Abstract
microRNA-1827 (miR-1827) is proposed to be enriched in exosomes from mesenchymal stem cells (MSCs-Exos). A recent study has addressed the suppressive effect of exosomes from human umbilical cord mesenchymal stem cells (hUC-MSCs-Exos) on colorectal cancer (CRC) metastasis. Hence, our study aims at investigating whether hUC-MSCs-Exos can modulate the liver metastasis in CRC by mediating miR-1827. Transmission electron microscopy (TEM) and nanoparticle tracking analysis (NTA) were used to identify hUC-MSCs-Exos. Using gain- and loss-of-function approaches, the expression of miR-1827 and succinate receptor 1 (SUCNR1) was altered. Consequently, the biological functions of CRC cells were assessed by CCK-8 and Transwell assays and macrophage M2 polarization was assayed by flow cytometry. Dual-luciferase reporter assay was applied to clarify interaction between miR-1827 and SUCNR1. CRC cells were incubated with hUC-MSCs-Exos and tumor-bearing mice were injected with hUC-MSCs-Exos to examine the effects on CRC cell growth and metastasis. SUCNR1, lowly expressed in CRC, could promote CRC cell growth and macrophage M2 polarization. miR-1827 could target SUCNR1 and hence suppress the progression and metastasis of CRC. hUC-MSCs-Exos carried miR-1827 to inhibit M2 macrophage polarization by downregulating SUCNR1 expression, and inhibited proliferating, migrating and invading properties of CRC cells. Furthermore, hUC-MSCs-Exos carrying miR-1827 blocked CRC liver metastasis in vivo. These findings indicate hUC-MSCs-Exos as an inhibitor of M2 macrophage polarization and liver metastasis in CRC through inducing miR-1827-targeted inhibition of SUCNR1. This provides a theoretical basis for understanding the mechanisms underlying Exos-based target therapy for CRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) has gradually become one of the most wide-spread cancers, taking up about 10% to all diagnosed cancers every year [1]. Despite advancement in new treatments, the cure rate and prognosis for people diagnosed with CRC have not been improved in the last decades [2]. The etiology of CRC has been identified to involve genetic and epigenetic alterations such as aberrant DNA methylation, dysregulation of microRNA (miRNAs) and noncoding RNAs, and changes in histone modification [3]. In addition to the genetic components, modifiable environmental components are also proved to play vital roles in the development of CRC, such as tobacco, alcohol intake, and physical activity [4]. Tumor metastasis is one of main factors inducing CRC-associated deaths and among which the liver metastasis is a frequent event [5]. In addition to surgical resection, radiotherapy in association with chemotherapy and immunotherapy, are rising as alternative treatment options for CRC patients with colorectal liver metastases [6, 7]. The development of novel and effective non-surgical treatments heavily relies on the understanding of the pathways related to initiation and progression of CRC [8].
Exosomes (Exos), tiny vesicles originated from different cell resources, are proposed as paracrine factors that can transmit biomolecules including miRNAs between cells [9, 10]. Exosomes from human umbilical cord-derived mesenchymal stem cells (hUC-MSCs-Exos) have been reported to inhibit CRC metastasis through carrying miRNAs [11]. miRNAs, dysregulated in tumor cells, can regulate the expression of oncogenes or anti-oncogenes in different types of cancer, whereby preventing or driving tumorigenesis [12, 13]. miR-1827 has been found to be lowly expressed in CRC, which is associated with poor CRC prognosis [14], yet no study has mentioned the relationship between miR-1827 and Exos. Also, miR-1827 is lowly expressed in lung and liver cancer cells, but upregulation of miR-1827 represents a promising anti-tumor target in these cancers [15, 16]. Succinate receptor 1 (SUCNR1) mutation is one of genetic mutations contributing to disease in rectal- and gastric cancer [17]. This gene has also been addressed to drive lung cancer metastasis by promoting macrophage polarization [18]. An existing has revealed the correlation between SUCNR1 and cAMP which is a key pathway involved in the progression of rectal cancer [19, 20]. Based on information from EVmiRNA database, miR-1827 was found a miRNA enriched in MSCs-Exos. As miR-1827 was identified to target SUCNR1 by in silico analysis, in this study, we perform in vivo rat models and in vitro cultures to investigate whether hUC-MSCs-Exos carry miR-1827 to modulate SUCNR1, affecting macrophage M2 polarization involved in colorectal liver metastasis.
Methods
In silico analysis
Differential analysis of CRC microarray GSE108153 (normal sample n = 21; CRC sample n = 21) in the GEO database was performed using the R language “limma” package, in which the expression data of miR-1827 was extracted and plotted in a box plot. The downstream genes of miR-1827 were predicted by the database TargetScan (Cumulative weighted context + + score<-0.2), miRWalk (bindingp > 0.9, accessibility < 0.01, au > 0.55), DIANA TOOLS (miTG score > 0.7), and RNA22. The target genes in the intersection were taken for KEGG pathway enrichment analysis by KOBAS. The expression profiles of COAD and READ and clinical data were downloaded from the TCGA database and analyzed using the GEPIA. The binding sites between miR-1827 and the key downstream genes were also predicted by the TargetScan.
Clinical sample collection
Cancer tissues and the adjacent normal tissues were collected from 28 CRC patients with liver metastases (aged 38–64 years with a mean age of 49.17 ± 5.14 years; 15 males and 13 females). The enrolled patients underwent palliative surgery (resection of only the primary tumors) in Guangdong Provincial People’s Hospital from December 2016 to June 2019. Among them, there were 12 cases of rectal cancer and 16 cases of colon cancer. All patients met the 2017 version of the Chinese CRC diagnostic criteria [21, 22], with tumor originated in the colorectum, a clear pathological diagnosis, aged > 18 years, and complete medical records. The CRC epithelial tissues and adjacent normal tissues were preserved in sterile tubes and kept in liquid nitrogen for subsequent experiments. All patients in this study provided informed consent and the study was approved by the clinical ethics committee of Guangdong Provincial People’s Hospital.
Cell culture
hUC-MSCs were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and cultured in serum-free medium (Gibco Company, Grand Island, NY) in a constant temperature incubator at 37 °C and 5% CO2. Four CRC cell lines (HCT-116, SW480, Caco-2, and HT-29) and the human colonic epithelial cell line HCoEpiC were purchased from the Cell Bank of the Chinese Academy of Sciences and cultured in Dulbecco’s modified eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in a constant temperature incubator at 37 °C and 5% CO2. The adherent cells were passaged and digested with 0.25% trypsin (Hyclone, Germany) and the log-phase cells were taken for the experiment [23, 24].
The purity of hUC-MSCs was detected by flow cytometry [25]. The hUC-MSCs were dispersed into single cell suspension (3 × 106 cells/mL). Antibodies used here were sourced from BD Farmingen. hUC-MSCs were labeled with antibodies CD34-FITC (555,821) and CD45-FITC (555,482), CD90-FITC (555,595), CD105-FITC (561,443) and incubated together at 4 °C for 30 min, followed by treatment with 4% paraformaldehyde (PFA, P804536, Maclin, Shanghai, China). The expression of the above-mentioned antibodies was detected by Accuri C6 Plus Flow cytometer (BD pharingen, USA).
Isolation of macrophages
The nude mice were euthanized and fixed. Their skin was scissored under aseptic conditions to expose the intact peritoneum [22, 26]. The peritoneal cavity was then filled with 5mL of pre-cooled serum-free DMEM, and the abdomen was gently rubbed for 5 min. The peritoneal cavity was repeatedly rinsed with a pipette to collect the lavage fluid. The harvested lavage fluid was injected into a centrifuge tube and centrifuged at 1,000 × g for 10 min, twice in succession, and the supernatant was discarded. After the cell precipitate was washed with DMEM, the cells were resuspended in DMEM with 10% serum. The cell suspension was seeded in 6 cm culture dishes and placed in a constant temperature incubator at 37℃ and 5% CO2 for 2 h. After 2 h, the culture supernatant was discarded, and the culture dishes were gently rinsed with DMEM with 10% 1–2 times, and the adhered cells were obtained as macrophages.
Isolation and identification of Exos
The FBS was ultracentrifuged at 100,000 × g for 18 h to delete Exos from the serum. When confluence reached approximately 80%, hUC-MSCs were rinsed twice with phosphate buffer saline (PBS) and incubated with 10% Exos-free FBS at 37 °C and 5% CO2 for 48 h. After incubation, the supernatant was centrifuged sequentially at 4 °C (× 300 g for 10 min; × 2,000 g for 15 min; × 5,000 g for 15 min). The precipitate was discarded after each centrifugation. The supernatant was centrifuged at ×12,000 g twice (30 and 70 min) at 4 °C, and the precipitate was collected. After differential centrifugation, the supernatant was ultracentrifuged at ×100,000 g for 70 min and the precipitate containing Exos was collected. Another round of ultracentrifugation at ×100,000 g for 70 min was performed after resuspension with PBS. Finally, the precipitate containing pure Exos was collected [27, 28].
A total of 20 µg of Exos was resuspended in 1 mL PBS and spun for 1 min to maintain uniform distribution. The nanoparticle tracking analysis (NTA) (NanoSight; Malvern Panalytical, Worcestershire, UK) was performed to directly measure the diameter and size distribution of the Exos. The particles were tracked and measured based on Brownian motion and diffusion coefficients. After isolation, Exos were diluted in 1 µmL filtered PBS. Control medium and filtered PBS served as controls. NTA measurement conditions were set to 23.75 ± 0.5 °C, 25 frames per second for 60 s. All samples had similar detection thresholds. Each reaction was repeated three times and the result was averaged [29].
The prepared Exos were fixed in 4% paraformaldehyde for 2 h at 4 °C and then fixed in 1% osmium tetroxide for 2 h after rinses. The Exos were routinely dehydrated with gradient ethanol and acetone and then treated with ethoxylated resin, paraffin-embedded and polymerised. Next, the Exos were cut into 0.5-µm semi-thin sections and visualized under a light microscope. After identification of the Exo location, 60 nm ultrathin sections were prepared for uranyl acetate and lead citrate staining. Microscopic observation was performed at 110 kV using a JEM 1230 transmission electron microscope (TEM) (JEOL USA Inc., Peabody, MA) [30].
Western blot assay was adopted to detect the expression of Exos surface protein markers CD9, CD81, CD63, Tsg101, and exosome negative protein marker Calnexin [31].
CRC cell uptake of Exos
Purified hUC-MSCs-Exos were tagged with PKH67 Green Fluorescent Kit (PKH67GL-1KT, Sigma-Aldrich, St.Louis, MO). Exos were resuspended in 1 mL Diluent C solution, and 4 µL PKH-67 ethanol dye solution was also replenished to the 1 mL Diluent C to prepare the dying solution (4 × 10− 6 M) which was mixed with the 1 mL Exos suspension for 5 min. The staining was terminated with 2 mL of 1% bovine serum albumin (BSA) for 1 min. The labelled Exos were ultracentrifuged at 100,000 × g for 70 min, re-centrifuged, and resuspended in 50 µL PBS. Then, PKH67-labelled Exos were incubated with CRC cells at 37 °C for 12 h. Cells were then fixed with 4% paraformaldehyde (PFA) and washed with PBS, while cell membranes were stained red with DiL (C1036, Beyotime Biotechnology, Shanghai, China). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The uptake of labeled Exos by CRC cells was observed using a fluorescent microscope (Zeiss LSM 800, Zeiss Public, Germany).
For study of the uptake of hUC-MSCs-Exos carrying Cy3-miR-1827 by CRC cells, hUC-MSCs were infected with Cy3-miR-1827 lentivirus (GenePharma, Shanghai, China) (serum-free medium). After 6 h, the cells were cultured with a renewed Exos-free 10% serum medium for 48 h. The cell supernatant was resuspended in PBS and added to CRC cells, which were fixed in 4% PFA in the same way as described above. After staining, the uptake of Exos (red light) by CRC cells carrying Cy3-miR-1827 was observed under fluorescence microscope (LSM710, Zeiss).
CRC cell transfection
When CRC cells reached 80-90% confluence, cell transfection was performed according to the lipofectamine 2000 instructions (11668-019, Invitrogen). Exos were isolated from untreated hUC-MSCs, hUC-MSCs transfected with NC mimic, and hUC-MSCs transfected with miR-1827 mimic, designated as hUC-MSCs-Exos, hUC-MSCs-Exos-NC mimic, and hUC-MSCs-Exos-miR-1827 mimic. CRC cells were treated with PBS, hUC-MSCs-Exos, hUC-MSCs-Exos-NC mimic, hUC-MSCs-Exos-miR-1827 mimic or transfected with short hairpin RNA against SUCNR1 (sh-SUCNR1), SUCNR1 overexpression vector (oe-SUCNR1), plasmids of miR-1827 mimic, miR-1827 inhibitor, and/or their corresponding negative controls (NC).
The final concentration of oe-SUCNR1 or sh-SUCNR1 used for transfection was 50 nM, and the shRNA sequences are shown in Table S1. NC-mimic, miR-1827 mimic, NC-inhibitor, and miR-1827 inhibitor were supplied by RiboBio (Guangzhou, China), and other above-mentioned plasmids and sequences were designed and sequenced by GeneChem (Shanghai, China) with pcDNA3.1 vector used for the construction of stable transfection vectors.
Cell counting kit-8 (CCK-8)
The proliferative capacity of the cells was assayed using the CCK8 kit (CA1210-100, Solarbio, Beijing, China). Briefly, cells were seeded in 96-well plates at 5 × 103 cells per well and cultured for 3 days. Each well was added with 10 µL CCK8 solution and incubated in an incubator for 2 h. Subsequently, the absorbance at 450 nm was measured using a microplate reader (BIO-RAD 680, BIO-RAD, Hercules, CA), and the optical density (OD) values were measured at different time points (0 h, 24 h, 48 h, 72 h), followed by plotting of cell proliferation curves.
Transwell assay
Transwell assay
An 8 μm multi-well Transwell chamber (Item No. 3452, NJXBio, Nanjing, China) with (for invasion assay) or without Matrigel (for migration assay) was utilized to assess cell migration and invasion abilities. Each well of the apical chamber was filled with 300 µL of serum-free DMEM containing 1 × 105 transfected cells/mL, while the basolateral chamber contained 700 µL of complete medium. The cells were incubated for 48 h, fixed in 4% PFA, and later stained with crystal violet. Cells retained in the apical chamber were removed with a cotton swab and imaged by light microscopy (Olympus, China). Five fields of view were randomly selected for counting, and the number of cells was expressed as the mean number.
Western blot assay
The total protein of cells or tissues was extracted using pre-cooled phenylmethanesulfonyl fluoride (PMSF)-containing radio immunoprecipitation assay (RIPA) lysis buffer (R0010, Solarbio) at 4 °C strictly according to the instructions. Each protein sample was separated by electrophoresis in 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to the polyvinylidene fluoride membrane (Millipore, Germany) by wet transfer method, which was blocked with 5% BSA at room temperature for 1 h. Diluted primary antibodies to SUCNR1 (NBP1-00861, 1:5000, Novus Biologicals, Beijing, China), ARG1 (9819, 1:5000, Cell Signaling, Danvers, MA), Bcl2 (ab182858, 1:2000, Abcam), PCNA (ab29, 1:1000, Abcam), Ki67 (ab16667, 1:1000, Abcam), MMP2 (ab92536, 1:1000, Abcam), MMP9 (ab283575, 1:1000, Abcam), N-cadherin (ab76011, 1:5000, Abcam), E-cadherin (ab231303, 1:1000, Abcam), Fizz1 (ab271225, 1:1000, Abcam), Mgl1 (PA5-120697, 1/1000, Invitrogen), GAPDH (ab8245, 1:1000, Abcam) were incubated with the membrane overnight at 4 °C in a shaker. The membrane was washed 10 min × 3 times with TBST. The horseradish peroxide (HRP)-labeled goat anti-rabbit secondary antibody to immunoglobulin G (ab6721, 1:5000, Abcam, Cambridge, UK) was added for 1 h incubation. The HRP substrate enhanced chemiluminescent luminescent solution was purchased from Lianshuo Biological (WBKLS0050, Shanghai, China). Finally, glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab8245, 1:5000, Abcam) was utilized as the internal reference and the relative protein expression was analyzed using a gel imaging analysis system (GIS-500, QMgene, Beijing, China) and Image J software.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from cells using a Trizol reagent (Invitrogen), while a NanoDrop spectrophotometer (Thermo) was utilized to analyze the amount of RNAs. The complementary DNA (cDNA) was obtained according to the instructions of miRNA Reverse Transcription Kit (B532451, Sangon Biotech) and cDNA Reverse Transcription Kit (B532445, Sangon Biotech), followed by amplification of gene fragments. miR-1827 reverse transcription primer was purchased from RiboBio (Guangzhou, China). The other target gene primers were synthesized by Takara (Dalian, China) (Table S2). The reaction system was prepared according to the instructions of the fluorescent quantitative PCR kit (AQ101-02, TransGene Biotech, Beijing, China). Fluorescent quantitative PCR was performed in an ABI ViiA 7 (DaanGene, Guangzhou, China) with GAPDH served as an internal reference. For miRNA detection, the reaction system was prepared according to the instructions of the miRNA fluorescence quantitative PCR kit (AQ101-02, TransGene Biotech), in which 1 µL of reverse transcribed cDNA was taken and the reaction amplification system was 10 µL, with U6 as internal reference. The relative quantification method (2−ΔΔCT method) was utilized to calculate the relative transcription level of the target gene.
Dual-luciferase reporter gene assay
The relationship between the SUCNR1 gene and miR-1827 was confirmed through the bioinformatics database microRNA.org. Human embryonic kidney (HEK)-293T cells were cultured in DMEM containing 10% FBS at 37 °C and 5% CO2. The luciferase reporter gene assay was used to verify whether the SUCNR1 mRNA 3’untranslated region (UTR) has a binding relationship with miR-1827. A dual luciferase reporter gene vector containing SUCNR1 3’UTR and mutants with mutated binding site to miR-1827 were constructed: pGLO-SUCNR1 wild type (Wt) and pGLO-SUCNR1 mutant type (Mut) (5’-CCAGCUGCCAUUGCAGACGGAUG-3’), respectively. The two reporter plasmids were co-transfected into HEK293 cells with miR-1827 mimic and pRL-TK (an internal reference plasmid expressing renilla luciferase), respectively. Cells were collected 48 h after transfection and relative luciferase activity values were determined using a dual luciferase reporter assay system (E1910; Promega Corporation, Madison, WI). Relative luciferase activity was expressed as the firefly luciferase activity/sea kidney luciferase activity.
Macrophage M2 polarization and flow cytometry
Peritoneal macrophages were seeded in 6-well plates at a density of 3 × 106 cells per well. Cells were subsequently treated with different conditioned media (Control, HT-29, HT-29-sh-NC, or HT-29-sh-SUCNR1) for 3 days. Cells were harvested and fixed by incubation in PBS containing 1% formaldehyde on ice for 15 min. Cells were then washed and blocked with 1% BSA on ice for 1 h. Next the cells were stained with anti-CD106 (Vcam1)-FITC (BD Biosciences, San Diego, CA), anti-CD11c-PE (BD Biosciences), and anti-CD11b-PerCP (Biolegend, Los Angles, CA) on ice. After 3 h of staining, cells were washed and resuspended in PBS. Flow cytometry was performed by a FACS Calibur (BD Biosciences) and analyzed using FlowJo software. For the analysis of macrophage M2 polarization at tumour sites, tumour tissues were lysed and macrophages were collected for subsequent experiments.
Xenograft tumors in nude mice
A total of 1 × 106 CRC cells were injected subcutaneously into BALB/c nude mice (Slac Laboratory Animal, Hunan, China) to construct a subcutaneous transplantation tumor model in nude mice. When the tumors reached about 100 mm3, the nude mice were untreated or injected with agomir NC, miR-1827 agomir, hUC-MSCs-Exos-NC mimic + oe-NC, hUC-MSCs-Exos-NC mimic + oe-SUCNR1, hUC-MSCs-Exos-miR-1827-mimic + oe-NC, or hUC-MSCs-Exos-miR-1827-mimic + oe-SUCNR1, with 10 mice in each treatment, 70 mice in total.
Oligonucleotides, plasmids and transfected plasmid complexes were diluted according to the instructions of En-transterTM-in vivo. Each mouse was locally injected with 3 µL PBS mixture containing 0.5 nM transfection content for consecutive 3 d at the site of the subcutaneous tumors. The control mice were locally injected with the same amount of PBS (3 µL) for consecutive 3 d at the site of the subcutaneous tumors. Operations were strictly in accordance with the instructions. All vector construction, sequencing and identification were carried out by GeneChem. After 5 weeks of treatment, nude mice were euthanized to obtain livers for observation of metastatic tumor nodules.
Statistical analysis
The SPSS version 21.0 (IBM SPSS Statistics, Chicago) was used for statistical analysis. Measurement data were expressed as mean ± standard deviation. Paired t-tests were conducted for comparison of paired data with normal distributions and homogeneous variances between two groups and unpaired t-tests for that of unpaired data. One-way analysis of variance (ANOVA) and Tukey’s post hoc test were utilized for multiple group comparisons. Data among groups at different time points were compared using repeated measures ANOVA with Bonferroni post hoc tests. Patient survival was assessed using Kaplan-Meier method and compared by Log-rank test. A p < 0.05 indicates a statistically significant difference.
Results
Significance of MSCs-derived Exos/miR-1827/SUCNR1 in CRC progression and patient prognosis
Low expression of miR-1827 in CRC tissues and cells was identified by differential analysis on microarray GSE108153 (Fig. 1 A). Our predictions from the EVmiRNA database revealed enrichment of miR-1827 in MSCs-derived Exos (Fig. 1B). Combined with the known knowledge previously reported, we hypothesized that MSCs-derived Exos carried miR-1827 to influence CRC growth and metastasis.
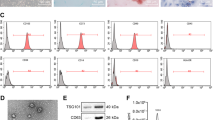
Bioinformatics analysis reveals a possible mechanism of miR-1827/SUCNR1 involved in CRC growth and metastasis. A, Expression of miR-1827 in microarray GSE108153 (42 samples including 21 normal samples and 21 CRC samples); the blue box on the left indicates the gene expression in normal samples and the red box on the right indicates the gene expression in CRC samples. B, EVmiRNA prediction of miR-1827 expression in various Exos. C, Venn diagram of downstream genes obtained from TargetScan, miRWalk, DIANA TOOLS, and RNA22 databases. D, KEGG enrichment analyses of the 36 intersection genes; the vertical axis indicates the enriched entry and the horizontal axis indicates the number of genes enriched; the bubble color (right color scale) indicates enrichment significance-logP values. E, Expression of SUCNR1 in the COAD and READ datasets from the TCGA database by GEPIA analysis; red boxes indicate cancer samples while gray boxes indicate normal samples. * indicates p < 0.05. F, Survival curves of SUCNR1 in the COAD and READ datasets from the TCGA database. G, TargetScan prediction of binding sites between miR-1827 and SUCNR1. RPM: reads per million mapped reads
Subsequently, 1747, 887, 1508, and 8785 downstream genes of miR-1827 were obtained by TargetScan, miRWalk, DIANA TOOLS, and RNA22 prediction, respectively, and 36 genes were found in the intersection (Fig. 1 C). Subsequent KEGG enrichment analysis by KOBAS revealed highest enrichment in the cAMP signaling pathway, where the enriched genes were CAMK2A, SUCNR1, and HCN4 (Fig. 1D, Supplementary Table S3). [20, 32] Through GEPIA analysis of the TCGA database COAD and READ datasets, we identified high expression of SUCNR1 in CRC (Fig. 1E), while CAMK2A and HCN4 are lowly expressed in CRC (Figure S1A, S1B). Since miRNAs always exerted inhibitory effect on the expression of target genes, SUCNR1 was selected as the object of our study. Combined analysis of COAD and READ clinical data revealed close association between high SUCNR1 expression with reduced survival of CRC patients (Fig. 1 F). Additionally, SUCNR1 contained miR-1827 binding sites as predicted by TargetScan (Fig. 1G). In summary, we hypothesized that MSCs-derived Exos carrying miR-1827 may affect CRC progression and patient prognosis by regulating SUCNR1 expression.
Overexpression of miR-1827 inhibits CRC growth and metastasis
The study focus was shifted on the role of miR-1827 in CRC considering its low expression in CRC [14]. miR-1827 expression was markedly enhanced by manipulation with miR-1827 mimic in CRC cells HT-29 (Fig. 2 A). Consequently, proliferative, migrating, and invading abilities of HT-29 cells were attenuated after re-expression of miR-1827 (Fig. 2B-D, Figure S2A, S2B).
Effect of miR-1827 on the growth and metastatic capacity of CRC cellsin vitroandin vivo.A, RT-qPCR detection of miR-1827 expression in HT-29 cells. B, CCK8 assay of proliferation ability of HT-29 cells. C, Transwell assay of migrating cells. D, Transwell assay of invading cells (* indicates p < 0.05 compared with NC mimic group). E, Expression of miR-1827 in tumor tissues of nude mice by RT-qPCR (10 mice per group). F, Representative images of xenograft tumor formation and folded graph of tumor volume change in nude mice. G, Tumor weight statistics of nude mice. H, Statistics of the number of tumor metastatic nodules in the liver of nude mice (* indicates p < 0.05 compared to agomir-NC group). All cellular experiments were repeated 3 times
Additional in vivo experiments were performed and the results exhibited that miR-1827 expression remained unaffected after local injection with agomir-NC at the site of tumors; whereas, miR-1827 expression in the tumor-bearing mice was elevated by local injection with agomir-miR-1827 at the site of tumors (Fig. 2E), resulting in significant reductions in tumor volume and weight (Fig. 2 F, 2G). Furthermore, 5 weeks later, elevated miR-1827 expression contributed to fewer metastatic tumor nodes in the liver of the tumor-bearing mice (Fig. 2 H). Thus, miR-1827 acted as an inhibitor of CRC growth and metastasis.
hUC-MSCs-Exos deliver miR-1827 to CRC cells and inhibit the malignant phenotypes of CRC cells
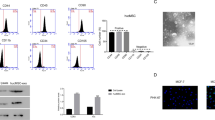
As EVmiRNA database revealed the presence of miR-1827 in MSCs-derived Exos, and hUC-MSCs-Exos are reported to curb CRC metastasis via carrying miRNAs [26]. Therefore, hUC-MSCs were utilized as the study target and Exos from which were isolated for subsequent experiments. We tested the purity of hUC-MSCs by flow cytometry prior to the experiment. The results of the flow cytometry exhibited that the positive rate of CD90 and CD105 in hUC-MSCs was more than 98%, while CD34 and CD45 were absent (Figure S3). It was indicated that the purity of hUC-MSCs was high and could be used for subsequent experiments. TEM and NTA supported the identification of Exos that most of the Exos were round- or oval-shaped, with a diameter of 30 ~ 150 nm (Fig. 3 A), and distributed within the range of 30 ~ 140 nm (Fig. 3B). Western blot identification of exosome markers showed that compared with cell lysate, CD9, CD63, CD81 and TGS101 were highly expressed in Exos, while Calnexin was basically not expressed (Fig. 3 C). The miR-1827 expression was abundant in hUC-MSCs-Exos relative to PBS (Fig. 3D). Also, miR-1827 expression was enhanced in the hUC-MSCs transfected with miR-1827 mimic but reduced in the hUC-MSCs transfected with miR-1827 inhibitor (Fig. 3E).
Effect of hUC-MSCs carrying miR-1827 on proliferation, migration and invasion of CRC cells. A, TEM identification of Exos structure. B, NTA of Exos diameter size. C, Western blot analysis to identify the expression of exosome-positive protein markers CD9, CD81, CD63, and Tsg101 and exosome-negative protein marker Calnexin. D, RT-qPCR detection of miR-1827 expression in PBS and hUC-MSCs-Exos (* indicates p < 0.05 compared with PBS). E, RT-qPCR detection of miR-1827 expression after hUC-MSCs transfected with miR-1827 mimic and miR-1827 inhibitor (* indicates p < 0.05 compared to hUC-MSCs-NC-mimic group; # indicates p < 0.05 compared to hUC-MSCs-miR-1827-mimic). F, fluorescence microscopy observation of PKH-67-labeled hUC-MSCs-Exos internalized by HT-29 cells. G, Cy3-miR-1827 content in the HT-29 cells co-cultured with hUC-MSCs-Exos. H, RT-qPCR detection of miR-1827 expression in HT-29 cells. I, CCK8 assay for proliferation of HT-29 cells. J, Transwell assay for number of migrated cells. K, The number of invading cells assessed by Transwell assay. L, Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin and E-cadherin protein levels measured by Western blot assay. (* indicates p < 0.05 compared to PBS groups). All experiments were repeated three times
Furthermore, we labeled hUC-MSCs-Exos with PKH-67 to investigate whether hUC-MSCs-Exos could enter into HT-29 cells. Fluorescence microscopic data exhibited no green fluorescence in the HT-29 cells treated with PBS or culture medium, but strong green fluorescence in the cytoplasm of the HT-29 cells incubated with hUC-MSCs-Exos (Fig. 3 F). This indicated that hUC-MSCs-Exos could be successfully internalized by HT-29 cells. Further, the miR-1827 in hUC-MSCs-Exos was labeled with Cy3, and the Cy3-miR-1827 content was observed by fluorescence microscopy after treatment of HT-29 cells with PBS, culture medium, and hUC-MSCs-Exos. Red fluorescence was visualized in more than 70% of the HT-29 cells treated with hUC-MSCs-Exos (manifested as high Cy3-miR-1827 content) rather than in PBS-treated cells (Fig. 3G). The above-mentioned results suggested that hUC-MSCs-Exos could carry miR-1827 into HT-29 cells.
Additionally, miR-1827 expression was interestingly raised in the HT-29 cells after incubation with hUC-MSCs-Exos instead of culture medium (Fig. 3 H). Afterwards, proliferative, migratory, and invasive potentials of HT-29 cells were restrained after incubation with hUC-MSCs-Exos (Fig. 3I-K, Figure S2C, S2D), corresponding to reductions in the protein levels of Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin as well as an elevation of E-cadherin protein level (Fig. 3 L). Taken together, hUC-MSCs-derived Exos deliver miR-1827 to CRC cells and inhibit their in vitro malignancy.
Upregulation of SUCNR1 in CRC and its contributory role in macrophage M2 polarization
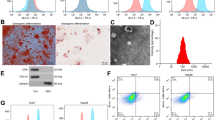
SUCNR1 was experimentally determined to be expressed highly in CRC tissues versus adjacent normal tissues (Fig. 4 A), as well as in four CRC cell lines (HCT-116, SW480, Caco-2, HT-29) versus normal colon cell line HCoEpiC (Fig. 4B). Among them, HT-29 cells with the highest SUCNR1 level were used in all subsequent studies.
Expression of SUCNR1 in CRC tissues and cells and its effect on macrophage M2 polarization and CRC cell migration and invasion capacities. A, RT-qPCR detection of SUCNR1 expression in CRC tissues (n = 28, * indicates p < 0.05 compared to adjacent normal tissues). B, RT-qPCR detection and Western blot analysis of SUCNR1 mRNA and protein expression in four CRC cell lines (HCT-116, SW480, Caco-2, and HT-29) and human normal colonic epithelial cell line HCoEpiC expression (* indicates p < 0.05 compared to the other four cell lines). C, RT-qPCR determination and Western blot analysis of SUCNR1 silencing efficiency in HT-29 cells with sh-SUCNR1 sequence (* indicates p < 0.05 compared to the sh-NC group). D, Western blot analysis of ARG1 expression in macrophages treated with CM from different HT-29 cells. E, RT-qPCR determination of Arg1 mRNA expression in macrophages treated with CM from different HT-29 cells. F, RT-qPCR determination and Western blot analysis of Fizz1 mRNA and protein expression in macrophages treated with CM from different HT-29 cells. G, RT-qPCR determination and Western blot analysis for Mgl1 mRNA and protein expression in macrophages treated with CM from different HT-29 cells. H, Quantitative analysis of M2 macrophages after treatment with CM from different HT-29 cells through Flow cytometry. (* indicates p < 0.05 compared to Ctrl-CM group; # indicates p < 0.05 compared to sh-NC-CM group). I, Transwell assay for number of migrating cells (* indicates p < 0.05 compared to sh-NC). J, Transwell assay for number of invading cells (* indicates p < 0.05 compared to sh-NC). K, Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin and E-cadherin protein levels measured by Western blot assay. (* indicates p < 0.05 compared to sh-NC). All experiments were repeated 3 times
To test the role of SUCNR1 in macrophage M2 polarization, SUCNR1 was silenced in the HT-29 cells using sh-SUCNR1-1, sh-SUCNR1-2, and sh-SUCNR1-3, with sh-SUCNR1-1 exhibiting the optimum silencing efficacy, which was selected for following experiments (Fig. 4 C). To examine the factors affecting macrophage polarization in CRC cells, we incubated nude mouse peritoneal macrophages with differentially treated HT-29 cell conditioned medium (CM) and analyzed the expression of the macrophage M2 polarization marker arginase 1 (ARG1). CM from the HT-29 cells and that from the HT-29 cells transfected with sh-NC resulted in elevated levels of ARG1 in macrophages, whereas ARG1 expression in macrophages was significantly lowered by incubation with CM from the SUCNR1-deficient HT-29 cells (Fig. 4D and E). Moreover, the levels of other macrophage M2 polarization markers (Fizz1 and Mgl1) and the number of tumor-associated macrophages (VCAM1 + CD11c + CD11blow-TAMs) in the macrophages showed elevations after incubation with CM from the HT-29 cells and that from the HT-29 cells transfected with sh-NC but reductions in response to incubation with CM from the SUCNR1-deficient HT-29 cells (Fig. 4 F-H). Transwell assays revealed that the migrating and invading abilities of HT-29 cells were inhibited after SUCNR1 silencing (Fig. 4I J, Figure S2E, S2F). Meanwhile, SUCNR1 deficiency caused reductions in the protein levels of Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin and an elevation of E-cadherin protein level (Fig. 4 K). The above results indicated that SUCNR1 could enhance macrophage M2 polarization, thus promoting CRC cell migration and invasion.
hUC-MSCs-Exos downregulates SUCNR1 by carrying miR-1827
As depicted in Fig. 1G, SUCNR1 had a miR-1827 binding site. Subsequently, this binding relationship was verified using a dual-luciferase reporter gene assay. The SUCNR1-Wt/miR-1827 mimic cotransfected group had significantly decreased luciferase activity, whereas, no significant difference existed in luciferase activity in the SUCNR1-Mut/miR-1827 mimic cotransfected group (Fig. 5 A). The results indicated that miR-1827 specifically bound to the SUCNR1 3’-UTR.
Targeted regulation of SUCNR1 by hUC-MSCs-Exos carrying miR-1827. A, Dual-luciferase reporter gene assay verified that miR-1827 could bind to SUCNR1 (* indicates p < 0.05 compared to NC-mimic group). B, After overexpression or silencing of miR-1827, RT-qPCR to detect transfection efficiency of miR-1827. C, SUCNR1 protein expression in HT-29 cells transfected with miR-1827 mimic or inhibitor (* indicates p < 0.05 compared to NC-mimic group; # indicates p < 0.05 compared to NC-inhibitor group). D, RT-qPCR to detect the expression of miR-1827 in HT-29 cells incubated with hUC-MSCs-Exos (* indicates p < 0.05 compared with the PBS + NC-inhibitor group; # indicates p < 0.05 compared with the hUC-MSCs-Exos + NC- inhibitor group). E, Protein expression of SUCNR1 in HT-29 cells after different treatments (* indicates p < 0.05 compared to PBS + NC-inhibitor group; # indicates p < 0.05 compared to hUC-MSCs-Exos + NC-inhibitor group). All experiments were repeated 3 times
In addition, miR-1827 mimic and miR-1827 inhibitor were successfully introduced into the HT-29 cells to elevate and reduce miR-1827 expression, respectively (Fig. 5B), consequently resulting in inhibition of SUCNR1 expression and promotion of SUCNR1 expression, respectively (Fig. 5 C).
Additionally, HT-29 cells were manipulated with NC-inhibitor or miR-1827 inhibitor and incubated with PBS or hUC-MSCs-Exos. Incubation of hUC-MSCs-Exos significantly increased miR-1827 expression but decreased that of SUCNR1 in the HT-29 cells manipulated with NC-inhibitor. However, miR-1827 expression was reduced while SUCNR1 expression increased by miR-1827 inhibitor in the HT-29 cells incubated with hUC-MSCs-Exos (Fig. 5D and E). Collectively, miR-1827 targeted SUCNR1 and downregulated its expression and hUC-MSCs-Exos can inhibit SUCNR1 expression via delivery of miR-1827.
hUC-MSCs-Exos carrying miR-1827 suppress SUCNR1 to inhibit macrophage M2 polarization
Our results above have demonstrated that inhibition of SUCNR1 attenuated macrophage M2 polarization and slowed down HT-29 cell migration. Therefore, we hypothesized that hUC-MSCs-Exos could carry miR-1827 to regulate SUCNR1, whereby inhibiting macrophage polarization and HT-29 cell migration. Firstly, HT-29 cells were manipulated with oe-NC or oe-SUCNR1 and incubated with hUC-MSCs-Exos-NC mimic or hUC-MSCs-Exos-miR-1827 mimic. Results demonstrated that oe-SUCNR1 did not alter the miR-1827 expression but induced a significant increase in the SUCNR1 expression in the HT-29 cells incubated with hUC-MSCs-Exos-NC mimic. Incubation of hUC-MSCs-Exos-miR-1827 mimic caused a significant increase in miR-1827 expression and a decrease in SUCNR1 expression in the HT-29 cells manipulated with oe-NC. Moreover, oe-SUCNR1 rescued the SUCNR1 expression without affecting the miR-1827 expression in the HT-29 cells incubated with hUC-MSCs-Exos-miR-1827 mimic (Fig. 6 A, 6B).
Effect of hUC-MSCs-Exos carrying miR-1827 on macrophage M2 polarization and CRC cell migration and invasion. A, RT-qPCR to detect the expression of miR-1827 in HT-29 cells (* indicates p < 0.05 compared to the hUC-MSCs-Exos-NC mimic group; # indicates p < 0.05 compared with hUC-MSCs-Exos-NC mimic + oe-SUCNR1 group. B, Western blot detection of SUCNR1 protein expression in HT-29 cells (* indicates p < 0.05 compared with hUC-MSCs-Exos-NC mimic group, # indicates p < 0.05 compared with hUC-MSCs-Exos-miR-1827 mimic + oe-NC group). C, CCK8 assay of proliferation ability of HT-29 cells. D, Number of migrating HT-29 cells by Transwell assay. E, Transwell assay of number of invading HT-29 cells. F, Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin and E-cadherin protein levels measured by Western blot assay. G, Western blot detection of ARG1 protein expression in macrophages treated with CM from different HT-29 cells. H, RT-qPCR detection of macrophage Fizz1 mRNA expression in macrophages treated with CM from different HT-29 cells. I, RT-qPCR detection of Mgl1 mRNA expression in macrophages treated with CM from different HT-29 cells. J, The percentages of M2-polarized macrophages treated with CM from different HT-29 cells (* indicates p < 0.05 compared to hUC-MSCs-Exos-NC mimic + CM group; # indicates p < 0.05 compared to hUC-MSCs-Exos-miR-1827 mimic + oe-NC + CM group). All experiments were repeated 3 times
In addition, function experiments revealed that SUCNR1 overexpression promoted proliferating, migrating, and invading properties of HT-29 cells incubated with hUC-MSCs-Exos-NC mimic, while incubation of hUC-MSCs-Exos-miR-1827 mimic inhibited these malignant functions of HT-29 cells manipulated with oe-NC. On the contrary, re-expression of SUCNR1 reversed the inhibition of these malignant functions of HT-29 cells induced by hUC-MSCs-Exos-miR-1827 mimic (Fig. 6 C-E, Figure S2G, 2 H). At protein levels, SUCNR1 overexpression upregulated Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin while downregulating E-cadherin in the HT-29 cells incubated with hUC-MSCs-Exos-NC mimic; incubation of hUC-MSCs-Exos-miR-1827 mimic resulted in decreased levels of Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin proteins and an elevated E-cadherin protein level. Furthermore, re-expression of SUCNR1 reversed the changes in the aforementioned proteins caused by hUC-MSCs-Exos-miR-1827 mimic (Fig. 6 F).
Furthermore, mouse macrophages were incubated with different CMs. In the presence of hUC-MSCs-Exos-NC mimic, oe-SUCNR1 promoted secretion of macrophage M2 polarization markers (ARG1, Fizz1 and Mgl1) and percentage of M2-polarized macrophages, while hUC-MSCs-Exos-miR-1827 mimic induced suppressive effects. oe-SUCNR1 reversed the increases in macrophage M2 polarization markers (Fig. 6G-I) and percentage of M2-polarized macrophages (Fig. 6 J) induced by hUC-MSCs-Exos-miR-1827 mimic. These data suggested that hUC-MSCs-Exos can carry miR-1827 to inhibit SUCNR1, delaying macrophage M2 polarization and CRC progression.
hUC-MSCs-Exos carrying miR-1827 inhibit macrophage M2 polarization and block colorectal liver metastasis in vivo
Finally, we examined the in vivo effect of hUC-MSCs-Exos carrying miR-1827 by regulating SUCNR1. There was no difference in the miR-1827 expression, while SUCNR1 expression was elevated in the tumor-bearing mice injected with hUC-MSCs-Exos-NC mimic + oe-SUCNR1 relative to the tumor-bearing mice injected with hUC-MSCs-Exos-NC mimic + oe-NC. However, miR-1827 expression increased and SUCNR1 expression decreased by injection with hUC-MSCs-Exos-miR-1827 mimic in the tumor-bearing mice injected with oe-NC. SUCNR1 was rescued by injection with oe-SUCNR1 in the tumor-bearing mice injected with hUC-MSCs-Exos-miR-1827 mimic (Fig. 7 A-B).
Effect of hUC-MSCs-Exos carrying miR-1827 on macrophage M2 polarization and liver metastasis in nude mice. A, RT-qPCR detection of miR-1827 expression in tumor tissues of nude mice (* indicates p < 0.05 compared to NC-mimic-Exos + oe-NC, # indicates p < 0.05 compared to NC-mimic-Exos + oe-SUCNR1). B, Western blot detection of SUCNR1 expression in tumor tissues of nude mice. C, Western blot detection of ARG1 protein expression in macrophages at tumor sites of nude mice. D, RT-qPCR for Arg1 mRNA expression in macrophages at tumor sites in nude mice. E, RT-qPCR for macrophage Fizz1 mRNA expression at tumor sites in nude mice. F, RT-qPCR for macrophage Mgl1 mRNA expression at tumor sites in nude mice. G, Flow cytometry to analyze the percentage of M2-polarized macrophages at tumor sites of nude mic. H, Representative images of xenograft tumor formation and folded line graph of tumor volume change in nude mice. I, Tumor weight statistics for each group of nude mice. J, Statistics on the number of tumor metastatic nodules in the liver of nude mice. K, Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin and E-cadherin protein levels in tumor tissues measured by Western blot assay. Ten mice per group. For panels B-J, * indicates p < 0.05 compared to NC-mimic-Exos + oe-NC, # indicates p < 0.05 compared to miR-1827 mimic-Exos + oe-NC. All cellular experiments were repeated 3 times
Then macrophages were collected from tumor sites of tumor-bearing mice and analyzed macrophage M2 polarization. Increased secretion of macrophage M2 polarisation markers (ARG1, Fizz1 and Mgl1) was detected after SUCNR1 overexpression in the tumor-bearing mice injected with hUC-MSCs-Exos-NC mimic. However, enhanced miR-1827 expression by hUC-MSCs-Exos-miR-1827 mimic resulted in reduced secretion of macrophage M2 polarization markers in the tumor-bearing mice injected with oe-NC. Furthermore, re-expression of SUCNR1 counteracted the reduced secretion of macrophage M2 polarization markers induced by hUC-MSCs-Exos-miR-1827 mimic (Fig. 7 C-F). In addition, consistent results were obtained regarding the percentage of macrophage M2 polarization numbers (Fig. 7G).
SUCNR1 overexpression led to increased tumor volume and weight in the tumor-bearing mice injected with hUC-MSCs-Exos-NC mimic. Enhanced miR-1827 expression by hUC-MSCs-Exos-miR-1827 mimic resulted in reduced tumor volume and weight in the tumor-bearing mice injected with oe-NC, which effect was negated by oe-SUCNR1 (Fig. 7 H-I). Livers were obtained from nude mice after 5 weeks to observe tumor metastasis. A significant increase in metastatic tumor nodes in the liver of tumor-bearing nude mice was detected in response to SUCNR1 overexpression in the presence of hUC-MSCs-Exos-NC mimic. However, miR-1827 enhancement by hUC-MSCs-Exos-miR-1827 mimic caused a decrease in metastatic tumor nodes in the liver of the tumor-bearing mice injected with oe-NC. However, oe-SUCNR1 reversed the inhibitory effects of hUC-MSCs-Exos-miR-1827 mimic on the metastatic tumor nodes in the liver (Fig. 7 J). Additionally, SUCNR1 overexpression led to upregulation of Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin as well as downregulation of E-cadherin protein level in the tumor-bearing mice injected with hUC-MSCs-Exos-NC mimic. Enhanced miR-1827 expression by hUC-MSCs-Exos-miR-1827 mimic resulted in reductions in Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin protein levels and an elevation in E-cadherin protein level in the tumor-bearing mice injected with oe-NC, which ipmact was counteracted by SUCNR1 re-expression (Fig. 7 K).
Based on these data, hUC-MSCs-Exos was suggested to carry miR-1827 to inhibit SUCNR1 expression, thus preventing macrophage M2 polarization and liver metastasis in vivo.
Discussion
Macrophage M2 polarization, which contributes to cancer metastasis, exerts an vital role in invasion and metastasis of CRC via facilitating angiogenesis and immunosuppression [33, 34], but the signaling mechanism underlying this process is unclear. In the present study, we explored the mechanism by which hUC-MSCs-Exos could affect macrophage M2 polarization and liver metastasis in CRC according to data from EVmiRNA database (Figure S5). Our study highlighted the contribution of SUCNR1 to macrophage M2 polarization in CRC and cancer progression. Additionally, miR-1827 was identified to target SUCNR1, whereby preventing liver metastasis in CRC, which may provide novel therapeutic targets against liver metastasis of CRC. Of great importance, our data provided evidence that hUC-MSCs-Exos could transmit miR-1827 into CRC cells to inhibit SUCNR1 expression whereby suppressing macrophage M2 polarization and liver metastasis.
We found an enrichment of miR-1827 in hUC-MSCs-Exos. A previous research has reported that miR-1827 is frequently down-regulated in human CRC while re-expression of miR-1827 restrains the growth of xenograft colorectal tumors [14]. Moreover, although the role of miR-1827 in CRC has not been fully elucidated, another study has shown that miR-1827 mimic could reduce the number of metastases and ectopic vessels in the zebrafish embryo model, and miR-1827 suppressed migration of lung adenocarcinoma cells [35]. Similarly, our experimental data showed that miR-1827 delivery by hUC-MSCs-Exos inhibited proliferative, migratory, and invading functions of CRC cells, and suppressed CRC growth and liver metastasis, as manifested by reduced tumor volume and weight as well as decreased metastatic nodules in livers which are indicators for alleviated CRC progression [36]. Also, miR-1827 carried by hUC-MSCs-Exos could reduce the expression of Bcl2, PCNA, Ki67, MMP2, MMP9, and N-cadherin while elevating E-cadherin, offering additional molecular evidence. Since we determined the tumor-suppressive role of miR-1827 in CRC, we further investigated the correlation between miR-1827 and hUC-MSCs-Exos. hUC-MSCs are reported to secret Exos that could repress cell proliferation and induce cell apoptosis in glioma [37]. hUCMSC-Exos have been frequently used in regenerative medicine and various diseases due to low immunogenicity and no tumorization [38]. Although miR-1827 could function to mediate the function of human bone marrow MSCs during osteogenic differentiation [39], if miR-1827 could be encapsulated in hUC-MSCs-Exos remains largely unknown. In our study, miR-1827 delivered by hUC-MSCs-Exos could exert tumor-suppressive effects on CRC, which was similar to the mechanism that human bone marrow MSCs-Exos suppress proliferation of acute myeloid leukemia cell line through the delivery of miR-222-3p [40]. Similar to a prior work stating that MSC-Exos attenuate myocardial Ischemia/Reperfusion injury in mice via shuttling miR-182 which modifies the polarization status of macrophages [41], we found that hUC-MSCs-Exos suppressed M2 polarization by carrying miR-1827. Accumulating evidence has verified the critical role of exosomal miRNAs in macrophage M2 polarization which contributes to CRC liver metastasis [42, 43]. In the present study, we demonstrated that hUC-MSCs-derived exosomal miR-1827 inhibited polarization of macrophages to M2 phenotype, which in turn curtailed liver metastasis of CRC.
After bioinformatics prediction, we further identified that SUCNR1 was negatively targeted by miR-1827. Succinate is a metabolic intermediate of the tricarboxylic acid cycle within host cells, and SUCNR1 has been demonstrated to enhance the intestinal inflammation [44]. Also, SUCNR1 is enriched in primary M2 macrophages and SUCNR1 can activate the Gq signaling to hyperpolarize the M2 phenotype of macrophages [45]. Our evidence showed that SUCNR1 was highly expressed in CRC cells, which was rarely reported yet. Furthermore, the study demonstrated that overexpressed SUCNR1 promoted M2 polarization, supported by high expression of Arg-1 and Fizz1. M2 macrophages are featured with high expression of mannose receptor CD206 and anti-inflammatory IL-10, together with a potent Arg-1 activity to boost angiogenesis and tumor progression [46]. SUCNR1 has been recently evidence to drive macrophage polarization, leading to lung cancer metastasis [17]. Both in vitro and in vivo experiments unraveled that suppressive role of SUCNR1 in preventing macrophage M2 polarization responsible for the anti-tumor effect of hUC-MSCs-Exos carrying miR-1827.
In summary, our data presented that hUC-MSCs secreted Exos, which carried and delivered miR-1827 into CRC cells to suppress the expression of SUCNR1, thereby inhibiting macrophage M2 polarization and further preventing CRC liver metastasis (Fig. 8). This study highlights exosomal miR-1827 as a valuable diagnostic biomarker and therapeutic target for metastasis of CRC. Due to limited time and funding, we failed to explore the possible downstream mechanisms of miR-1827/SUCNR1-mediated macrophage M2 polarization in colorectal liver metastasis. At present, there are still many questions that need to be addressed regarding the clinical use of hUC-MSC-derived Exos in the treatment of malignant tumors. Our findings are still at the basic research stage and more in-depth studies are needed to clarify the mechanism of hUC-MSC-derived Exos, and more animal and clinical trials are required to verify its safety and efficacy in order to promote the research and application of hUC-MSC-derived Exos in the field of tumor biotherapy.
Data Availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB (2019) Colorectal cancer. Lancet 394:1467–1480. https://doi.org/10.1016/S0140-6736(19)32319-0
Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T (2015) Colorectal cancer. Nat Rev Dis Primers 1:15065. https://doi.org/10.1038/nrdp.2015.65
Okugawa Y, Grady WM, Goel A (2015) Epigenetic alterations in Colorectal Cancer: emerging biomarkers. Gastroenterology 149:1204–1225. e1212. https://doi.org/10.1053/j.gastro.2015.07.011
Song M, Chan AT (2019) Environmental factors, gut microbiota, and Colorectal Cancer Prevention. Clin Gastroenterol Hepatol 17:275–289. https://doi.org/10.1016/j.cgh.2018.07.012
Hong Y, Rao Y (2019) Current status of nanoscale drug delivery systems for colorectal cancer liver metastasis. Biomed Pharmacother 114:108764. https://doi.org/10.1016/j.biopha.2019.108764
Chakedis J, Schmidt CR (2018) Surgical Treatment of Metastatic Colorectal Cancer. Surg Oncol Clin N Am 27:377–399. https://doi.org/10.1016/j.soc.2017.11.010
Elias D, Vigano L, Orsi F, Scorsetti M, Comito T, Lerut J, Cosola D, Torzilli G (2016) New Perspectives in the treatment of colorectal metastases. Liver Cancer 6:90–98. https://doi.org/10.1159/000449492
Koveitypour Z, Panahi F, Vakilian M, Peymani M, Seyed Forootan F, Nasr Esfahani MH, Ghaedi K (2019) Signaling pathways involved in colorectal cancer progression. Cell Biosci 9:97. https://doi.org/10.1186/s13578-019-0361-4
O’Driscoll L (2015) Expanding on exosomes and ectosomes in cancer. N Engl J Med 372:2359–2362. https://doi.org/10.1056/NEJMcibr1503100
Maia J, Caja S, Strano Moraes MC, Couto N, Costa-Silva B (2018) Exosome-based cell-cell communication in the Tumor Microenvironment. Front Cell Dev Biol 6:18. https://doi.org/10.3389/fcell.2018.00018
Chen X, Zhang S, Du K, Zheng N, Liu Y, Chen H, Xie G, Ma Y, Zhou Y, Zheng Y, Zeng L, Yang J, Shen L (2021) Gastric cancer-secreted exosomal X26nt increases angiogenesis and vascular permeability by targeting VE-cadherin. Cancer Sci 112:1839–1852. https://doi.org/10.1111/cas.14740
Lin S, Gregory RI (2015) MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15:321–333. https://doi.org/10.1038/nrc3932
Rupaimoole R, Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 16:203–222. https://doi.org/10.1038/nrd.2016.246
Zhang C, Liu J, Tan C, Yue X, Zhao Y, Peng J, Wang X, Laddha SV, Chan CS, Zheng S, Hu W, Feng Z (2016) microRNA-1827 represses MDM2 to positively regulate tumor suppressor p53 and suppress tumorigenesis. Oncotarget 7:8783–8796. https://doi.org/10.18632/oncotarget.7088
Guo X, Wang Z, Sun Q, Sun C, Hua H, Huang Q (2020) The inhibitory effect of microRNA-1827 on anoikis resistance in lung adenocarcinoma A549 cells via targeting caveolin-1. Acta Biochim Biophys Sin (Shanghai) 52:1148–1155. https://doi.org/10.1093/abbs/gmaa102
Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Mao Y (2020) Circular RNA hsa_circ_0003141 promotes tumorigenesis of hepatocellular carcinoma via a miR-1827/UBAP2 axis. Aging 12:9793–9806. https://doi.org/10.18632/aging.103244
Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, Yeh CC, Peng YJ, Kuo YY, Wen HT, Lin HC, Hsiao CW, Wu KK, Kung HJ, Hsu YJ, Kuo CC (2020) Cancer-Derived Succinate promotes macrophage polarization and Cancer metastasis via succinate receptor. Mol Cell 77:213–227e215. https://doi.org/10.1016/j.molcel.2019.10.023
Lu Y, Hou K, Li M, Wu X, Yuan S (2020) Exosome-delivered LncHEIH promotes gastric Cancer progression by upregulating EZH2 and stimulating methylation of the GSDME promoter. Front Cell Dev Biol 8:571297. https://doi.org/10.3389/fcell.2020.571297
Hogberg C, Gidlof O, Tan C, Svensson S, Nilsson-Ohman J, Erlinge D, Olde B (2011) Succinate independently stimulates full platelet activation via cAMP and phosphoinositide 3-kinase-beta signaling. J Thromb Haemost 9:361–372. https://doi.org/10.1111/j.1538-7836.2010.04158.x
Kong X, Ai G, Wang D, Chen R, Guo D, Yao Y, Wang K, Liang G, Qi F, Liu W, Zhang Y (2019) PDE4 and Epac1 synergistically promote rectal Carcinoma via the cAMP pathway. Anal Cell Pathol (Amst) 20197145198. https://doi.org/10.1155/2019/7145198
Yi H, Li XH, Yi B, Zheng J, Zhu G, Li C, Li MY, Zhang PF, Li JL, Chen ZC, Xiao ZQ (2010) Identification of Rack1, EF-Tu and Rhodanese as aging-related proteins in human colonic epithelium by proteomic analysis. J Proteome Res 9:1416–1423. https://doi.org/10.1021/pr9009386
Wu S, Chen H, Han N, Zhang C, Yan H (2019) Long noncoding RNA PVT1 silencing prevents the development of Uveal Melanoma by impairing MicroRNA-17-3p-Dependent MDM2 Upregulation. Invest Ophthalmol Vis Sci 60:4904–4914. https://doi.org/10.1167/iovs.19-27704
Pisano A, Grinan-Lison C, Farace C, Fiorito G, Fenu G, Jimenez G, Scognamillo F, Pena-Martin J, Naccarati A, Proll J, Atzmuller S, Pardini B, Attene F, Ibba G, Solinas MG, Bernhard D, Marchal JA, Madeddu R (2020) The inhibitory role of mir-486-5p on CSC phenotype has diagnostic and prognostic potential in Colorectal Cancer. Cancers (Basel) 12. https://doi.org/10.3390/cancers12113432
Lai HT, Tseng WK, Huang SW, Chao TC, Su Y (2020) MicroRNA-203 diminishes the stemness of human colon cancer cells by suppressing GATA6 expression. J Cell Physiol 235:2866–2880. https://doi.org/10.1002/jcp.29192
Li J, Huang J, Chen L, Ren W, Cai W (2022) Human umbilical cord mesenchymal stem cells contribute to the reconstruction of bladder function after acute spinal cord injury via p38 mitogen-activated protein kinase/nuclear factor-kappa B pathway. Bioengineered 13:4844–4856. https://doi.org/10.1080/21655979.2022.2036397
Li T, Wan Y, Su Z, Li J, Han M, Zhou C (2021) Mesenchymal stem cell-derived exosomal microRNA-3940-5p inhibits colorectal Cancer metastasis by targeting integrin alpha6. Dig Dis Sci 66:1916–1927. https://doi.org/10.1007/s10620-020-06458-1
Liao W, Ning Y, Xu HJ, Zou WZ, Hu J, Liu XZ, Yang Y, Li ZH (2019) BMSC-derived exosomes carrying microRNA-122-5p promote proliferation of osteoblasts in osteonecrosis of the femoral head. Clin Sci (Lond) 133:1955–1975. https://doi.org/10.1042/CS20181064
Qu Y, Zhang Q, Cai X, Li F, Ma Z, Xu M, Lu L (2017) Exosomes derived from mir-181-5p-modified adipose-derived mesenchymal stem cells prevent liver fibrosis via autophagy activation. J Cell Mol Med 21:2491–2502. https://doi.org/10.1111/jcmm.13170
Fang T, Lv H, Lv G, Li T, Wang C, Han Q, Yu L, Su B, Guo L, Huang S, Cao D, Tang L, Tang S, Wu M, Yang W, Wang H (2018) Tumor-derived exosomal mir-1247-3p induces cancer-associated fibroblast activation to foster lung metastasis of liver cancer. Nat Commun 9:191. https://doi.org/10.1038/s41467-017-02583-0
Fang X, Xu X, Lin X, Liu R (2019) Downregulated spinal IRF8 and BDNF in NAC are involved in neuropathic pain-induced depression relief via pulsed radiofrequency on dorsal root ganglion in rat SNI model. Brain Res Bull 146:192–200. https://doi.org/10.1016/j.brainresbull.2019.01.008
Zhang X, Han C, Du B, Nan, Zhang W, He G (2022) Isolation and identification of adipose stem cell exosomes and the study of its potential as drug delivery carrier in Vitro. Appl Biochem Biotechnol 194:2594–2603. https://doi.org/10.1007/s12010-022-03835-6
Xian D, Zhao Y (2019) LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med 23:3808–3823. https://doi.org/10.1111/jcmm.14071
Yang M, Liu J, Piao C, Shao J, Du J (2015) ICAM-1 suppresses tumor metastasis by inhibiting macrophage M2 polarization through blockade of efferocytosis. Cell Death Dis 6:e1780. https://doi.org/10.1038/cddis.2015.144
Cortese N, Soldani C, Franceschini B, Barbagallo M, Marchesi F, Torzilli G, Donadon M (2019) Macrophages in Colorectal Cancer Liver Metastases. Cancers (Basel) 11. https://doi.org/10.3390/cancers11050633
Ho CS, Noor SM, Nagoor NH (2018) MiR-378 and MiR-1827 regulate Tumor Invasion, Migration and Angiogenesis in Human Lung Adenocarcinoma by Targeting RBX1 and CRKL, respectively. J Cancer 9:331–345. https://doi.org/10.7150/jca.18188
Hua F, Shang S, Yang YW, Zhang HZ, Xu TL, Yu JJ, Zhou DD, Cui B, Li K, Lv XX, Zhang XW, Liu SS, Yu JM, Wang F, Zhang C, Huang B, Hu ZW (2019) TRIB3 interacts with beta-catenin and TCF4 to increase stem cell features of Colorectal Cancer Stem cells and tumorigenesis. Gastroenterology 156:708–721e715. https://doi.org/10.1053/j.gastro.2018.10.031
Hao SC, Ma H, Niu ZF, Sun SY, Zou YR, Xia HC (2019) hUC-MSCs secreted exosomes inhibit the glioma cell progression through PTENP1/miR-10a-5p/PTEN pathway. Eur Rev Med Pharmacol Sci 23:10013–10023. https://doi.org/10.26355/eurrev_201911_19568
Yaghoubi Y, Movassaghpour A, Zamani M, Talebi M, Mehdizadeh A, Yousefi M (2019) Human umbilical cord mesenchymal stem cells derived-exosomes in diseases treatment. Life Sci 233:116733. https://doi.org/10.1016/j.lfs.2019.116733
Liu L, Zeng D, Chen Y, Zhou J, Liao Y, Shi B (2020) Microarc oxidation surface of titanium implants promote osteogenic differentiation by activating ERK1/2-miR-1827-Osterix. In Vitro Cell Dev Biol Anim 56:296–306. https://doi.org/10.1007/s11626-020-00444-7
Wang J, Li P, Xu X, Zhang B, Zhang J (2020) MicroRNA-200a inhibits inflammation and atherosclerotic lesion formation by disrupting EZH2-Mediated methylation of STAT3. Front Immunol 11:907. https://doi.org/10.3389/fimmu.2020.00907
Zhao J, Li X, Hu J, Chen F, Qiao S, Sun X, Gao L, Xie J, Xu B (2019) Mesenchymal stromal cell-derived exosomes attenuate myocardial ischaemia-reperfusion injury through mir-182-regulated macrophage polarization. Cardiovasc Res 115:1205–1216. https://doi.org/10.1093/cvr/cvz040
Wang D, Wang X, Si M, Yang J, Sun S, Wu H, Cui S, Qu X, Yu X (2020) Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett 474:36–52. https://doi.org/10.1016/j.canlet.2020.01.005
Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, Zhang Z, Cai S, Xu Y, Li X, He X, Zhong X, Li G, Chen Z, Li D (2020) Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol 13:156. https://doi.org/10.1186/s13045-020-00991-2
Connors J, Dawe N, Van Limbergen J (2018) The role of Succinate in the regulation of intestinal inflammation. Nutrients 11. https://doi.org/10.3390/nu11010025
Trauelsen M, Hiron TK, Lin D, Petersen JE, Breton B, Husted AS, Hjorth SA, Inoue A, Frimurer TM, Bouvier M, O’Callaghan CA, Schwartz TW (2021) Extracellular succinate hyperpolarizes M2 macrophages through SUCNR1/GPR91-mediated gq signaling. Cell Rep 35:109246. https://doi.org/10.1016/j.celrep.2021.109246
Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A (2018) Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol 233:6425–6440. https://doi.org/10.1002/jcp.26429
Acknowledgements
Not applicable.
Funding
This study was supported by National Natural Science Foundation of China (82102497), Guangdong Medical Research Fund(A2020032), Guangdong Basic and Applied Basic Research Fund Project (2020A1515110633), Guangzhou Science and Technology Plan Project (202102020151), Research Fund for the Doctoral Program of GDPH (2020bq02,2020bq03).
Author information
Authors and Affiliations
Contributions
JRC and JHM conceived and designed research JRC, JHM and JX Lin performed experiments ZYL, CFY, RA and JX Lai analyzed data DDZ, YMG and BG interpreted results of experiments; ZYL and CFY prepared figures JRC, ZYL, CFY, DDZ, RA and JX Lai drafted manuscript JHM, JX Lin, YMG, BG and LXC edited and revised manuscript JRC, ZYL, CFY, JHM, JX Lin, DDZ, RA, JX Lai, YMG and BG approved final version of manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no conflicts of interest to disclose with respect to this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jierong Chen, Ziyue Li, Caifeng Yue, Jianhong Ma are regarded as co-first authors.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, J., Li, Z., Yue, C. et al. Human umbilical cord mesenchymal stem cell-derived exosomes carrying miR-1827 downregulate SUCNR1 to inhibit macrophage M2 polarization and prevent colorectal liver metastasis. Apoptosis 28, 549–565 (2023). https://doi.org/10.1007/s10495-022-01798-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-022-01798-x