Abstract
Prodigiosin, a secondary metabolite isolated from marine Vibrio sp., has antimicrobial and anticancer properties. This study investigated the cell death mechanism of prodigiosin in glioblastoma. Glioblastoma multiforme (GBM) is an aggressive primary cancer of the central nervous system. Despite treatment, or standard therapy, the median survival of glioblastoma patients is about 14.6 month. The results of the present study clearly showed that prodigiosin significantly reduced the cell viability and neurosphere formation ability of U87MG and GBM8401 human glioblastoma cell lines. Moreover, prodigiosin with fluorescence signals was detected in the endoplasmic reticulum and found to induce excessive levels of autophagy. These findings were confirmed by observation of LC3 puncta formation and acridine orange staining. Furthermore, prodigiosin caused cell death by activating the JNK pathway and decreasing the AKT/mTOR pathway in glioblastoma cells. Moreover, we found that the autophagy inhibitor 3-methyladenine reversed prodigiosin induced autophagic cell death. These findings of this study suggest that prodigiosin induces autophagic cell death and apoptosis in glioblastoma cells.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Prodigiosin, a bioactive secondary metabolite from marine Vibrio sp., has been shown to exhibit antibacterial, antimalarial, antibiotic, immunosuppressive, and antineoplastic actives [1]. In addition, prodigiosin meets Lipinski’s rule of five, which are rules for evaluating orally active drugs in humans [2]. Prodigiosin has be reported to exhibit potent apoptotic activity in human cancer cell lines including lymphocytic leukemia [3], melanoma [4], hematopoietic [5], breast [6], colorectal [7], and gastric [8] cancer cells. However, the cell death mechanism of prodigiosin glioblastoma remains unknown.
Glioblastoma multiforme (GBM) is the most aggressive and common malignancy of the central nervous system. It is characterized by a heterogeneous population of cells that are highly angiogenetic, infiltrative, and resistant to chemotherapy and radio therapy [9]. The median survival of primary GBM patients is approximately 14.6 months, despite patients receiving aggressive treatment with surgery, radiotherapy, and few alkylating chemotherapy drugs such as temozolomide (TMZ) [10]. Although TMZ is the primary chemotherapeutic drug for current GBM therapy, more than 90% of patients show no response after the second cycle of chemotherapy [11]. Approximately 40% of GBM patients exhibit an upregulated epidermal growth factor receptor (EGFR) pathway [12], and phosphatase and tensin homologue (PTEN) mutations, which are correlated with a poor prognosis, occur in 5–40% of these patients [13]. EGFR and PTEN mutations, which are correlated with resistance to downstream EGFR signaling, activate the phosphoinositide 3-kinase (PI3K)/AKT/mechanistic target of repamycin (mTOR) and induce cell proliferation, survival, growth, protein synthesis, and autophagy [14]. Spontaneous upregulation of the PI3K/AKT/mTOR pathway has been observed in GBM tumours [15]. Moreover, the phosphorylation level of AKT is highly correlated with a poor prognosis in some cancer patients who receive radiotherapy [16]. Therefore, EGFR and its downstream signaling pathways are critical targets for designing GBM drugs.
Autophagy is an important mechanism which facilitates cell survival under endoplasmic reticulum (ER) stress such as changes in environmental conditions, nutrient depletion, and toxicity. It helps cells digest aggregated or misfolded proteins and damaged organelles and recycle cellular components. Internal or external cellular stressors cause the accumulation of toxic unfolded or misfolded proteins in the ER, inducing the unfolded protein response (UPR), autophagy, or mitochondrial biogenesis [17]. To investigate autophagy, some studies have proposed using the autophagic tumor stroma model of cancer. According to this model, for cancer cells to maintain growth, they recycle cellular components through autophagy [18]. However, excessive autophagosomes formation without phagocyte participation has been observed in cells undergoing characterized autophagic cell death [19]. Moreover, autophagic cell death and apoptosis are achieved through parallel pathways, with mutual influence [20]. Briefly, persistent or intense ER stress mediates autophagic cell death. Failure of UPR-induced mechanisms leads to the conversion of autophagy to cell death [21] by triggering the C/EBP homologous protein (CHOP), which transcribes cell death-related genes [22] and activates c-Jun N-terminal kinase (JNK) signaling [23]. In addition, some reports have suggested that autophagic cell death maybe an alternative pathway for cells with defective apoptosis [24, 25]. Thus, autophagic cell death is a therapeutic target for anticancer agent resistance.
In this paper, we summarize the experimental results from an investigation of the mechanism of prodigiosin as an anticancer agent. We found that prodigiosin was localized to ER and induced ER stress and large-scale autophagic vacuolization. We also found that it induced autophagic cell death by activating the JNK pathway, and 3-methyladenine (3-MA), an autophagy inhibitor, could reverse the induced cell death in glioblastoma cell lines. Prodigiosin also significantly downregulated the AKT/mTOR pathway and induced caspase-3 activity. Moreover, treatment with prodigiosin inhibited neurosphere formation at a concentration much lower than that of TMZ. Neurosphere formation is a rapid method for predicting the clinical outcome of malignant glioma. This finding indicates that prodigiosin induces autophagic cell death and apoptosis and is thus a potential therapeutic agent for glioblastoma.
Materials and methods
Compound
Marine-derived prodigiosin was provided by Ping-Jyun Sung (National Museum of Marine Biology and Aquarium, Taiwan). It was isolated from gram-negative γ-proteobacteria, Vibrio sp. C1-TDSG02-1. This bacterial strain was collected from Siaogang Harbor, Taitung, in Eastern Taiwan at a water depth of 17 m. The strain C1-TDSG02-1 was 99.0% identical to Vibrio sp. BL-182 (Genbank accession no. AY663829.1) on the basis of the 16S rDNA gene sequence. Extraction of the culture broth (8.0 L) with ethyl acetate (EtOAc, 4 × 8.0 L) yielded 45.7 g of crude extract. An EtOAc layer was separated on silica gel followed by elution chromatography with a mixture of n-hexane/EtOAc (stepwise, pure n-hexane, pure EtOAc) to yield 16 subfractions. Fraction 6 was chromatographed on silica and eluted using a mixture of n-hexane/actone (stepwise, 10:1, pure actone) to afford 11 subfractions. Fraction 6–5 was chromatographed on silica and eluted using a mixture of n-hexane/actone (4:1) to afford prodigiosin (1.94 g). Prodigiosin was isolated as a red powder that gave an [M+H]+ ion peak at 324 m/z in the electrospray-ionization mass spectrometry spectra. A nuclear magnetic resonance (NMR) spectrum of pure prodigiosin is shown in Fig. S1. A stock solution of 100 mM prodigiosin was prepared in dimethyl sulfoxide (DMSO) and stored at − 20 °C.
Reagents
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were purchased from SERVA (Heidelberg, Germany); TMZ and 3-metyladenine (3-MA) were purchased from Cayman (MI, USA); NPE nuclear isolation medium-4,6-diamidino-2-phenylindole dihydrochloride (NIM-DAPI) were purchased from Beckman Coulter Inc. (CA, USA). Tween-20, TritonX-100, DMSO, acridine orange, 4′,6-diamidino-2-phenylindole (DAPI), basic fibroblast growth factor (bFGF), epidermal growth factor (EGF), bovine serum albumin (BSA), Bafilomycine A1 and Wortmannin were purchased from Sigma-Aldrich (MO, USA). caspase-3 colorimetric assay kit was obtained from Abcam (MA, USA). DC™ protein assay kits, iScriptTM cDNA Synthesis Kit (Bio-Rad, CA, US). Acridine orange, TRIzol Reagent, minimum essential medium eagle alpha modifications (alpha-MEM), Roswell Park Memorial Institute medium (RPMI) 1640, heat-inactivated fetal bovine serum (FBS), Penicillin–Streptomycin (P/S) (10,000 U/mL), and B-27 supplements were purchased from Thermo Fisher Scientific (CA, USA).
Antibodies
β-actin, p-AKT (Ser473), AKT, p-S6 (Ser235/236), S6, p-mTOR (Ser2448), PARP, p-JNK (Thr183/Tyr185), and JNK antibodies were purchased from Cell Signaling Technology (MA, USA). LC3A/B, mTOR and calnexin antibodies were from Abcam (MA, USA); p62/SQSTM1 antibody was from Proteintech (IL, USA); LC3B antibody was from Abgent (CA, USA). Alexa Fluor® 647 goat anti-rabbit IgG Ab and Alexa Fluor® 633 goat anti-mouse IgG antibodies were from Thermo Fisher Scientific (CA, USA).
Cell culture
The human brain malignant glioma U87MG and GBM8401 [26] lines were purchased from the Food Industry Research and Development Institute (Hsinchu, Taiwan). The human glioblastoma cancer cell lines U87MG maintained at alpha-MEM and GBM8401 maintained at RPMI 1640 medium. All mediums content 10% FBS and 50 U/mL P/S. Cell lines were cultured under a humidified atmosphere consisting of 5% CO2 mixed in 95% air at 37 °C. These cell lines were subcultured every 2–3 days up to the 10th passage. These cells were used in the following experiments.
Neurosphere formation
For in vitro culture, the glioblastoma cell line U87MG or GBM8401 was incubated in alpha-MEM or RPMI 1640, respectively. To promote sphere formation B-27 supplements, 20 ng/mL EGF and 20 ng/mL bFGF added to the serum-free culture medium [27]. 5000 cells were cultured in 10 mL culture medium on a 10 cm2 dish. Spheres were cultured in an ultra-low adherent dish containing prodigiosin or TMZ during sphere formation and were filtered through a 100 µm filter at 7 days after culturing. The number of spheres was observed and counted by under a microscope (Leica, Wetzlar, Germany).
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) assay for cell viability analysis
The cell viability of the U87MG and GBM8401 cell lines was determined using an MTT assay. Cells at a density of 2 × 104/well were seeded onto a 96-well plate. On the next day, various concentrations of drugs were added. At the end of the exposure time, 20 µL of 5 mg/mL MTT was added to each well and incubated at 37 °C for 4 h. The medium was carefully removed, and 200 µL of DMSO was added to each well. Absorbance at 570 nm was read on an enzyme-linked immunosorbent assay reader (Epoch, BioTek, VT, USA). IC50 was calculated using GraphPad Prism version 5.
Acridine orange staining
Cells were seeded onto round coverslips on 12-well plates overnight in 5% CO2 mixed in 95% air at 37 °C. On the next day, prodigiosin was added to each well under each condition. After prodigiosin treatment, acridine orange stain was immediately added at a final concentration of 2 µg/mL and was incubated for 10 min at 37 °C. Observation at emission wavelengths of 515 nm (green fluorescence) and 650 nm (red fluorescence), on a Leica DM-6000B fluorescence microscope (Leica, Wetzlar, Germany). Images were captured using SPOT Xploer integrating camera (Diagnostic Instruments, MI, USA).
Flow cytometry analysis of cell cycle
U87MG and GBM8401 cells at 5 × 105 cells/well were seeded onto 6-well plates and were treated with the indicated concentrations of prodigiosin for 24 and 48 h. Cells were washed twice with phosphate-buffered saline (PBS) and were resuspended in PBS. Moreover, for each sample, 1 × 105 cells in 100 µL PBS were stained with 100 µL of NIM-DAPI. Nuclei were analyzed using a Cell Lab Quanta™ SC flow cytometer equipped with 350-nm ultraviolet laser (Beckman Coulter Inc., CA, USA). The data were analyzed using Kaluza® Flow analysis software.
Apoptosis detection using caspase-3 activity assay
Caspase-3 activity was assessed according to manufacturer’s protocol. GBM8401 cells were treated with 0, 1, 5, 10 µM prodigiosin for 24 h. After treatments, cells were trypsinized and centrifuged a 1000×g rpm for 5 min. Cell pellet were lysed with 150 µL of pre-cooled commercial lysis buffer on ice for 10 min then centrifuged at 13,000×g rpm for 1 min at 4 °C. The protein concentration of each sample was measured by DC™ protein assay kits. Activity assay was reacted in 200 µL Reaction buffer with 10 µL of caspase-3 substrate (DEVD-pNA) and 10 mM DTT were added to 100 µg proteins and incubated at 37 °C for 2 h. The substrate emission (p-NA) was measured using ELISA plate reader at 405 nm.
Signaling pathways detection using western blotting
U87MG and GBM8401 cells were treated at the indicated time points of prodigiosin for different concentration depending on the experiment. Supernatants were collected, and cells were washed with PBS before addition of RIPA lysis buffer contented cOmplete ULTRA protease inhibitor cocktail tablets (Roche Diagnostics, Mannheim, Germany). The immunoreactive bands were visualized by Immobilon Western Chemiluminescent HRP Substrate (Merck Millipore, MA, United States). The images were visualized using the UVP BioChemi Imaging System (UVP, CA, USA), and relative densitometric quantification was performed using LabWorks 4.0 software (UVP, CA, USA).
ER stress related genes detection using semiquantitative real-time PCR
Total RNA was extracted from U87MG and GBM8401 cells using TRIzol Reagent according to the manufacturer’s instructions. The concentration, yield, and quality control indices of total RNA will base on the absorbance at 260 and 280 nm by spectrophotometer. Equal amounts of total RNA were reversely transcribed into single-strand cDNA using the iScriptTM cDNA Synthesis Kit. First-strand cDNA was synthesized by using 1 µg of total RNA, 4 µL of 5X iScript reaction mix (containing both oligo(dT) primers and random hexamers, reaction buffer with dNTP), iScript reverse transcriptase, and nuclease-free water in a total volume of 20 µL. The reaction was carried out at 25 °C for 50 min, 42 °C for 30 min, and be terminated by deactivation of the enzyme at 85 °C for 5 min. Genes were detected by semiquantitative real-time PCR. 100 ng cDNA per sample was amplified by specific 2× PCR Super Master Mix (TOOLS, Taipei, Taiwan) on TProfessional TRIO thermocycler (Biometria, Göttingen, Germany). The PCR program are: 95 °C for 1 min, 28 cycles of 95 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min, followed by 5 min at 72 °C. The PCR products were loaded onto DNA view-stained (TOOLS, Taipei, Taiwan) 2% agarose gels in TBE. The specific primer sequences used in this work were listed in Supplementary information (Table S1).
Light chain 3 puncta observation using fluorescence microscopy
Glioblastoma cell lines were seeded onto a glass slide and were treated with various concentrations of prodigiosin, depending on the experiment, at the indicated time points. These cells on the glass slide were fixed in cold methanol on ice for 15 min. Cells were permeabilized by treatment with 0.1% TritonX-100 in PBS on ice for 10 min and were blocked with 1% BSA and 0.1% Tween-20 in PBS at room temperature for 1 h. Moreover, cells were incubated with the LC3B antibody in TTBS (diluted in buffer containing 0.1% BSA and 0.1% Tween-20) overnight at 4 °C. Cells were then incubated with the secondary fluorescent antibodies of Alexa Fluor® 647 goat anti-rabbit IgG for 60 min at room temperature. Nuclei were stained with DAPI for 10 min at room temperature. Between each step, cells were washed with PBS. At the final step, glass slides were mounted using Dako Fluorescent Mounting Medium (Dako North America, CA, USA). Fluorescence images were scanned using a Leica TCS SP5II fluorescence microscope (Leica, Wetzlar, Germany) and captured using a SPOT Xplorer integrated camera (Diagnostic Instruments Inc., MI, USA).
Observation of prodigiosin localization to the ER using confocal microscopy
The same processes and reagents mentioned in the earlier text were applied to glioblastoma cells on the glass slide. Cells were incubated with calnexin Ab overnight at 4 °C. To avoid detecting fluorescence at the wavelength of prodigiosin, cells were incubated with the secondary fluorescent antibodies of Alexa Fluor® 647 goat anti-rabbit IgG for 60 min at room temperature. Confocal images were recorded using the Leica TCS SP5II equipped with Leica HyD (Hybrid Detector). The exposure times were identical for all cell samples on the same microscope slide.
Statistical analyses
Final quantitative data are presented as the mean ± standard error. Data were analyzed using the two-tailed Student’s t test with P < 0.05 as the level of statistical significance.
Results
Prodigiosin induces cell death and morphological changes in human glioblastoma cells
Glioblastoma cells were grown to approximately 70% confluence. Subsequently, the MTT assay was used to determine the cell viability of the human glioblastoma cell lines U87MG and GBM8401 and the human fetal glial cell line SVGp12 (Fig. 1a); the cells were treated with various concentrations of the marine-derived compound prodigiosin for 24 and 48 h. The half-maximal inhibitory concentrations (IC50) values obtained at 24 h of treatment with prodigiosin in U87MG, GBM8401, and SVGp12 are shown in Fig. 1b. Prodigiosin significantly reduced cell viability in a dose-dependent manner. Microscopic observation revealed that prodigiosin induced the formation of large swollen vacuoles in the cytosol of U87MG and GBM8401 cells (Fig. 1c). These swollen vacuoles were mainly autophagic vacuoles. To analyze autophagy, we used acridine orange to stain acidic vesicular organelles (AVOs), a morphological characteristic of autophagy that includes autolysosomes [28]. The nucleolus and cytoplasm of in the U87MG and GBM8401 cells were detected as green fluorescence signals. In prodigiosin-treated U87MG and GBM8401 cells (Fig. 1d), AVOs were detected as concentrated red fluorescence signals.
Prodigiosin affects the viability of glioblastoma cell lines. a U87MG, GBM8401 and SVGp12 cells were treated with 0, 0.3125, 0.625, 1.25, 2.5, 5, 10, 20, and 40 µM prodigiosin for 24 and 48 h. Cell viability was determined using an MTT assay. Data are presented as the mean ± SD (n = 6). *P < 0.05 relative to controls. b IC50 value from U87MG, GBM8401, and SVGp12 cell after 24 h of treatment with prodigiosin. c The morphology of U87MG, GBM8401 and SVGp12 cells after treated with prodigiosin for 24 h, compared with the controls. Cells were analyzed using phase-contrast microscopy. Scale bars, 50 µm. d U87MG and GBM8401 cells were stained with 2 µM acridine orange for 15 min and were immediately analyzed using fluorescence microscopy after treatment with 10 µM prodigiosin for 24 h. Lysosome-related structures could be observed as small orange bodies, and DNA was detected as green fluorescence signals. Scale bars, 100 µm
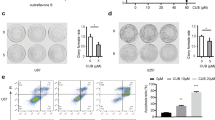
Prodigiosin reduces growth of spheres formed from glioblastoma cells
Neurosphere formation is a rapid method for predicting the clinical treatment outcome of malignant glioma [29]. We examined the efficacy of prodigiosin and TMZ for neurosphere formation from glioblastoma cell suspensions. As shown in Fig. 2a, 200 µM TMZ significantly reduced the size and number of neurospheres (P < 0.05; Fig. 2b). No neurosphere formation was observed under the 0.1 µM prodigiosin treatment condition. These results indicated that prodigiosin depletes the self-renewing glioblastoma population at a concentration much lower than that of TMZ.
Neurosphere derived from prodigiosin- and TMZ-treated glioblastoma cells. Photomicrographs of neurospheres expanded at 7 days after the transfer of cells to neurosphere-formation medium (serum free alpha-minimum essential medium containing bFGF/EGF and B-27 supplement). a U87MG and GBM8401 cells were analyzed using microscopy at ×100 magnification. b Numbers of sphere (> 100 µm) per well. Scale bars, 200 µm. Values are expressed as the mean ± SD (n = 3). *P < 0.05 relative to control
Prodigiosin induces apoptosis in glioblastoma cells
To determine the apoptotic effect of prodigiosin in glioblastoma cells, we used flow cytometry to detect the incomplete DNA content of nuclei (Fig. 3a). Prodigiosin dose-dependently increased the population of U87MG (Fig. 3b) and GBM8401 (Fig. 3c) cells with sub-G1 DNA content and decreased the population of these cells with G2/M content. Moreover, treatment with prodigiosin for 24 h significantly increased the activity of caspase-3, the primary activator of apoptosis, in a dose dependent manner in U87MG (Fig. 3d) and GBM8401 (Fig. 3e) cells. Cleaved apoptotic protein poly (ADP-ribose) polymerase (PARP) was increased in prodigiosin-treated glioblastoma cells (Fig. 3f).
Prodigiosin affects the cell cycle and induces apoptosis in glioblastoma cell lines. a Cell cycle analysis of U87MG and GBM8401 cells treated with 5 and 10 µM prodigiosin for 24 and 48 h. The cell cycle distribution of cell cycle was analyzed using DAPI staining and flow cytometry. Percentage of sub-G1, G1, S, and G2/M fractions for b U87MG and c GBM8401 cells. Caspase-3 activity were assayed using 100-µg cell lysates of d U87MG and e GBM8401 cells treated with 0, 1, 5, and 10 µM prodigiosin for 24 h. Values are expressed as the mean ± SD (n = 3). *P < 0.05 relative to controls. f Western blot analysis results of PARP and cleaved-PARP in U87MG and GBM8401 cells subjected to 1, 5, and 10 µM prodigiosin treatment for 24 h. Values are expressed as the mean ± SD. *P < 0.05 relative to controls
Prodigiosin is strongly correlated with calnexin, an ER marker, and induces ER stress and autophagy in glioblastoma cells
The ER is a major intracellular organelle for protein synthesis, folding, modification, and transportation. Calnexin, an ER membrane-bound chaperone protein, is an ER stress-related UPR protein that is activated in cells in response to biochemical, pathological, and physiological stimuli. In this study, prodigiosin was found to be colocalized and strongly correlated with calnexin, an ER marker [30]. Furthermore, it showed moderate correlation with mitochondria marker cytochrome c, but very weak correlation with ribosome marker S6 in the cytosol of GBM8401 cells. The quantification of colocalization quantification was performed using Pearson’s correlation coefficient [31], and the description of strength correlation was in accordance with Evans’s guide [32] (Fig. 4a). Splicing of X-box binding protein-1 (sXBP1) mRNA is a marker of ER stress [33]. Binding immunoglobulin protein (BiP)/GRP78 is a central regulator that targets misfolded proteins for degradation, and it regulates stress-induced autophagy [34]. The expression of CHOP, a proapoptotic factor mainly induced by ER stress, is mediated by UPR, and CHOP induces apoptosis [35]. In our study, semiquantitative polymerase chain reaction results (Fig. 4b) revealed that sXBP1, BiP/GRP78, and CHOP mRNAs were upregulated in prodigiosin-treated cells. Additionally, western blot analysis showed increased BiP/GRP78, as well as CHOP protein (Fig. 4c). LC3 is an accepted specific marker for detecting autophagy [36]. The immunostaining results showed LC3 puncta formation in prodigiosin-treated GBM8401 cells, but prodigiosin was not colocalized with LC3 protein in the cytosol (Fig. 4d). Moreover, western blot analysis results revealed that LC3-II/I and phosphor-JNK increased after prodigiosin treatment (Fig. 4e). JNK activation has been recognized as an essential component of autophagic cell death [37]. Therefore, these results demonstrate that prodigiosin is located in the ER and induces autophagy and, possibly, autophagic cell death in glioblastoma cells.
Prodigiosin induces ER stress and autophagy in glioblastoma cell lines. a Confocal images of organelle markers and prodigiosin in prodigiosin-treated GBM8401 cells for 4 h. The colocalization of prodigiosin (red signals) with calnexin, cytochrome c, and S6 (green signals). Scale bars, 20 µm. The plot shows Pearson’s correlation coefficients for the colocalization of prodigisin and organelle markers. Data were taken frome at least 10 cells. Bars indicate mean ± SD. *P < 0.05 relative to the calnexin group. b Spliced XBP1, binding immunoglobulin protein/GRP78, and C/EBP homologous protein (CHOP) mRNA expression increased after 5 µM prodigiosin treatment. Expression levels were examined using semiquantitative reverse transcription polymerase chain reaction. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. c Western blot analysis of the expression of ER stress proteins BiP/GRP78 and CHOP. β-Actin was used as a loading control. d Autophagy was analyzed by performing immunofluorescence staining of LC3 protein (green signal), and nuclei were labeled using DAPI (blue signal). Prodigiosin was detected at the excitation and emission wavelengths of 540 and 605 nm, respectively (red signal). Cells were observed using fluorescence microscopy. e Western blot analysis was performed to study the expression of autophagy-related proteins pJNK, JNK, and LC3 in U87MG and GBM8401 cells treated with 5 µM prodigiosin for 15, 30, 60, 90, and 120 min. β-actin was used as a loading control
Prodigiosin suppresses AKT/mTOR signaling and affects autophagic cell death-related protein in glioblastoma cell lines
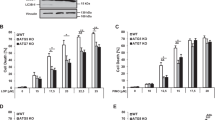
Many reports have indicated that AKT pathway inhibition and downstream mTOR complex 1 (mTORC1) inactivation are the major cellular responses that initiate autophagy and apoptosis [38]. Moreover, p62 inhibition has been found to result in the information of misregulated autophagosomes and the induction of autophagic cell death in carcinoma cells [39]. The apoptotic effector Bax is also involves in the process of lysosome membrane premeabilization and autophagic cell death [40]. To assess the effect of prodigiosin on the AKT/mTOR pathway, we used U87MG and GBM8401, which are glioblastoma cell lines with constitutive AKT activation. Western blot analysis results revealed that prodigiosin treatment time-dependently decreased AKT, mTOR, and S6 phosphorylation in both U87MG and GBM8401 glioblastoma cells within 24 h (Fig. 5a). In addition, western blot analysis results showed that p62 level decreased, whereas the Bax and LC3-II/LC3-I ratio increased (Fig. 5b), in prodigiosin-treated glioblastoma cells. Collectively, these results showed that prodigiosin not only induced autophagy through suppressing the AKT/mTOR pathway, but also affected proteins related to autophagic cell death in both U87MG and GMB8401 glioblastoma cell lines.
Effect of prodigiosin on critical effectors of the PI3K/AKT/mTOR pathway in glioblastoma cells. U87MG and GBM8401 cells were treated with 5 µM prodigiosin for 0, 2, 4, 8, 16, and 24 h. a Western blot analysis was performed to study the expression of pAkt, p-mTOR, and pS6. β-Actin was used as a loading control. Quantification results of p-AKT/AKT, p-mTOR/mTOR, and p-S6/S6. b Western blot analysis was performed to study the expression of p62, LC3, and Bax. β-Actin was used as a loading control. Quantification results of p62/β-Actin, LC3-II/LC3-I, and Bax/β-Actin. Values are expressed as the mean ± SD. *P < 0.05 relative to controls
Effect of autophagy inhibitors on prodigiosin-induced autophagic cell death
Autophagic cell death is a programmed cell death mechanism that induces large-scale autophagic vacuolization in the cytoplasm [41]. To demonstrate that prodigiosin induces cell death through autophagy, we pretreated GBM8401 cells with 3-MA, an autophagy inhibitor, for 2 h, and we then treated the cell with prodigiosin for 24 h. Immunocytochemistry confirmed that 0.5 mM 3-MA prevented the LC3 puncta accumulation induced by 5 µM prodigiosin (Fig. 6a). MTT assay results demonstrated that the autophagy inhibitor 3-MA significantly increased prodigiosin-induced cytotoxicity (Fig. 6b). Moreover, a cell cycle analysis using flow cytometry found reduced sub-G1 populations in GBM8401 cells pretreated with 3-MA (Fig. 6c), Bafilomycine A1, or Wortmannin (Fig. S2). The dosages of autophagy inhibitors did not cause significant cell death in GBM8401 cells (Fig. S3). Therefore, these results reveal that prodigiosin induces, at least in part, glioblastoma cell death through autophagy.
Effects of the autophagy inhibitor 3-MA on prodigiosin-induced cell death. a GBM8401 cells were pretreated with 0.5 mM 3-MA for 2 h before treatment with 5 µM prodigiosin for 2 h. Autophagy was analyzed by performing immunofluorescence staining of LC3 protein (green signal), and nuclei were labeled using DAPI (blue signal). Prodigiosin was detected at the excitation and emission wavelengths of 540 and 605 nm, respectively (red signal). Scale bars, 50 µm. b MTT assay results of GBM8401 cells treated with 5 µM prodigiosin with or without 0.5 mM 3-MA. c Flow cytometry was used to determine cell cycle phases of 5 µM prodigiosin-treated GBM8401 cells pretreated with or without 3-MA (0.5 mM or 1 mM). The cell cycle distribution was analyzed using DAPI staining and flow cytometry
Discussion
Prodigiosin is a secondary metabolite that is also a fluorescent pigment produced by both gram-negative and -positive bacteria. The prodigiosin used in this study was derived from cultured marine Vibrio sp. The antibiotic activity of prodigiosin was first detected in 1946 [42]. It has been reported that prodigiosin exerts a higher antibacterial activity against gram-positive bacterial species than against gram-negative bacterial species, and this antibacterial activity is mediated through the inhibition of topoisomerase IV and DNA gyrase [1]. Moreover, prodigiosin has been reported to cause mitochondrial dysfunction in Trypanosoma cruzi, leading to cell death; T. cruzi is the protozoan parasite that induces Chagas disease. Prodigiosin has been observed to exhibit anticancer activity in several cancers. It triggers mitochondrial apoptosis [43] and antimetastatic effects [44] in melanoma cancer cells. It also causes DNA fragmentation in haematopoietic cancer cells, leading to apoptosis. Moreover, its anticancer activity is mediated through the inhibition of Wnt/β-catenin signaling [6], inducing p21WAF1/CIP1 expression [45] in breast cancer cells. Until now, studies have revealed that apoptosis is the overall mechanism for the anticancer activity of prodigiosin against several human cancers. However, no study has investigated the cell death mechanism of prodigiosin in glioblastoma cell lines. In the present study, we focused on the cell death mechanisms of prodigiosin in glioblastoma cells.
In our current study, we used in vitro neurosphere formation model for demonstrating the effect of cancer stem-like cells (CSCs). Sphere formation ability provides insights into malignant properties such as tumour initiation, metastasis, self-renewal, treatment resistance, and recurrence [46]. Thus, in vitro sphere formation assay is a rapid method for predicting the efficacy of cancer therapeutic agents [47]. Some studies have indicated that autophagic process may be a critical factor for CSCs and chemotherapeutic agent resistance [48]. However, autophagy-related cell death may be induced depending on different types of stress, cells, and stimuli. For example, Delta-24-RGD causes autophagic cell death in brain tumour stem cell lines derived from human GBM surgical specimens by mediating the formation of numerous autophagic vacuoles [49]. In the present study, prodigiosin totally inhibited neurosphere formation at a concentration of 0.1 µM, which is much lower than 200 µM of the first-line GBM chemotherapy drug TMZ (Fig. 2). Moreover, 100 µM TMZ led to autophagy-related cell death in the U373 glioblastoma cell line, and 3-day treatment with TMZ caused 70% cell death [50]. According to our results, 5 µM prodigiosin induces the formation of numerous autophagic vacuoles within 15 min and cell death within 24 h in the U87MG and GBM8401 cell lines (Fig. 1). Thus, the results strongly demonstrate that prodigiosin has higher efficiency for inhibiting the growth of glioblastoma cells and its CSCs than does TMZ.
The ER is the largest organelle in the cytosol of eukaryotic cells and is a main intracellular organelle for protein synthesis, folding, and transport. It is also a major site of calcium storage, carbohydrate metabolism, and lipid synthesis. To the best of our knowledge, the current study is the first to report that prodigiosin is located in the ER and is colocalized with calnexin, an ER membrane-bound chaperone protein, in GBM8401 glioblastoma cells (Fig. 4a). We hypothesized that prodigiosin passes through the cell membrane and is then transported to the ER, causing severe ER stress. ER stress is an important cellular response that facilitates cell survival by inducing autophagy for the digestion of damaged organisms or proteins. Generally, autophagy is a pro-survival pathway induced in cells under stresses such as hypoxia, starvation, toxicity, or nutrient depletion [51]. Moreover, autophagy is an important cellular response for maintaining the functional structure of the ER [52]. Generally, autophagy has both positive and negative effects. For example, when cancer cells are exposed to different levels of stresses, the cellular response of ER stress confers stress tolerance, enabling cell survival [53]. However, several studies have indicated that excessive levels of autophagy cause autophagic cell death through mechanisms other than apoptosis and necrosis cell death. In our study, we observed that the accumulation of LC3 puncta, an autophagy marker, increased within minutes after prodigiosin treatment (Fig. 4e). Furthermore, we detected upregulated ER stress markers BiP/GRP78 and sXBP1 expression in both U87MG and GBM8401 glioblastoma cells (Fig. 4b, c). These findings demonstrated that prodigiosin is located in the ER and triggers ER stress and autophagy.
Approximately 88% of GBM patients exhibit an upregulated PI3K/AKT/mTOR pathway, which is highly related to PTEN mutations or the loss of heterozygocity [54, 55]. This signaling pathway is the classic regulatory pathway that negatively regulates autophagy and apoptosis [56]. Many studies have indicated that the PI3K/AKT/mTOR pathway is an important target for GBM treatment. Until now, only one anti brain tumour agent (i.e., Afinitor [everolimus]) targeting AKT/mTOR in brain tumour cells has been approved by the U.S. Food and Drug Administration. mTORC1, which is activated downstream of PI3K, is involved in the inhibition of autophagy and the upregulation of protein/nucleotide synthesis for proliferation, mitochondrial biogenesis, and lipid synthesis. mTORC2 regulates the phosphorylation of AKT, which is related to cell survival, actin organization, and K+ excretion [57]. Using in silicon model, a previous study predicted that prodigiosin interacts with mTOR and inhibits mTOR in insulin-stimulated melanoma cells [4]. Our current results indicate that prodigiosin decreased the abnormally upregulated AKT/mTOR signaling pathway and downstream S6 in both U87MG and GBM8401 glioblastoma cells. Moreover, we observed that prodigiosin caused large-scale autophagic vacuolization, resulting in the formation of numerous AVOs in the cytosol (Fig. 1d), and prodigiosin induced apoptosis through caspase-3 activation and PARP cleavage (Fig. 3f) in prodigiosin-treated cells. The ER stress-induced apoptosis marker CHOP also increased in prodigiosin-treated glioblastoma cells (Fig. 4b, c). Moreover, flow cytometry results revealed that prodigiosin induced cell cycle arrest in the sub-G1 phase (Fig. 3a, b, c). Consequently, these results strongly demonstrate that prodigiosin is a candidate therapeutic compound for decreasing abnormally activated AKT/mTOR signaling and inducing apoptosis in glioblastoma cells.
Excessive levels of cellular autophagy promote autophagic cell death. The total area of large-scale autophagic vacuolization in the cytoplasm is larger than the resultant vacuolated appearance [58, 59]. To clarify apoptosis and autophagic cell death, according to the Nomenclature Committee on Cell Death, autophagic cell death describes cell death that is suppressed by the inhibition of the autophagy pathway. Moreover, apoptosis resistance is one of major reasons for chemotherapeutic agent resistance in malignant cancer. Therefore, autophagic cell death may be a strategy for treating apoptosis-resistant cancers [60]. The JNK signaling transduction pathway, a mitogen-activated protein kinase signaling pathway, is a crucial autophagic cell death pathway. It is highly correlated with the regulation of autophagy [61], cell growth, inflammation, and apoptosis in response to stress [62]. The activated JNK pathway induces autophagy through the inactivation of the Bcl-2 antiapoptotic protein, leading to its disassociation from Beclin-1, an autophagic protein that initiates the formation of the pre-autophagosomal structure [63]. Etoposide is a chemotherapy medication which induces both apoptosis and autophagic cell death. In Shimizu et al.’s study, etoposide activated JNK signaling in Bax/Bak double-knockout mouse embryonic fibroblasts, which are an apoptosis resistance model. Moreover, autophagy inhibitor 3-MA decreased phospho c-Jun and reversed cell survival in etoposide-induced cell death in this model [37]. In our results, the induction of LC3 puncta and then JNK signaling following cell death signaling were found in prodigiosin-treated U87MG and GBM8401 glioblastoma cells (Fig. 4e). We also demonstrated that 3-MA decreased LC3 puncta numbers and reversed prodigiosin-induced autophagic cell death in prodigiosin-treated GBM8401 glioblastoma cells (Fig. 6). Collectively, these findings indicate that prodigiosin collaboratively causes autophagic cell death and apoptosis in glioblastoma cells. Consistent with our results, several small compounds, such as dehydroepiandrosterone [64] and polyphyllin VII [65], have been reported to induce autophagic cell death and apoptosis simultaneously. However, the autophagic cell death mechanism requires further investigation to identify specific makers for differentiating the mechanisms of apoptosis.
p53 is an important tumour suppressor gene involved in many central cellular processes. Approximately 30–50% of patients with primary high-grade glioma exhibit mutant p53 [66]. Mutant p53 may not only lack tumour suppression activity but also increase oncogenic characteristics such as increased invasion, migration, proliferation, anoikis, colony formation, genomic instability, propagation, and chemoresistance [67, 68]. Importantly, mutant p53 has been reported to be highly related to the poor prognosis of glioblastoma patients undergoing TMZ treatment, reducing their overall survival [69]. In addition, it inhibits autophagy by reducing the adenosine monophosphate-activated protein kinase pathway, stimulating the AKT/mTOR pathway and the antiautophagic hypoxia inducible factor-1 protein [70]. We found that 48-h treatment with relatively low concentrations (0.3125–1.25 µM) of prodigiosin led to a higher cytotoxicity rate in p53 mutant GBM8401 than in p53 wild-type U87MG. Prodigiosin has been found to upregulate the proapoptotic protein p73 and disrupts the interaction between mutant p53 in p53 double-mutant SW480 colorectal carcinoma cells [71]. Our results indicated that the inhibition rate of prodigiosin (5 µM) for the AKT/mTOR pathway is faster in p53 mutant GBM8401 cells than in p53 wild-type U87MG cells (Fig. 5). Therefore, we infer that prodigiosin induces cell death in mutant-p53 glioblastoma to a higher extent.
In summary, this study is the first to report that prodigiosin is located in the ER and induces ER stress, apoptosis and autophagic cell death in human glioblastoma cells (Fig. 7). Our data indicate that prodigiosin induces autophagic cell death through ER stress, activating the JNK pathway and excessive levels of autophagy and simultaneously decreasing the AKT/mTOR signaling pathway, causing apoptosis. The autophagy inhibitor 3-MA can reverse prodigiosin-induced cell death. Our results also demonstrate that prodigiosin inhibits the number of neurospheres formed from glioblastoma cells. Altogether, these results indicate that prodigiosin is a probable cancer therapeutic agent for both apoptosis and autophagic cell death and is thus an attractive candidate for cancer therapy. In conclusion, autophagic cell death and apoptosis are simultaneously induced by prodigiosin.
References
Darshan N, Manonmani H (2015) Prodigiosin and its potential applications. J Food Sci Technol 52(9):5393–5407
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23(1–3):3–25. https://doi.org/10.1016/S0169-409X(96)00423-1
Campas C, Dalmau M, Montaner B, Barragan M, Bellosillo B, Colomer D, Pons G, Pérez-Tomás R, Gil J (2003) Prodigiosin induces apoptosis of B and T cells from B-cell chronic lymphocytic leukemia. Leukemia 17(4):746–750. https://doi.org/10.1038/sj.leu.2402860
Espona-Fiedler M, Soto-Cerrato V, Hosseini A, Lizcano J, Guallar V, Quesada R, Gao T, Pérez-Tomás R (2012) Identification of dual mTORC1 and mTORC2 inhibitors in melanoma cells: prodigiosin vs. obatoclax. Biochem Pharmacol 83(4):489–496. https://doi.org/10.1016/j.bcp.2011.11.027
Montaner B, Navarro S, Piqué M, Vilaseca M, Martinell M, Giralt E, Gil J, Pérez-Tomás R (2000) Prodigiosin from the supernatant of Serratia marcescens induces apoptosis in haematopoietic cancer cell lines. Br J Pharmacol 131(3):585–593. https://doi.org/10.1038/sj.bjp.0703614
Wang Z, Li B, Zhou L, Yu S, Su Z, Song J, Sun Q, Sha O, Wang X, Jiang W (2016) Prodigiosin inhibits Wnt/β-catenin signaling and exerts anticancer activity in breast cancer cells. Proc Natl Acad Sci 113(46):13150–13155. https://doi.org/10.1073/pnas.1616336113
Prabhu VV, Hong B, Allen JE, Zhang S, Lulla AR, Dicker DT, El-Deiry WS (2016) Small-molecule prodigiosin restores p53 tumor suppressor activity in chemoresistant colorectal cancer stem cells via c-Jun-mediated ∆Np73 inhibition and p73 activation. Cancer Res 76(7):1989–1999. https://doi.org/10.1158/0008-5472.CAN-14-2430
Tomás RP, Ruir CD, Montaner B (2001) Prodigiosin induces cell death and morphological changes indicative of apoptosis in gastric cancer cell line HGT-1. Histol Histopathol 16(2):415–421. https://doi.org/10.1038/sj.cdd.4400987
Ramirez YP, Weatherbee JL, Wheelhouse RT, Ross AH (2013) Glioblastoma multiforme therapy and mechanisms of resistance. Pharmaceuticals 6(12):1475–1506. https://doi.org/10.3390/ph6121475
Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466. https://doi.org/10.1016/S1470-2045(09)70025-7
Oliva CR, Nozell SE, Diers A, McClugage SG, Sarkaria JN, Markert JM, Darley-Usmar VM, Bailey SM, Gillespie GY, Landar A (2010) Acquisition of temozolomide chemoresistance in gliomas leads to remodeling of mitochondrial electron transport chain. J Biol Chem 285(51):39759–39767. https://doi.org/10.1074/jbc.M110.147504
Hatanpaa KJ, Burma S, Zhao D, Habib AA (2010) Epidermal growth factor receptor in glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 12(9):675–684. https://doi.org/10.1593/neo.10688
Yang Y, Shao N, Luo G, Li L, Zheng L, Nilsson-Ehle P, Xu N (2010) Mutations of PTEN gene in gliomas correlate to tumor differentiation and short-term survival rate. Anticancer Res 30(3):981–985. https://doi.org/10.1038/sj.onc.1201756
Taylor E, Furnari TB, Cavenee FK W (2012) Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets 12(3):197–209. https://doi.org/10.2174/156800912799277557
Annovazzi L, Mellai M, Caldera V, Valente G, Tessitore L, Schiffer D (2009) mTOR, S6 and AKT expression in relation to proliferation and apoptosis/autophagy in glioma. Anticancer Res 29(8):3087–3094. https://doi.org/10.18632/oncotarget.10995
Rahaman SO, Harbor PC, Chernova O, Barnett GH, Vogelbaum MA, Haque SJ (2002) Inhibition of constitutively active Stat3 suppresses proliferation and induces apoptosis in glioblastoma multiforme cells. Oncogene 21(55):8404–8413. https://doi.org/10.1038/sj.onc.1206047
Senft D, Ze’ev AR (2015) UPR, autophagy, and mitochondria crosstalk underlies the ER stress response. Trends Biochem Sci 40 (3):141–148. https://doi.org/10.1016/j.tibs.2015.01.002
Martinez-Outschoorn UE, Whitaker-Menezes D, Pavlides S, Chiavarina B, Bonuccelli G, Trimmer C, Tsirigos A, Migneco G, Witkiewicz AK, Balliet RM (2010) The autophagic tumor stroma model of cancer or “battery-operated tumor growth” a simple solution to the autophagy paradox. Cell cycle 9(21):4297–4306. https://doi.org/10.4161/cc.9.21.13817
Eisenberg-Lerner A, Bialik S, Simon H-U, Kimchi A (2009) Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 16(7):966–975. https://doi.org/10.1038/cdd.2009.33
Tsujimoto Y, Shimizu S (2005) Another way to die: autophagic programmed cell death. Cell Death Differ 12:1528–1534. https://doi.org/10.1038/sj.cdd.4401777
Sano R, Reed JC (2013) ER stress-induced cell death mechanisms. Biochim Biophys Acta 1833(12):3460–3470. https://doi.org/10.1038/cdd.2010.181
Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11(4):381–389. https://doi.org/10.1007/978-94-007-4351-9_15
Jang G-H, Lee M (2014) BH3-mimetic gossypol-induced autophagic cell death in mutant BRAF melanoma cells with high expression of p21 Cip1. Life Sci 102(1):41–48. https://doi.org/10.1016/j.lfs.2014.02.036
Li J-R, Cheng C-L, Yang W-J, Yang C-R, Ou Y-C, Wu M-J, Ko J-L (2014) FIP-gts potentiate autophagic cell death against cisplatin-resistant urothelial cancer cells. Anticancer Res 34(6):2973–2983. https://doi.org/10.3109/07357907.2011.629379
Sui X, Chen R, Wang Z, Huang Z, Kong N, Zhang M, Han W, Lou F, Yang J, Zhang Q (2013) Autophagy and chemotherapy resistance: a promising therapeutic target for cancer treatment. Cell Death Dis 4(10):e838. https://doi.org/10.1038/cddis.2013.350
Lee WH, Yeh MY, Tu YC, Han SH, Wang YC (1988) Establishment and characterization of a malignant glioma cell line, GBM8401/TSGH, NDMC. J Surg Oncol 38(3):173–181. https://doi.org/10.1002/jso.2930380309
Hong X, Chedid K, Kalkanis SN (2012) Glioblastoma cell line-derived spheres in serum‑containing medium versus serum-free medium: a comparison of cancer stem cell properties. Int J Oncol 41(5):1693–1700. https://doi.org/10.3892/ijo.2012.1592
Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3(5):452–460. https://doi.org/10.4161/auto.4451
Panosyan EH, Laks DR, Masterman-Smith M, Mottahedeh J, Yong WH, Cloughesy TF, Lazareff JA, Mischel PS, Moore TB, Kornblum HI (2010) Clinical outcome in pediatric glial and embryonal brain tumors correlates with in vitro multi-passageable neurosphere formation. Pediatr Blood Cancer 55(4):644–651. https://doi.org/10.1002/pbc.22627
Müller-Taubenberger A, Lupas AN, Li H, Ecke M, Simmeth E, Gerisch G (2001) Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J 20(23):6772–6782
Dunn KW, Kamocka MM, McDonald JH (2011) A practical guide to evaluating colocalization in biological microscopy. Am J Physiol-Cell Physiol 300(4):C723–C742
Evans JD (1996) Straightforward statistics for the behavioral sciences. Brooks/Cole, Pacific Grove
van Schadewijk A, van’t Wout EF, Stolk J, Hiemstra PS (2012) A quantitative method for detection of spliced X-box binding protein-1 (XBP1) mRNA as a measure of endoplasmic reticulum (ER) stress. Cell Stress Chaperones 17(2):275–279. https://doi.org/10.1007/s12192-011-0306-2
Li J, Ni M, Lee B, Barron E, Hinton D, Lee A (2008) The unfolded protein response regulator GRP78/BiP is required for endoplasmic reticulum integrity and stress-induced autophagy in mammalian cells. Cell Death Differ 15(9):1460–1471. https://doi.org/10.1038/cdd.2008.81
Liu K, Shi Y, Guo X, Wang S, Ouyang Y, Hao M, Liu D, Qiao L, Li N, Zheng J (2014) CHOP mediates ASPP2-induced autophagic apoptosis in hepatoma cells by releasing Beclin-1 from Bcl-2 and inducing nuclear translocation of Bcl-2. Cell Death Dis 5(7):e1323. https://doi.org/10.1038/cddis.2014.276
Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19(21):5720–5728. https://doi.org/10.1093/emboj/19.21.5720
Shimizu S, Konishi A, Nishida Y, Mizuta T, Nishina H, Yamamoto A, Tsujimoto Y (2010) Involvement of JNK in the regulation of autophagic cell death. Oncogene 29(14):2070–2082
Eberhart K, Oral O, Gozuacik D (2013) Induction of autophagic cell death by anticancer agents. Autophagy. https://doi.org/10.1016/B978-0-12-405530-8.00013-3
Nihira K, Miki Y, Ono K, Suzuki T, Sasano H (2014) An inhibition of p62/SQSTM1 caused autophagic cell death of several human carcinoma cells. Cancer Sci 105(5):568–575. https://doi.org/10.1111/cas.12396
Karch J, Schips TG, Maliken BD, Brody MJ, Sargent MA, Kanisciak O, Molkentin JD (2017) Autophagic cell death is dependent on lysosomal membrane permeability through Bax and Bak. eLife. https://doi.org/10.7554/eLife.30543
Liu Y, Levine B (2015) Autosis and autophagic cell death: the dark side of autophagy. Cell Death Differ 22(3):367–376. https://doi.org/10.1038/cdd.2014.143
Lichstein HC, Van De Sand VF (1946) The antibiotic activity of violacein, prodigiosin, and phthiocol. J Bacteriol 52(1):145. https://doi.org/10.3181/00379727-72-17533
Hosseini A, Espona-Fiedler M, Soto-Cerrato V, Quesada R, Pérez-Tomás R, Guallar V (2013) Molecular interactions of prodiginines with the BH3 domain of anti-apoptotic Bcl-2 family members. PLoS ONE 8(2):e57562. https://doi.org/10.1371/journal.pone.0057562
Zhang J, Shen Y, Liu J, Wei D (2005) Antimetastatic effect of prodigiosin through inhibition of tumor invasion. Biochem Pharmacol 69(3):407–414. https://doi.org/10.1016/j.bcp.2004.08.037
Soto-Cerrato V, Viñals F, Lambert JR, Pérez-Tomás R (2007) The anticancer agent prodigiosin induces p21 WAF1/CIP1 expression via transforming growth factor-beta receptor pathway. Biochem Pharmacol 74(9):1340–1349. https://doi.org/10.1016/j.bcp.2007.07.016
Alison MR, Lim SM, Nicholson LJ (2011) Cancer stem cells: problems for therapy? J Pathol 223(2):148–162. https://doi.org/10.1634/stemcells.2006-0136
Chen Y-C, Ingram PN, Fouladdel S, McDermott SP, Azizi E, Wicha MS, Yoon E (2016) High-throughput single-cell derived sphere formation for cancer stem-like cell identification and analysis. Sci Rep. https://doi.org/10.1038/srep27301
Ojha R, Bhattacharyya S, Singh SK (2015) Autophagy in cancer stem cells: a potential link between chemoresistance, recurrence, and metastasis. BioRes Open Access 4(1):97–108. https://doi.org/10.1089/biores.2014.0035
Jiang H, Gomez-Manzano C, Aoki H, Alonso MM, Kondo S, McCormick F, Xu J, Kondo Y, Bekele BN, Colman H (2007) Examination of the therapeutic potential of Delta-24-RGD in brain tumor stem cells: role of autophagic cell death. J Natl Cancer Inst 99(18):1410–1414. https://doi.org/10.1093/jnci/djm102
Kanzawa T, Germano I, Komata T, Ito H, Kondo Y, Kondo S (2004) Role of autophagy in temozolomide-induced cytotoxicity for malignant glioma cells. Cell Death Differ 11(4):448–457. https://doi.org/10.1038/sj.cdd.4401359
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333(1–2):169–174. https://doi.org/10.1016/0014-5793(93)80398-E
Bernales S, Schuck S, Walter P (2007) ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 3(3):285–287. https://doi.org/10.4161/auto.3930
Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gélinas C, Fan Y (2006) Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell 10(1):51–64. https://doi.org/10.1016/j.ccr.2006.06.001
McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N, Aldape K (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068. https://doi.org/10.1038/nature07385
Carico C, Nuño M, Mukherjee D, Elramsisy A, Dantis J, Hu J, Rudnick J, John SY, Black KL, Bannykh SI (2012) Loss of PTEN is not associated with poor survival in newly diagnosed glioblastoma patients of the temozolomide era. PLoS ONE 7(3):e33684. https://doi.org/10.1371/journal.pone.0033684
Gozuacik D, Kimchi A (2004) Autophagy as a cell death and tumor suppressor mechanism. Oncogene 23(16):2891–2906. https://doi.org/10.1038/sj.onc.1207521
Fantus D, Rogers NM, Grahammer F, Huber TB, Thomson AW (2016) Roles of mTOR complexes in the kidney: implications for renal disease and transplantation. Nat Rev Nephrol. https://doi.org/10.1038/nrneph.2016.108
Gozuacik D, Kimchi A (2007) Autophagy and cell death. Curr Top Dev Biol 78:217–245. https://doi.org/10.4161/auto.20669
Schweichel JU, Merker HJ (1973) The morphology of various types of cell death in prenatal tissues. Teratology 7(3):253–266. https://doi.org/10.1002/tera.1420070306
Eberhart K, Oral O, Gozuacik D (2013) Autophagy: Chap. 13. Induction of autophagic cell death by anticancer agents. Elsevier Inc. Chapters, New York
Ogata M, Hino S-i, Saito A, Morikawa K, Kondo S, Kanemoto S, Murakami T, Taniguchi M, Tanii I, Yoshinaga K (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26(24):9220–9231. https://doi.org/10.1128/MCB.01453-06
Leppä S, Bohmann D (1999) Diverse functions of JNK signaling and c-Jun in stress response and apoptosis. Oncogene. https://doi.org/10.1038/sj.onc.1203173
Wei Y, Pattingre S, Sinha S, Bassik M, Levine B (2008) JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Molecular Cell 30(6):678–688. https://doi.org/10.4161/auto.20586
Vegliante R, Desideri E, Di Leo L, Ciriolo MR (2016) Dehydroepiandrosterone triggers autophagic cell death in human hepatoma cell line HepG2 via JNK-mediated p62/SQSTM1 expression. Carcinogenesis. https://doi.org/10.1093/carcin/bgw003
Zhang C, Jia X, Wang K, Bao J, Li P, Chen M, Wan J-B, Su H, Mei Z, He C (2016) Polyphyllin VII induces an autophagic cell death by activation of the JNK pathway and inhibition of PI3K/AKT/mTOR pathway in HepG2 cells. PLoS ONE 11(1):e0147405. https://doi.org/10.1371/journal.pone.0147405
Louis DN, Von Deimling A, Chung RY, Rubio M-P, Whaley JM, Eibl RH, Ohgaki H, Wiestler OD, Thor AD, Seizinger BR (1993) Comparative study of p53 gene and protein alterations in human astrocytic tumors. J Neuropathol Exp Neurol 52(1):31–38. https://doi.org/10.1097/00005072-199301000-00005
Strano S, Dell’Orso S, Di Agostino S, Fontemaggi G, Sacchi A, Blandino G (2007) Mutant p53: an oncogenic transcription factor. Oncogene 26(15):2212–2219. https://doi.org/10.1038/sj.onc.1210296
Muller PA, Vousden KH (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25(3):304–317. https://doi.org/10.1016/j.ccr.2014.01.021
Wang X, Chen J-x, Liu J-p, You C, Liu Y-h, Mao Q (2014) Gain of function of mutant TP53 in glioblastoma: prognosis and response to temozolomide. Ann Surg Oncol 21(4):1337–1344. https://doi.org/10.1245/s10434-013-3380-0
Cordani M, Butera G, Pacchiana R, Donadelli M (2016) Molecular interplay between mutant p53 proteins and autophagy in cancer cells. Biochim Biophys Acta. https://doi.org/10.1016/j.bbcan.2016.11.003
Hong B, Prabhu VV, Zhang S, van den Heuvel APJ, Dicker DT, Kopelovich L, El-Deiry WS (2014) Prodigiosin rescues deficient p53 signaling and antitumor effects via upregulating p73 and disrupting its interaction with mutant p53. Cancer Res 74(4):1153–1165. https://doi.org/10.1158/0008-5472.CAN-13-0955
Acknowledgements
This research was supported by Ministry of Science and Technology, Taiwan (105-2320-B-182A-015), National Research Program for Biopharmaceuticals, Taiwan (105-2325-B-110-001), and Chang Gung Memorial Hospital, Taiwan (CMRPG8D0821).
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, SY., Chen, NF., Kuo, HM. et al. Prodigiosin stimulates endoplasmic reticulum stress and induces autophagic cell death in glioblastoma cells. Apoptosis 23, 314–328 (2018). https://doi.org/10.1007/s10495-018-1456-9
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-018-1456-9