Abstract
Daily activity times and circadian rhythms of crickets have been a subject of behavioral and physiological study for decades. However, recent studies suggest that the underlying molecular mechanism of cricket endogenous clocks differ from the model of circadian rhythm generation in Drosophila. Here we examine the circadian free-running periods of walking and singing in two Hawaiian swordtail cricket species, Laupala cerasina and Laupala paranigra, that differ in the daily timing of mating related activities. Additionally, we examine variation in sequence and daily cycling of the period (per) gene transcript between these species. The species differed significantly in free-running period of singing, but did not differ significantly in the free-running period of locomotion. Like in Drosophila, per transcript abundance showed cycling consistent with a role in circadian rhythm generation. The amino acid differences identified between these species suggest a potential of the per gene in interspecific behavioral variation in Laupala.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Circadian rhythms, endogenously generated oscillations occurring on an approximately 24 h scale, have been found in organisms ranging from eubacteria to animals, plants and fungi (Dunlap 1999). The endogenous clock underlying circadian oscillation is an adaptation that enables the organism to predict daily and seasonal changes in the external environment (Yerushalmi and Green 2009). The circadian clock allows an organism to maintain circadian cycles, or free-run, in the absence of external cues. Much of our understanding of circadian clocks comes from Drosophila. In 1971, Konopka and Benzer reported three mutations of an X-linked gene, later named period (per), that alter the endogenous circadian free-running periods of flies. This work opened the door for subsequent investigations of the molecular basis of circadian rhythms. The primary model of a circadian clock based on Drosophila consists of interlocked transcriptional/translational feedback loops, the period/timeless (per/tim) loop and the clock/cycle (clk/cyc) loop, with other genes involved in regulating and entraining cycling (Hardin 2005; Yu and Hardin 2006; Weber 2009). Circadian cycling is maintained by an approximately 24 h cycle of protein expression, phosphorylation, trafficking between cytoplasm and nucleus, and degradation (Edery et al. 1994; Myers et al. 1996; Lee et al. 1998; Kloss et al. 2001; Shafer et al. 2002). Mammals possess a similar feedback clock mechanism, comprised of a set of genes partially overlapping with those of the Drosophila clock (Yu and Hardin 2006). Studies in a wide variety of organisms including cyanobacteria (Huang et al. 1991; Kondo et al. 1994), Neurospora cressa (Gardner and Feldman 1980; Bianchi 1964), several plants (Engelmann et al. 1974; Lecharny and Wagner 1984; Millar and Kay 1991), mammals (Refinetti 1996; Zheng et al. 1999; Liu et al. 2007), and a wide array of insects (Mazor and Dunkelblum 2005; Guldemond et al. 1994; Shimizu et al. 1997; Nowosielski and Patton 1963; Moriyama et al. 2009; Mazzoni et al. 2002; Lin et al. 2002) have further elucidated the behavioral and molecular basis of circadian rhythms.
Here we investigated circadian rhythms as well as sequence variation and transcription of per in two species of the Hawaiian swordtail crickets genus Laupala. Given the commonly observed daily cycling in cricket song, circadian rhythms of crickets have been a frequent subject of study, from early behavioral and physiological studies (Nowosielski and Patton 1963; Loher 1972, 1974; Sokolove 1975; Sokolove and Loher 1975; Wiedenmann and Loher 1984; Abe et al. 1997) to more recent molecular analyses (Hassaneen et al. 2011; Shao et al. 2008a, b; Moriyama et al. 2008; Abdelsalam et al. 2008; Lupien et al. 2003; Shao et al. 2006; Danbara et al. 2010). Like other organisms, crickets possess an endogenous circadian clock (Loher 1972, 1974; Abe et al. 1997); however, there are physiological differences in the clock of crickets relative to Drosophila. For example, the daily variation in expression of circadian clock proteins, including PER, CLK, TIM, which occurs in Drosophila (Dunlap 1999; Hardin 2005; Yu and Hardin 2006; Rosato et al. 2006), appears to be lacking in crickets (Shao et al. 2006, 2008a, b; Lupien et al. 2003). Additionally, CLK is the only one of these proteins which has been observed to translocate into the nucleus in crickets (Shao et al. 2008a). However, it was recently demonstrated that per transcript abundance undergoes daily variation in Gryllus bimaculatus and that inhibition of PER expression leads to arrhythmia (Moriyama et al. 2008, 2009). In contrast, while the tim transcript also cycles daily, inhibition of TIM expression does not induce arrhythmia (Danbara et al. 2010). Thus, many of the genetic underpinnings of circadian rhythms are shared among crickets and Drosophila, but substantial differences exist in the molecular mechanisms leading to those rhythms. However, a clear understanding of the circadian mechanism of crickets has not been elucidated.
The genus Laupala is particularly exciting for the study of temporal variation. Closely related species in the genus differ in several temporal characters, providing a compelling system of comparative analyses. The most conspicuous character differentiating the species is pulse rate of the male calling song, a sexually selected trait that is genetically regulated (Otte 1994; Shaw 1996, 2000a). Females show phonotactic preference for conspecific pulse rates (Shaw 2000b; Oh et al. 2012), demonstrating that pulse rate may function as an interspecific mating barrier. Species of Laupala have also been shown to display daily behavioral rhythms whereby males sing and courting pairs mate at predictable times during daylight hours (Danley et al. 2007; Fergus et al. 2011). Moreover, significant differences have been observed in the daily timing of singing and of mating between the sympatric species Laupala cerasina and Laupala paranigra, with L. paranigra exhibiting later timing of both activities (Danley et al. 2007; Fergus et al. 2011). This divergence in the timing of daily activity may help ameliorate interspecific acoustic interference between singing males and/or reduce the likelihood of interspecific mating.
In this study we examined the daily timing of singing and locomotion in visually and acoustically isolated males to verify the later activity phase of L. paranigra relative to L. cerasina. We then tested the hypothesis that these behaviors exhibit a free-running circadian rhythm in the absence of external cues, or zeitgebers. We test the hypothesis that L. paranigra has a longer free-running period than L. cerasina, inspired by the suggestion that later activity patterns correspond to longer free-running periods (Hamblen–Coyle et al. 1992; Miyatake et al. 2002). Additionally, we cloned and sequenced the per transcript from both species and measured transcript abundance at different times throughout the day to measure circadian expression. Our results suggest that daily cycling of the per transcript is consistent with a role in circadian rhythms, and that endogenous circadian rhythms may be pleiotropically linked to temporal variation on an ultradian scale.
Methods
Experimental animals
Laupala cerasina were collected from Kalopa State Park (20°02′N, 155°26′W) and L. paranigra were collected from Kaiwiki Road (19°45′N, 155°10′W) in 2005 from the Big Island of Hawaii. Fifth generation lab reared individuals were maintained at 19.4–20.1 °C under a 12:12 light:dark (L:D) cycle. Crickets were provided Cricket Feed (Fluker Farms, Port Allen, LA) and water on moistened Kimwipes. Care was taken to prevent sibling matings to reduce inbreeding, thus maintaining some genetic diversity in the laboratory populations. Adult males were used in all parts of this study. The mean adult ages (days past final molt) on the initial trial day in this study were 41.5 ± 18.9 days for L. cerasina and 41.3 ± 9.3 days for L. paranigra.
Daily timing and free-running periods
The songs of 26 L. cerasina and 23 L. paranigra adult male crickets were digitally recorded at temperatures between 19.4° and 20.8 °C and pulse rates, in pulses per second (PPS), were measured. Both song recording and pulse rate measurements were performed using Raven Interactive Sound Analysis Software (Cornell Lab of Ornithology, Ithaca, NY). To determine the song pulse rates, five pulse periods were measured (±0.5 ms) and averaged for each individual. The inverse of the mean period was calculated to produce the pulse rate. A regression analysis was used to assess the relationship between pulse rates and recording temperatures.
To measure daily activity times and endogenous free-running circadian periods, the males from which songs had been recorded were placed into individual, visually and acoustically isolated chambers with moistened Kimwipes and methylparaben treated food (Fig. 1). Each chamber contained an omnidirectional condenser microphone (Model 270-090, RadioShack Corporation, Fort Worth, TX) and an infrared photomicrosensor (Omron Industrial Automation, Schaumburg, IL) to detect motion. Each chamber was placed in a larger enclosure made of Quiet Barrier HD sound-insulating acrylic (American Micro Industries, Inc, Chambersburg, PA). Both the inner and outer chambers had 0.25 cm thick sound-insulating acrylic glass windows to allow light into the chambers. Upto six chambers were run simultaneously. The sound attenuation between chambers was estimated at over 100 dB at 5 kHz (the approximate carrier frequency of Laupala song) while the maximum observed volume of a Laupala song is under 80 dB (DJF, personal observation).
Song and locomotor activity were recorded continuously with the omnidirectional microphone and infrared photosensor, respectively, transmitted via a DI-720-USB Data Acquisition System (DATAQ Instruments, Inc, Akron, OH) to a computer that recorded the data using WinDaq Lite data acquisition software. The data were collected in one second bins by using the intelligent oversampling feature of WinDaq to sample all the inputs at 240 Hz and record either the maximum sample for the sound data or the mean of the samples for the locomotion data at one second intervals. By recording in this manner, song pulses, which are less than one second, are still detectable in the one-second bins. Short motion signals, such as an antenna moving past the infrared sensor, do not produce signals that differ substantially above background electrical noise. The crickets were recorded in the chambers for three days under 12:12 L:D conditions before switching to constant light (L:L) for at least 10 days. Because Laupala are known to sing and mate during the day (Danley et al. 2007; Fergus et al. 2011) we chose to use constant light to increase the likelihood of activity throughout the experiment. The light reaching the inner chambers in both L:D and L:L was low light (<15 lux). Though the light reaching the inner chambers was dim, lower light levels are sufficient to entrain daily rhythms in Drosophila (Bachleitner et al. 2007). Temperatures during the trials remained between 19.7 and 20.2 °C, and did not vary on a daily cycle during the constant lighting conditions.
To examine the daily timing of singing and locomotor activity we estimated the proportion of occurrence of each activity within each individual. For statistical analysis, the 24 h L:D cycle was divided into four bins of 6 h (ZT 0–6, 6–12, 12–18, and 18–24), and individual data collected over 2 days of the L:D period were grouped into the four bins. The total song or locomotion activity was calculated for each 6 hour bin and normalized to the maximal activity for each individual. Student’s t tests were used to compare activities between species for each of the 6 hour periods. For visualization purposes, we calculated the mean activity for each species in 0.5 h bins, normalized to the maximal activity for each species, and plotted this against zeitgeber time (ZT).
For each individual, we estimated the endogenous free-running periods separately for the song and locomotion data using the Lomb–Scargle periodogram method (Ruf 1999) with a resolution of 0.1 h. When the Lomb–Scargle method was not able to identify a significant free-running period estimate those data were excluded from further analyses; this situation occurred among seven of 92 total individual estimates. Double-plotted actograms were produced to visualize the data. To determine whether song and locomotion may be regulated by different circadian pacemakers we used a correlation analysis and a paired t test to verify that the free-running periods of song and locomotion are positively correlated and did not differ significantly from one another within individuals. Regression analyses were used to determine whether free-running period is related to the adult age (number of days past final molt) of the crickets or the start date of the trial, a factor that would account for any effects of temperature variation during rearing or trials. Possible interspecific differences in the free-running periods were examined using t-tests assuming unequal variances. An analysis of covariance (ANCOVA) was performed to examine song pulse rate as a function of circadian free-running period, species, and the interaction between free-running period and species. Regression analyses were performed with L. paranigra data to further examine the significant relationship between song pulse rate and circadian free-running period identified in this species. The statistical software JMP (SAS Institute, Cary, NC) was used to conduct all statistical tests.
Identification and sequencing of per
To identify the Laupala per sequence we isolated total RNA from the head of a L. cerasina male using the PureLink Total RNA Purification System (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized from RNA with Superscript II (Invitrogen, Carlsbad, CA). We designed degenerate PCR primers (GGATACAGAATGGTCCGCTTTYRTNAAYCC and CGAAGAATCGCTGAATGTTTTCRTTRTARTT) based on regions of high amino acid conservation between the cricket G. bimaculatus and other insects using the CODEHOP method. Using these primers we performed PCR and amplified a 470 bp region of the per transcript from the cDNA. This fragment was cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) and sequenced. Based on the initial sequence, rapid amplification of cDNA ends (RACE) was performed from L. cerasina head total RNA using a Generacer kit (Invitrogen, Carlsbad, CA) to amplify and sequence the 5′ and 3′ portions of the transcript.
To obtain high quality sequences from both species, total RNA was isolated from heads of L. cerasina and L. paranigra males, and reverse transcribed into first-strand cDNA as above. PCR primers were designed based on the RACE sequences to amplify most of the per transcript. The forward primer, ACGCATGTGTCAAGAGCTGA, binds to a site within the 5′ untranslated region (UTR) and the reverse primer, ATGGGTCGAGGTTTGTTGTG, binds near the 3′ end of the coding region. First-strand cDNA products from each species were used as templates for PCR reactions with Platinum Taq HiFi polymerase (Invitrogen, Carlsbad, CA), producing products of approximately 3 200 bp. Each PCR reaction was performed in duplicate so that each portion of the PCR product was amplified, cloned, and sequenced at least twice from each species, with most regions being sequenced several times to reduce the presence of PCR and sequencing errors. Because there was no suitable 3′ UTR priming site, the 3′ end of the transcript was amplified using 3′ RACE in both species. The sequences were assembled into contiguous sequences (contigs), aligned, and analyzed using BioEdit sequence analysis software (Ibis Therapeutic, Carlsbad, CA), ClustalW (Thompson et al. 1994), and NetPhos 2.0 phosphorylation site prediction software (Blom et al. 1999). The BLOSUM 62 matrix was used for determining amino acid similarities. The deduced amino acid sequences of Laupala PER were compared with the PER sequences from G. bimaculatus (GenBank Accession: BAG48878) and Drosophila melanogaster (Accession: AAF45804).
Two potential alternative splice sites were identified within the coding region of per based on presence or absence of regions of DNA within different clones. To make sure these potential alternative isoforms were not artifacts of cloning we verified alternative splicing by splice-site specific PCR with primer pairs flanking each of the putative splice sites (Table 1). If a site is not alternatively spliced, PCR with flanking primers is expected to produce a single band, whereas two bands are expected if alternative splicing does occur. We performed the PCRs on cDNA from heads of both L. cerasina and L. paranigra using the primers flanking each of the putative splice sites, as well as an apparently non-spliced control region of per as a control.
Analysis of daily per cycling
To determine whether daily cycling of the per transcript occurs in Laupala and whether there are differences between the species, we performed reverse-transcription quantitative PCR (qPCR) with total RNA from the heads of L. cerasina and L. paranigra males maintained under 12:12 L:D and sacrificed at each of six times of day. Heads were used because evidence suggests that the master circadian pacemaker in crickets is located in the optic lobes (Loher 1974; Sokolove and Loher 1975; Tomioka and Chiba 1992; Abe et al. 1997). Three males from both species were sacrificed at each of the following times: ZT 3, 9, 11, 13, 15 and 21 (ZT 0 = lights on). The time course focused heavily around lights off because per transcript abundance is expected to peak around this time (Moriyama et al. 2008). Crickets were sacrificed by CO2 anesthesia, decapitated, the heads submerged in RNAlater (Ambion, Inc, Austin, TX), and stored at 4 °C until all tissues were collected and could be processed. Total RNA was extracted from each of the heads using the PureLink kit, and quantified using a nanodrop 1 000 spectrophotometer (Thermo Scientific, Waltham, MA). Two μg of each RNA sample was treated with DNase (Turbo DNA-free; Ambion, Inc) in 50 μl reactions according to the manufacturer’s protocol to remove contaminating DNA. The RNA samples were reverse transcribed with random hexamer primers using AffinityScript reverse transcriptase (Stratagene, La Jolla, CA).
We performed qPCR using the relative standard method with Power SYBR PCR Master Mix (Applied Biosystems Inc, Foster City, CA) and a 7 900 HT Sequence Detection System (Applied Biosystems Inc, Foster City, CA). The per primers used were ATGGGTGGTTCACAGTCTCCACAT and TGACCAGGAAGAAGCATGCCAGTA, with 16S rRNA as an internal control. For the standard curves, cDNA samples were pooled and diluted to produce a curve ranging from 0.25× to 4× the amount used in the experimental reactions. We standardized the per and 16S control results to their respective standard curves, and then normalized the per results to the 16S internal control for that sample. The per transcript abundances normalized to the peak abundance were plotted against time using SigmaPlot 10 (SPSS Inc, Chicago, IL). A two-way ANOVA was used to test for statistically significant daily variation or interspecific differences.
Results
Daily timing and free-running periods
There are significant differences in the timing of daily activities between L. cerasina and L. paranigra under L:D conditions (Fig. 2). L. cerasina performed a significantly greater portion of their song activity in the first 6 h of light (t = 3.96, df = 33, p < 0.01), while L. paranigra sang primarily during the last 6 h of light (t = 4.54, df = 33, p < 0.01). The locomotion data indicate that L. paranigra are less active early in the dark phase than they are late in the dark phase, with a gradual increase in activity as the dark phase progresses. During two time periods, the first 6 h of light (t = 2.55, df = 33, p = 0.016) and the first 6 h of dark (t = 5.66, df = 33, p < 0.01), L. cerasina were significantly more likely than L. paranigra to show locomotory activity, with no significant differences at other times of day.
Daily song and locomotor activity. The normalized activity of L. cerasina and L. paranigra under L:D conditions are plotted against ZT hour. The horizontal lines with asterisks above the graphs indicate significant interspecific difference in activity over the 6 hour period. (* p < 0.05; ** p < 0.01)
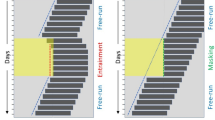
Males of each species exhibited endogenous circadian free-running periods for both singing and locomotion over a minimum of 10 days of constant light (Fig. 3). Using Lomb–Scargle periodogram analyses, we identified significant free-running periods at the α = 0.01 level in 81 and 96 % of L. cerasina for song and locomotion respectively and in 100 and 95 % of L. paranigra for song and locomotion. A paired t test showed no difference in individual free-running periods as estimated by singing versus by locomotion (t = 0.37, df = 38, p = 0.71). Additionally, free-running period estimates based on song were positively correlated with free-running period estimates based on locomotion for both L. cerasina (p < 0.01, r = 0.63) and L. paranigra (p < 0.01, r = 0.67).
Representative double-plotted actograms of locomotor and song activity. The white and gray bars and ZT hour indicate the timing of the initial L:D regime and arrows indicate the change to constant low light. Each actogram displays data from one individual. All individuals had free-running periods greater that 24 h for both locomotion and song, with L. cerasina generally having a longer free-running period than L. paranigra. The free-running periods for these individuals are shown below each actogram
Based on regression analyses, the ages of the crickets used in this study had no significant effect on the free-running estimates based on song or locomotion for either species (song: L. cerasina, p = 0.65; L. paranigra, p = 0.90; locomotion: L. cerasina, p = 0.79; L. paranigra, p = 0.09). There was a significant negative relationship between the trial start day and the free-running period estimate based on song in L. paranigra (p = 0.013, R 2 = 0.29), with individuals in later trials having shorter free-running periods than those in earlier trials. However, no significant relationship was found in the locomotion estimate from L. paranigra (p = 0.11) or either of the L. cerasina free-running period estimates (song: p = 0.23; locomotion p = 0.50).
The mean free-running period estimates based on both song and locomotion were longer in L. cerasina than in L. paranigra. The estimates based on song were significantly different between species (F 1,39 = 9.60, p < 0.01) while the difference based on locomotion was not statistically significant (F 1,42 = 0.28, p = 0.60). The estimated interspecific difference in free-running periods ranged from 0.12 h (locomotion) to 0.73 h (song; Table 2).
Males’ songs were recorded and the pulse rates were measured for both species (Table 2). Regression analyses showed no significant relationships between song pulse rate and trial start date (L. cerasina: p = 0.75; L. paranigra: p = 0.25), age (L. cerasina: p = 0.66; L. paranigra: p = 0.48), or song recording temperature (L. cerasina: p = 0.24; L. paranigra: p = 0.28). ANCOVAs demonstrated statistically significant interactions between species and free-running periods, measured from song or from locomotion, on song pulse rates (song*species: F 1,37 = 4.41, p = 0.043; locomotion*species: F 1,40 = 4.83, p = 0.034). These significant interactions indicate that the relationship between pulse rate and free-running period differed by species. To this end, we found significant relationships between song pulse rate and free-running period of both song (p = 0.011, R 2 = 0.31) and locomotion (p = 0.011, R 2 = 0.33) in L. paranigra, with longer free-running periods corresponding to slower pulse rates (Fig. 4). We did not see a relationship between song pulse rate and free-running period in L. cerasina (song: p = 0.36; locomotion: p = 0.30).
Linear regressions of L. paranigra song pulse rate by endogenous circadian free-running period of song (closed circle, solid line) and of locomotion (open circles, dashed line). The data show a significant negative relationship between song pulse rate and the free-running periods of song and locomotion
Identification and sequencing of per
We obtained the 3 770 bp full length sequence of the L. cerasina per transcript (GenBank accession No. GU053569) and 3 702 bp of the L. paranigra per transcript (GenBank accession No. GU053570), which included all but the end of the 5′ untranslated region. The nucleotide sequences are 99.6 % identical between species, with only 14 interspecific nucleotide substitutions out of 3 702 bp. The deduced PER amino acid sequences had eight non-synonymous (amino acid coding) substitutions. Two predicted serine phosphorylation sites were differentially present/absent between the L. paranigra and L. cerasina sequences.
Laupala PER amino acid sequences differed substantially from both G. bimaculatus and D. melanogaster (Table 3; Fig. 5). The divergence between G. bimaculatus and Laupala resulted largely from regions of low sequence identity interspersed with regions of high identity (Fig. 5). Amino acid divergence within Laupala, and between Laupala and G. bimaculatus, primarily occurred outside of the functional domains as identified in G. bimaculatus (Moriyama et al. 2008).
Alignment of L. cerasina (Lc), L. paranigra (Lp), and G. bimaculatus (Gb; BAG48878) PER proteins. Alternative splice sites are italicized and underlined. Functional protein domains (PAS-A, PAS-B, NLS, and CLD), based on (Moriyama et al. 2008), are indicated below the alignment. Arrows designate variable predicted phosphorylation sites while the other Laupala amino acid substitutions are marked with a triangle
Two potential alternative splice sites were detected in the process of cloning and sequencing the per transcript (Fig. 6). The upstream alternative splice site (splice site 1) is 75 bp, and the downstream alternative splice site (splice site 2) is 123 bp in length. PCR products corresponding to both the spliced and non-spliced sizes were observed in the heads of L. cerasina and L. paranigra, while the control PCR of a non-spliced region showed a single band in each species. The larger band at splice site one in L. paranigra, for which we did not get sequence data, may represent a PCR artifact or an additional splice isoform. These results confirm the alternative splicing of per in Laupala.
Alternative splicing of the per transcript. a The schematic representation of the per transcript demonstrates the location of primers (arrows) relative to the alternative splice sites. Expected PCR product sizes with and without the alternative exons (gray regions) are given below each site. b The PCR results which show bands at the expected sizes indicate that both sites are alternatively spliced in the heads of L. cerasina (left) and L. paranigra (right)
Analysis of daily per cycling
Transcript abundance of per varied significantly through the day (F 5,24 = 4.41, p < 0.01, Fig. 7). As expected, the per transcript was most abundant close to lights off (ZT: 12), with the peak observation occurring 1 h before lights off (ZT 11) in both L. cerasina and L. paranigra. The two species did not differ in relative per abundance across times (F 1,24 = 0.15, p = 0.70).
qPCR of per transcript abundance in the heads of L. cerasina and L. paranigra throughout the day. The white and gray bar at the top indicates periods of light and dark. The results demonstrate daily cycling of per transcript abundance in both species, but no interspecific difference in timing. The data points are offset slightly for ease of viewing
Discussion
Daily activity times and free-running periods
Circadian rhythms are an important adaptation (Yerushalmi and Green 2009) that allow an organism to anticipate and prepare for daily and seasonal environmental variation. Tremendous effort has gone into examining circadian rhythms in insects (Sandrelli et al. 2008; Helfrich-Forster 2005; Tomioka and Abdelsalam 2004; Shirasu et al. 2003; Giebultowicz 2000; Helfrich-Forster et al. 1998; Chiba and Tomioka 1987), with Drosophila providing the most well characterized molecular model of a circadian clock (Konopka and Benzer 1971; Hardin 2005; Yu and Hardin 2006; Weber 2009; Edery et al. 1994; Sandrelli et al. 2007; Ewer et al. 1992; Tauber et al. 2007; Rieger et al. 2006). However, even before the first clock mutant was reported in D. melanogaster, researchers had been investigating daily variation and circadian rhythms in crickets (Nowosielski and Patton 1963; Nowosielski and Patton 1964; Alexander and Meral 1967; McFarlane 1968). The circadian mechanism in crickets appears to differ from that of Drosophila, both in the molecular basis of the endogenous clock as well as the localization of the circadian pacemaker neurons (Sokolove and Loher 1975; Abe et al. 1997; Shao et al. 2006, 2008a, b; Abdelsalam et al. 2008; Lupien et al. 2003; Shiga et al. 1999). Understanding both the behavioral variation in circadian rhythms among crickets and mechanisms underlying cricket circadian clocks will provide insight into the evolution and plasticity of circadian rhythm generation, as well as further our understanding of the molecular basis of circadian timing.
In this study we found significant differences in the daily timing of both song activity and locomotion under L:D conditions between L. cerasina and L. paranigra. For both activities the onset of activity is earlier in L. cerasina than in L. paranigra, consistent with previous reports of daily timing in these species (Danley et al. 2007; Fergus et al. 2011). Interestingly, the interspecific differences in activity times appear to result from both a shift in the timing of activity as a whole as well as variation in the underlying temporal pattern of activity. Most notably, while the locomotion activity of L. cerasina is high immediately at lights off and remains high throughout the dark phase, the locomotion activity of L. paranigra appears to increase throughout the dark phase to peak just before lights on (Fig. 2b).
We have demonstrated free-running periods for both song and locomotion activity in Laupala. The positive correlations and lack of significant differences within species between the free-running period of song and of locomotion is consistent with previous work from Teleogryllus commodus suggesting a single circadian pacemaker regulates the timing of both song and locomotion in crickets (Sokolove 1975).
There was a statistically significant difference in the circadian free-running period of song, but not of locomotion, between L. cerasina and L. paranigra. The observed difference of approximately one half-hour in the mean free-running periods of song is smaller than the roughly 2 h difference in mating times observed between these species (Danley et al. 2007; Fergus et al. 2011) or the 10 h difference seen here in peak song time (Fig. 2a). However, the timings of singing, locomotion, and mating are not likely to scale exactly with one another due to other non-circadian temporal regulation and zeitgeber cues. More notably, the direction of the difference in free-running periods is the opposite of what we hypothesized. In D. melanogaster and Bactrocera cucurbitae, flies with longer free-running periods have later phases of locomotor activity and mating than flies with short free-running periods (Hamblen–Coyle et al. 1992; Miyatake et al. 2002). While L. cerasina has a longer free-running period than L. paranigra, singing, mating, and locomotion occurs earlier in L. cerasina than L. paranigra. Thus, though song and locomotion are under circadian regulation, the basis of the difference in daily timing between the two Laupala species may be independent of the variation in circadian rhythms, or the neural or molecular mechanism regulating both behaviors is different than that seen in other species.
We observed a significant negative relationship between free-running periods and song pulse rate in L. paranigra. This relationship is consistent with the relationships observed in T. oceanicus (Lupien 1998), D. melanogaster (Kyriacou and Hall 1980) and B. cucurbitae (Miyatake and Kanmiya 2004), but was not seen in L. cerasina. It is important to note that the relationship between pulse rate and song free-running period in L. paranigra may be confounded by the significant effect of trial start date on song free-running period. However, song pulse rate was not significantly related to trial start date, and a significant negative relationship also exists between song pulse rate and locomotion free-running period, which was not significantly related to trial start day. Furthermore, variation of up to 0.5 °C during the rearing of the crickets is unlikely to have substantially influenced the results because of the small amplitude of this variation and the tightly regulated temperature during the experimental trials. Thus the relationship between free-running period and pulse rate in L. paranigra does not appear to be explained by inter-trial variation and may represent a shared underlying mechanism with other species.
Extrapolating the best-fit line of the pulse rate regression for L. paranigra to the mean song rate of L. cerasina (2.38 pps) would predict an endogenous circadian free-running period of less than 3 h for L. cerasina. Such a short free-running period would certainly be maladaptive. If there is a common genetic basis to the variation in song and circadian rhythms, there is likely a constraint on the extent to which the gene(s) affecting these traits can co-vary.
Analysis of the period gene
In D. melanogaster circadian free-running period and modulation of courtship song interpulse interval are both affected by mutations of the per gene (Kyriacou and Hall 1980). Moreover, natural variation in per accounts for differences between Drosophila species in courtship and the daily timing of locomotion and mating (Wheeler et al. 1991; Tauber et al. 2003; Petersen et al. 1988). While PER protein abundance has not been shown to cycle in crickets (Lupien et al. 2003; Shao et al. 2006), per transcript abundance has been found to cycle on a daily basis in the cricket G. bimaculatus and was important for proper expression of circadian rhythms in that species (Moriyama et al. 2008, 2009). The temporal variation in circadian rhythms, daily activity, and song pulse rate observed in Laupala make per a strong candidate for Laupala behavioral variation.
Using qPCR we tested the hypothesis that per transcript abundance varies across the day. Previous studies suggest that longer free-running periods correlate with a later occurrence of circadian regulated events under L:D conditions (reviewed in Miyatake 2002). Counter to this, L. cerasina has been found to have a longer free-running period (this study) but earlier occurrences of locomotor, courtship, and mating times compared to L. paranigra (Fergus et al. 2011). Because of this apparent inconsistency we examined the phase timing of per transcript accumulation in L. cerasina and L. paranigra. We confirmed that there is daily cycling of the per transcript in the heads of both species with peaks around lights off, similar to what was observed in G. bimaculatus (Moriyama et al. 2008). This is consistent with a role for per in maintaining circadian rhythms. However, there was no significant difference between L. paranigra and L. cerasina in the temporal expression of per transcripts under L:D conditions. We did observe a tendency for transcript abundance to rise and fall more quickly in L. cerasina than in L. paranigra, which may suggest an earlier shift in cycling in L. cerasina. Such a phase shift in per accumulation, if real, is consistent with the earlier activity times of daily behaviors in L. cerasina and is consistent with a more complex relationship between longer free-running periods and later activity times under entrained conditions.
We observed interspecific sequence variation in the per transcript, which may play a role in behavioral differences between the species. There are eight amino acids differences between these species, two of which alter predicted serine phosphorylation sites within the PER protein. According to the Drosophila model, PER phosphorylation is involved in setting the pace of the circadian clock by inducing degradation of PER (Zheng and Sehgal 2008; Bae and Edery 2006). It has been proposed that cycling of phosphorylation of clock proteins, as opposed to cycling of protein abundance, may be key to maintaining the circadian clock (Zheng and Sehgal 2008). Such a model may be consistent with the circadian clock of crickets, in which circadian clock protein levels are constant throughout the day (Shao et al. 2006, 2008b; Lupien et al. 2003). In addition to the changes in putative phosphorylation sites, two non-conserved amino acid changes between L. cerasina and L. paranigra occurred within the PAS-B domain, a protein interaction domain. One of these PAS-B substitutions is between a negatively charged, polar residue (glutamate; L. cerasina) and a neutral, non-polar residue (valine; L. paranigra) while the other substitution is between a neutral residue (threonine; L. cerasina) and a positively charged residue (histidine; L. paranigra). Such differences in polarity and charge in the protein binding site could alter protein binding affinities and may thus have consequences for PER function. The remainder of the amino acid substitutions fall outside of known functional domains and are not predicted to alter phosphorylation; however, most of these substitutions are also non-conserved and may affect the overall conformation or function of the protein.
In addition to the molecular variation and cycling identified in Laupala per, we found two alternative splice sites of the transcript. Both of these sites are alternatively spliced in the heads of both species examined here. Alternative splicing of a single gene produces multiple isoforms with different properties. The potential function of the alternative splice isoforms of per is intriguing. Though the total abundance of PER protein has not been found to cycle throughout the day in crickets, the cycling of different splice forms has not been examined. Cycling of alternative splice isoforms may be key to maintaining circadian cycling in crickets.
Our results suggest that the variation in daily behavior observed between L. cerasina and L. paranigra is not due to a simple shift in endogenous circadian timekeeping. The complex nature of the daily temporal variation suggests that it is likely the result of multiple genetic factors. Laupala provides a tractable system with which to investigate temporal variation and elucidate the genetic underpinnings of temporal regulation. The variation observed in the per gene demonstrates the potential for a role of per in behavioral variation, and provides the foundation for further analyses of this candidate gene.
References
Abdelsalam S, Uemura H, Umezaki Y, Saifullah ASM, Shimohigashi M, Tomioka K (2008) Characterization of PDF-immunoreactive neurons in the optic lobe and cerebral lobe of the cricket, Gryllus bimaculatus. J Insect Physiol 54(7):1205–1212
Abe Y, Ushirogawa H, Tomioka K (1997) Circadian locomotor rhythms in the cricket, Gyrllodes sigillatus. I. Localization of the pacemaker and the photoreceptor. Zool Sci 14(5):719–727
Alexander RD, Meral GH (1967) Seasonal and daily chirping cycles in the northern spring and fall field crickets, Gryllus veletis and G. pennsylvanicus. Ohio J Sci 67(4):200–209
Bachleitner W, Kempinger L, Wulbeck C, Rieger D, Helfrich-Forster C (2007) Moonlight shifts the endogenous clock of Drosophila melanogaster. Proc Natl Acad Sci USA 104(9):3538–3543
Bae K, Edery I (2006) Regulating a circadian clock’s period, phase and amplitude by phosphorylation: insights from Drosophila. J Biochem 140(5):609–617
Bianchi DE (1964) Endogenous circadian rhythm in Neurospora crassa. J Gen Microbiol 35(3):437–445
Blom N, Gammeltoft S, Brunak S (1999) Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 294(5):1351–1362
Chiba Y, Tomioka K (1987) Insect circadian activity with special reference to the localization of the pacemaker. Zool Sci 4(6):945–954
Danbara Y, Sakamoto T, Uryu O, Tomioka K (2010) RNA interference of timeless gene does not disrupt circadian locomotor rhythms in the cricket Gryllus bimaculatus. J Insect Physiol 56(12):1738–1745
Danley PD, de Carvalho TN, Fergus DJ, Shaw KL (2007) Reproductive asynchrony and the divergence of Hawaiian crickets. Ethology 113(12):1125–1132
Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96(2):271–290
Edery I, Zwiebel LJ, Dembinska ME, Rosbash M (1994) Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA 91(6):2260–2264
Engelmann W, Maurer A, Muhlbach M, Johnsson A (1974) Action of lithium ions and heavy-water in slowing circadian rhythms of petal movement in Kalanchoe. J Interdiscip Cycle Res 5(3–4):199–205
Ewer J, Frisch B, Hamblencoyle MJ, Rosbash M, Hall JC (1992) Expression of the period clock gene within different cell-types in the brain of Drosophila adults and mosaic analysis of these cells influence on circadian behavioral rhythms. J Neurosci 12(9):3321–3349
Fergus DJ, de Carvalho TN, Shaw KL (2011) Genetic regulation of differential timing in courtship and mating of the Hawaiian cricket Laupala. Behav Genet 41(4):607–614
Gardner GF, Feldman JF (1980) The Frq locus in Neurospora crassa––a key element in circadian clock organization. Genetics 96(4):877–886
Giebultowicz JM (2000) Molecular mechanism and cellular distribution of insect circadian clocks. Annu Rev Entomol 45:769–793
Guldemond JA, Tigges WT, Devrijer PWF (1994) Circadian-rhythm of sex-pheromone production and male activity of coexisting sibling species of Cryptomyzus aphids (Homoptera, Aphididae). Eur J Entomol 91(1):85–89
Hamblen–Coyle MJ, Wheeler DA, Rutila JE, Rosbash M, Hall JC (1992) Behavior of period-altered circadian rhythm mutants of Drosophila in light-dark cycles (Diptera, Drosophilidae). J Insect Behav 5(4):417–446
Hardin PE (2005) The circadian timekeeping system of Drosophila. Curr Biol 15(17):R714–R722
Hassaneen E, Sallam AE, Abo-Ghalia A, Moriyama Y, Karpova SG, Abdelsalam S, Matsushima A, Shimohigashi Y, Tomioka K (2011) Pigment-dispersing factor affects nocturnal activity rhythms, photic entrainment, and the free-running period of the circadian clock in the cricket Gryllus bimaculatus. J Biol Rhythms 26(1):3–13
Helfrich-Forster C (2005) Organization of endogenous clocks in insects. Biochem Soc Trans 33:957–961
Helfrich-Forster C, Stengl M, Homberg U (1998) Organization of the circadian system in insects. Chronobiol Int 15(6):567–594
Huang TC, Lay KC, Tong SR (1991) Resetting the endogenous circadian N2-fixing rhythm of the prokaryote Synechococcus Rf-1. Bot Bul Acad Sin 32(2):129–133
Kloss B, Rothenfluh A, Young MW, Saez L (2001) Phosphorylation of PERIOD is influenced by cycling physical associations of DOUBLE-TIME, PERIOD, and TIMELESS in the Drosophila clock. Neuron 30(3):699–706
Kondo T, Tsinoremas NF, Golden SS, Johnson CH, Kutsuna S, Ishiura M (1994) Circadian clock mutants of cyanobacteria. Science 266(5188):1233–1236
Konopka RJ, Benzer S (1971) Clock mutants of Drosophila melanogaster. Proc Natl Acad Sci USA 68(9):2112–2116
Kyriacou CP, Hall JC (1980) Circadian-rhythm mutations in Drosophila melanogaster affect short-term fluctuations in the male’s courtship song. Proc Natl Acad Sci USA 77(11):6729–6733
Lecharny A, Wagner E (1984) Stem extension rate in light grown plants-evidence for an endogenous circadian rhythm in Chenopodium rubrum. Physiol Plant 60(3):437–443
Lee CG, Bae KH, Edery I (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation, and interactions with the PER–TIM complex. Neuron 21(4):857–867
Lin GGH, Liou RF, Lee HJ (2002) The period gene of the German cockroach and its novel linking power between vertebrate and invertebrate. Chronobiol Int 19(6):1023–1040
Liu C, Li SM, Liu TH, Borjigin J, Lin JD (2007) Transcriptional coactivator PGC––1a integrates the mammalian clock and energy metabolism. Nature 447(7143):477–481
Loher W (1972) Circadian control of stridulation in the cricket Teleogryllus commodus walker. J Comp Physiol 79(2):173–190
Loher W (1974) Circadian control of spermatophore formation in the cricket Teleogryllus commodus walker. J Insect Physiol 20(7):1155–1172
Lupien M (1998) Correlation between ultradian and circadian rhythms in the cricket, Teleogryllus oceanicus: potential role for the period gene. Department of Biology, McGill University, Montreal, p 66
Lupien M, Marshall S, Leser W, Pollack GS, Honegger HW (2003) Antibodies against the PER protein of Drosophila label neurons in the optic lobe, central brain, and thoracic ganglia of the crickets Teleogryllus commodus and Teleogryllus oceanicus. Cell Tissue Res 312(3):377–391
Mazor M, Dunkelblum E (2005) Circadian rhythms of sexual behavior and pheromone titers of two closely related moth species Autographa gamma and Cornutiplusia circumflexa. J Chem Ecol 31(9):2153–2168
Mazzoni CJ, Gomes CA, Souza NA, de Queiroz RG, Justiniano SCB, Ward RD, Kyriacou CP, Peixoto AA (2002) Molecular evolution of the period gene in sandflies. J Mol Evol 55(5):553–562
McFarlane JE (1968) Diel periodicity in spermatophore formation in house cricket Acheta domesticus (L). Can J Zool 46(4):695–698
Millar AJ, Kay SA (1991) Circadian control of cab gene-transcription and messenger-RNA accumulation in Arabidopsis. Plant Cell 3(5):541–550
Miyatake T (2002) Pleiotropic effect, clock genes, and reproductive isolation. Popul Ecol 44(3):201–207
Miyatake T, Kanmiya K (2004) Male courtship song in circadian rhythm mutants of Bactrocera cucurbitae (Tephritidae:Diptera). J Insect Physiol 50(1):85–91
Miyatake T, Matsumoto A, Matsuyama T, Ueda HR, Toyosato T, Tanimura T (2002) The period gene and allochronic reproductive isolation in Bactrocera cucurbitae. Proc R Soc Lond B Biol Sci 269(1508):2467–2472
Moriyama Y, Sakamoto T, Karpova SG, Matsumoto A, Noji S, Tomioka K (2008) RNA interference of the clock gene period disrupts circadian rhythms in the cricket Gryllus bimaculatus. J Biol Rhythms 23(4):308–318
Moriyama Y, Sakamoto T, Matsumoto A, Noji S, Tomioka K (2009) Functional analysis of the circadian clock gene period by RNA interference in nymphal crickets Gryllus bimaculatus. J Insect Physiol 55(5):396–400
Myers MP, Wager–Smith K, Rothenfluh–Hilfiker A, Young MW (1996) Light-induced degradation of TIMELESS and entrainment of the Drosophila circadian clock. Science 271(5256):1736–1740
Nowosielski JW, Patton RL (1963) Studies on circadian rhythm of the house cricket Gryllus domesticus L. J Insect Physiol 9(4):401–404
Nowosielski JW, Patton RL (1964) Daily fluctuation in blood sugar concentration of house cricket Gryllus domesticus L. Science 144(361):180–181
Oh KP, Fergus DJ, Grace JL, Shaw KL (2012) Interspecific genetics of speciation phenotypes: song and preference coevolution in Hawaiian crickets. J Evol Biol 25(8):1500–1512
Otte D (1994) The Crickets of Hawaii. The Orthopterist’s Society, Philadelphia
Petersen G, Hall JC, Rosbash M (1988) The period gene of Drosophila carries species-specific behavioral instructions. EMBO J 7(12):3939–3947
Refinetti R (1996) Ultradian rhythms of body temperature and locomotor activity in wild-type and tau-mutant hamsters. Anim Biol 5(3):111–115
Rieger D, Shafer OT, Tomioka K, Helfrich–Forster C (2006) Functional analysis of circadian pacemaker neurons in Drosophila melanogaster. J Neurosci 26(9):2531–2543
Rosato E, Tauber E, Kyriacou CP (2006) Molecular genetics of the fruit-fly circadian clock. Eur J Hum Genet 14(6):729–738
Ruf T (1999) The Lomb–Scargle periodogram in biological rhythm research: analysis of incomplete and unequally spaced time-series. Biol Rhythm Res 30(2):178–201
Sandrelli F, Tauber E, Pegoraro M, Mazzotta G, Cisotto P, Landskron J, Stanewsky R, Piccin A, Rosato E, Zordan M, Costa R, Kyriacou CP (2007) A molecular basis for natural selection at the timeless locus in Drosophila melanogaster. Science 316(5833):1898–1900
Sandrelli F, Costa R, Kyriacou CP, Rosato E (2008) Comparative analysis of circadian clock genes in insects. Insect Mol Biol 17(5):447–463
Shafer OT, Rosbash M, Truman JW (2002) Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci 22(14):5946–5954
Shao QM, Sehadova H, Ichihara N, Sehnal F, Takeda M (2006) Immunoreactivities to three circadian clock proteins in two ground crickets suggest interspecific diversity of the circadian clock structure. J Biol Rhythms 21(2):118–131
Shao QM, Bembenek J, Trang LTD, Hiragaki S, Takeda M (2008a) Molecular structure, expression patterns, and localization of the circadian transcription modulator CYCLE in the cricket Dianemobius nigrofasciatus. J Insect Physiol 54(2):403–413
Shao QM, Hiragaki S, Takeda M (2008b) Co-localization and unique distributions of two clock proteins CYCLE and CLOCK in the cephalic ganglia of the ground cricket, Allonemobius allardi. Cell Tissue Res 331(2):435–446
Shaw KL (1996) Polygenic inheritance of a behavioral phenotype:interspecific genetics of song in the Hawaiian cricket genus Laupala. Evolution 50(1):256–266
Shaw KL (2000a) Further acoustic diversity in Hawaiian forests: two new species of Hawaiian cricket (Orthoptera : Gryllidae : Trigonidiinae : Laupala). Zool J Linn Soc-Lond 129(1):73–91
Shaw KL (2000b) Interspecific genetics of mate recognition: inheritance of female acoustic preference in Hawaiian crickets. Evolution 54(4):1303–1312
Shiga S, Numata H, Yoshioka E (1999) Localization of the photoreceptor and pacemaker for the circadian activity rhythm in the band-legged ground cricket, Dianemobius nigrofasciatus. Zool Sci 16(2):193–201
Shimizu T, Miyatake T, Watari Y, Arai T (1997) A gene pleiotropically controlling developmental and circadian periods in the melon fly, Bactrocera cucurbitae (Diptera:Tephritidae). Heredity 79:600–605
Shirasu N, Shimohigashi Y, Tominaga Y, Shimohigashi M (2003) Molecular cogs of the insect circadian clock. Zool Sci 20(8):947–955
Sokolove PG (1975) Locomotory and stridulatory circadian rhythms in the cricket, Teleogryllus commodus. J Insect Physiol 21(3):537–558
Sokolove PG, Loher W (1975) Role of eyes, optic lobes, and pars intercerebralis in locomotory and stridulatory circadian rhythms of Teleogryllus commodus. J Insect Physiol 21(4):785–799
Tauber E, Roe H, Costa R, Hennessy JM, Kyriacou CP (2003) Temporal mating isolation driven by a behavioral gene in Drosophila. Curr Biol 13(2):140–145
Tauber E, Zordan M, Sandrelli F, Pegoraro M, Osterwalder N, Breda C, Daga A, Selmin A, Monger K, Benna C, Rosato E, Kyriacou CP, Costa R (2007) Natural selection favors a newly derived timeless allele in Drosophila melanogaster. Science 316(5833):1895–1898
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal-W:improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tomioka K, Abdelsalam S (2004) Circadian organization in hemimetabolous insects. Zool Sci 21(12):1153–1162
Tomioka K, Chiba Y (1992) Characterization of an optic lobe circadian pacemaker by in situ and in vitro recording of neural activity in the cricket, Gryllus bimaculatus. J Comp Physiol A 171(1):1–7
Weber F (2009) Remodeling the clock:coactivators and signal transduction in the circadian clockworks. Naturwissenschaften 96(3):321–337
Wheeler D, Kyriacou C, Greenacre M, Yu Q, Rutila J, Rosbash M, Hall J (1991) Molecular transfer of a species-specific behavior from Drosophila simulans to Drosophila melanogaster. Science 251(4997):1082–1085
Wiedenmann G, Loher W (1984) Circadian control of singing in crickets––2 different pacemakers for early-evening and before-dawn activity. J Insect Physiol 30(2):145–151
Yerushalmi S, Green RM (2009) Evidence for the adaptive significance of circadian rhythms. Ecol Lett 12(9):970–981
Yu W, Hardin PE (2006) Circadian oscillators of Drosophila and mammals. J Cell Sci 119(23):4793–4795
Zheng XZ, Sehgal A (2008) Probing the relative importance of molecular oscillations in the circadian clock. Genetics 178(3):1147–1155
Zheng B, Larkin DW, Albrecht U, Zhong Sheng S, Sage M, Eichele G, Cheng Chi L, Bradley A (1999) The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature 400(6740):169–173
Acknowledgments
DJF was supported by an NIH training Grant (No. 5T32GM007469 to the Cornell graduate field of Neurobiology and Behavior). Parts of this work were supported by an NSF doctoral dissertation improvement grant (IOS0709993). We would like to thank Chris Wiley, Holly Menninger, Chris Ellison, and Biz Turnell for valuable discussion and feedback.
Author information
Authors and Affiliations
Corresponding author
Additional information
Edited by Charalambos Kyriacou
Rights and permissions
About this article
Cite this article
Fergus, D.J., Shaw, K.L. Circadian Rhythms and Period Expression in the Hawaiian Cricket Genus Laupala . Behav Genet 43, 241–253 (2013). https://doi.org/10.1007/s10519-012-9576-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10519-012-9576-4