic among family members would violate this.
We believe the proposed method will provide a useful tool for researchers interested in the complexity of intra-familial dynamics, allowing investigations of research questions that may otherwise be difficult to study.
Data availability
Not applicable.
Code availability
Code for estimating the model is available at https://github.com/espenmei/trio
References
Bates TC, Maher S, Medland SE, McAloney K, Wright MJ, Hansell NK, Kendler KS, Martin NG, Gillespie NA (2018) The nature of nurture: using a virtual-parent design to test parenting effects on children’s educational attainment in genotyped families. Twin Res Hum Genet 21(2):73–83
Bijma P (2014) The quantitative genetics of indirect genetic effects: a selective review of modelling issues. Heredity 112(1):61–69
Dyrdal GM, Røysamb E, Nes RB, Vittersø J (2011) Can a happy relationship predict a happy life? A populationbased study of maternal well-being during the life transition of pregnancy, infancy, and toddlerhood. J Happiness Stud 12(6):947–962
Eaves LJ, Pourcain BS, Smith GD, York TP, Evans DM (2014) Resolving the effects of maternal and offspring genotype on dyadic outcomes in genome wide complex trait analysis (“M-GCTA”). Behav Genet 44(5):445–455
Elks CE, Den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJF, Ong KK (2012) Variability in the heritability of body mass index: a systematic review and metaregression. Front Endocrinol 3:29
Evans DM, Moen G-H, Hwang L-D, Lawlor DA, Warrington NM (2019) Elucidating the role of maternal environmental exposures on offspring health and disease using two-sample Mendelian randomization. Int J Epidemiol 48(3):861–875
Helgeland Ø, Vaudel M, Juliusson PB, Holmen OL, Juodakis J, Bacelis J, Jacobsson B, Lindekleiv H, Hveem K, Lie RT et al (2019) Genome-wide association study reveals dynamic role of genetic variation in infant and early childhood growth. Nat Commun 10(1):1–10
Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, Benonisdottir S, Oddsson A, Halldorsson BV, Masson G et al (2018) The nature of nurture: effects of parental genotypes. Science 359(6374):424–428
Laurin C, Cuellar-Partida G, Hemani G, Smith GD, Yang J, Evans DM (2018) Partitioning phenotypic variance due to parent-of-origin effects using genomic relatedness matrices. Behav Genet 48(1):67–79
Lunde A, Melve KK, Gjessing HK, Skjærven R, Irgens LM (2007) Genetic and environmental in uences on birth weight, birth length, head circumference, and gestational age by use of population-based parent-offspring data. Am J Epidemiol 165(7):734–741
Lynch M, Walsh B et al (1998) Genetics and analysis of quantitative traits, vol 1. Sinauer, Sunderland
Maes HHM, Neale MC, Eaves LJ (1997) Genetic and environmental factors in relative body weight and human adiposity. Behav Genet 27(4):325–351
Magnus P (1984) Causes of variation in birth weight: a study of offspring of twins. Clin Genet 25(1):15–24
Magnus P, Birke C, Vejrup K, Haugan A, Alsaker E, Daltveit AK, Handal M, Haugen M, Høiseth G, Knudsen GP et al (2016) Cohort profile update: the Norwegian mother and child cohort study (MoBa). Int J Epidemiol 45(2):382–388
McAdam AG, Dany G, Wilson AJ (2014) The effects of others’ genes: maternal and other indirect genetic effects. Quant Genet Wild. https://doi.org/10.1093/acprof:oso/9780199674237.003.0006
McAdams TA, Neiderhiser JM, Rijsdijk FV, Narusyte J, Lichtenstein P, Eley TC (2014) Accounting for genetic and environmental confounds in associations between parent and child characteristics: a systematic review of children-of-twins studies. Psychol Bull 140(4):1138
Neale MCCL, Cardon LR (2013) Methodology for genetic studies of twins and families, vol 67. Springer, New York
Neale MC, Hunter MD, Pritikin JN, Zahery M, Brick TR, Kirkpatrick RM, Estabrook R, Bates TC, Maes HH, Boker SM (2016) OpenMx 2.0: extended structural equation and statistical modeling. Psychometrika 81(2):535–549. https://doi.org/10.1007/s11336-014-9435-8
Polderman TJC, Benyamin B, De Leeuw CA, Sullivan PF, Van Bochoven A, Visscher PM, Posthuma D (2015) Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat Genet 47(7):702
Qiao Z, Zheng J, Helgeland Ø, Vaudel M, Johansson S, Njølstad PR, Smith GD, Warrington NM, Evans DM (2020) Introducing M-GCTA a software package to estimate maternal (or paternal) genetic effects on offspring phenotypes. Behav Genet 50(1):51–66
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna. https://www.R-project.org/
Røysamb E, Vittersø J, Tambs K (2014) The relationship satisfaction scale-psychometric properties
South SC, Krueger RF, Elkins IJ, Iacono WG, McGue M (2016) Romantic relationship satisfaction moderates the etiology of adult personality. Behav Genet 46(1):124–142
Visscher PM, Hemani G, Vinkhuyzen AAE, Chen G-B, Lee SH, Wray NR, Goddard ME, Yang J (2014) Statistical power to detect genetic (co) variance of complex traits using SNP data in unrelated samples. PLoS Genet. https://doi.org/10.1371/journal.pgen.1004269
Warrington NM, Beaumont RN, Horikoshi M, Day FR, Helgeland Ø, Laurin C, Bacelis J, Peng S, Hao K, Feenstra B et al (2019) Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat Genet 51(5):804–814
Yang J, Bakshi A, Zhu Z, Hemani G, Vinkhuyzen AAE, Lee SH, Robinson MR, Perry JRB, Nolte IM, van Vliet-Ostaptchouk JV et al (2015) Genetic variance estimation with imputed variants finds negligible missing heritability for human height and body mass index. Nat Genet 47(10):1114
Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW et al (2010) Common SNPs explain a large proportion of the heritability for human height. Nat Genet 42(7):565
Yang J, Hong Lee S, Goddard ME, Visscher PM (2011) GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88(1):76–82
Yang J, Hong Lee S, Wray NR, Goddard ME, Visscher PM (2016) GCTA-GREML accounts for linkage disequilibrium when estimating genetic variance from genome-wide SNPs. Proc Natll Acad Sci 113(32):E4579–E4580
Yang J, Lee SH, Goddard ME, Visscher PM (2013) Genome-wide complex trait analysis (GCTA): methods, data analyses, and interpretations. Genome-wide association studies and genomic prediction. Springer, New York, pp 215–236
Yang J, Zeng J, Goddard ME, Wray NR, Visscher PM (2017) Concepts, estimation and interpretation of SNP-based heritability. Nat Genet 49(9):1304
York TP, Eaves LJ, Lichtenstein P, Neale MC, Svensson A, Latendresse S, Långström N, Strauss III JF (2013) Fetal and maternal genes’ in uence on gestational age in a quantitative genetic analysis of 244,000 Swedish births. Am J Epidemiol 178(4):543–550
York TP, Strauss JF, Neale MC, Eaves LJ (2009) Estimating fetal and maternal genetic contributions to premature birth from multiparous pregnancy histories of twins using MCMC and maximum-likelihood approaches. Twin Res Hum Genet 12(4):333–342
Young AI (2019) Solving the missing heritability problem. PLoS Genet 15(6):e1008222
Young AI, Benonisdottir S, Przeworski M, Kong A (2019) Deconstructing the sources of genotype–phenotype associations in humans. Science 365(6460):1396–1400
Young AI, Frigge ML, Gudbjartsson DF, Thorleifsson G, Bjornsdottir G, Sulem P, Masson G, Thorsteinsdottir U, Stefansson K, Kong A (2018) Relatedness disequilibrium regression estimates heritability without environmental bias. Nat Genet 50(9):1304–1310
Zhu Z, Bakshi A, Vinkhuyzen AAE, Hemani G, Lee SH, Nolte IM, van Vliet-Ostaptchouk JV, Snieder H, Esko T, Milani L et al (2015) Dominance genetic variation contributes little to the missing heritability for human complex traits. Am J Hum Genet 96(3):377–385
Acknowledgements
The Norwegian Mother, Father and Child Cohort Study is supported by the Norwegian Ministry of Health and Care Services and the Ministry of Education and Research. We are grateful to all the participating families in Norway who take part in this on-going cohort study. This publication is a part of the project “Intergenerational Transmission of Internalizing and Externalizing Psychopathological Spectra: A Genome-Wide Complex Trait Study” supported by the Research Council of Norway (262177). Espen Moen Eilertsen and Eivind Ystrom was supported by the Norwegian Research Council (262177 and 288083). Laurie Hannigan was supported by a grant from the South-Eastern Norway Regional Health Authority (2018059). Alexandra Havdahl was supported by the South-Eastern Norway Regional Health Authority (2018058 and 2020022). Tom A. McAdams was supported by a Sir Henry Dale Fellowship, jointly funded by the Wellcome Trust and the Royal Society (107706/Z/15/Z) and the Norwegian Research Council (288083). Eshim S. Jami was supported by the European Union’s Horizon 2020 research and innovation programme, Marie Sklodowska Curie Actions (721567). This work was partly supported by the Research Council of Norway through its Centres of Excellence funding scheme, project number 262700. We thank the Norwegian Institute of Public Health (NIPH) for generating high-quality genomic data. This research is part of the HARVEST collaboration, supported by the Research Council of Norway (229624). We further thank the Center for Diabetes Research, the University of Bergen for providing genotype data and performing quality control and imputation of the data funded by the ERC AdG project SELECTionPREDISPOSED, Stiftelsen Kristian Gerhard Jebsen, Trond Mohn Foundation, the Research Council of Norway, the Novo Nordisk Foundation, the University of Bergen, and the Western Norway health Authorities (Helse Vest).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Espen Moen Eilertsen, Eshim Shahid Jami, Tom A. McAdams, Laurie J. Hannigan, Alexandra S. Havdahl, Per Magnus, David M. Evans, and Eivind Ystrom declare that they have no conflict of interest.
Human and Animal Rights
This study was performed in line with the principles of the Declaration of Helsinki. Approval was granted by the Regional Committee for Medical Research Ethics.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Stacey Cherny.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eilertsen, E.M., Jami, E.S., McAdams, T.A. et al. Direct and Indirect Effects of Maternal, Paternal, and Offspring Genotypes: Trio-GCTA. Behav Genet 51, 154–161 (2021). https://doi.org/10.1007/s10519-020-10036-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-020-10036-6