Abstract
In this study, we demonstrated that survivin downregulation with TRAIL expression greatly enhanced the cytotoxic death of pancreatic cancer cells after gemcitabine treatment. Using real-time RT-PCR, we analyzed five survivin shRNAs to identify the best target sequence for suppression of human survivin, with the goal of treating gemcitabine-resistant pancreatic cancer cells. Survivin shRNA 5, corresponding to target 5, showed the greatest reduction in survivin mRNA levels. Furthermore, combined treatment with survivin shRNA-expressing adenovirus with gemcitabine plus TRAIL decreased uncleaved PARP and increased consequent PARP cleavage, which was correlated with the greatest levels of survivin downregulation and cell death. These results indicate that survivin functions as a common mediator of gemcitabine- and TRAIL-induced cell death. Using a nude mouse model implanted with MiaPaCa-2 pancreatic cancer cells, we observed tumor regression induced by an oncolytic adenovirus expressing survivin shRNA and TRAIL plus gemcitabine. Together, our findings provide a strong rationale for treating pancreatic cancer patients with both gemcitabine and oncolytic adenovirus armed with survivin shRNA and TRAIL.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gemcitabine has been used as an anticancer drug to treat advanced pancreatic cancer patients. Although it has proven effective as a standard chemotherapy agent, the response rate remains at 5.4 %, and the 5-year survival rate is extremely poor due to the development of intrinsic or acquired resistance of pancreatic cancer to gemcitabine [1–4]. Currently, agents that either enhance the effects of gemcitabine or overcome chemoresistance to the drug are urgently needed to treat pancreatic cancer. Recently, nab-paclitaxel plus gemcitabine was reported to significantly improve overall survival, progression-free survival, and response rates in patients with metastatic pancreatic cancer [5, 6]. Nevertheless, overall prognosis remains poor for pancreatic cancer patients, which motivates us to provide more effective therapeutic modalities. In the present study, we show that survivin downregulation, with the help of TRAIL, counteracts gemcitabine-induced acquired resistance by enhancing cancer-cell death.
Survivin, a structurally-unique IAP (inhibitor of apoptosis protein)-family protein, contains a single baculovirus IAP repeat (BIR) domain and lacks a carboxy-terminal RING finger motif [7]. Survivin is a bifunctional protein that regulates cell division and suppresses apoptosis without direct suppression of caspase activity [8, 9]. Most importantly, survivin is expressed in the majority of cancers and developing fetal tissue, though it is rarely expressed in normal healthy adult tissue [7, 8, 10–12], making it a potential target for anticancer therapy [13, 14]. To date, there are many reports using antisense oligonucleotides, ribozymes, or siRNA to attenuate survivin expression and inhibit tumor-cell growth in vitro and in vivo [8, 12, 15–20]. For therapeutic applications, adenoviral vectors are efficient gene delivery systems for delivering siRNA. Oncolytic adenovirus, which originated from E1B-55 kDa-deleted adenovirus, has been used to mediate survivin knockdown in colon cancer [20]. However, use of this adenovirus for pancreatic cancer cells has not been reported. Moreover, survivin inhibition by gemcitabine treatment is not sufficient for pancreatic cancer-cell death, as less than 70 % cytotoxicity is observed using survivin siRNA plus gemcitabine in both PANC-1 and BxPC3 cells [12]. On the other hand, less than 40 % cytotoxicity is observed when survivin is downregulated in YM155 in MiaPaCa-2 cells [13]. Agents that either enhance the effects of gemcitabine or overcome chemoresistance to the drug are needed for pancreatic cancer treatment [2, 21].
Meanwhile, TRAIL has been identified as a tumor-selective, apoptosis-inducing cytokine with minimum toxicity to normal cells [22]; however, cancer cells frequently develop resistance to TRAIL by overexpressing either FLIP, Bcl2/Bcl-xL, or survivin [23–27]. Intriguingly, TRAIL also causes G2/M arrest and an increase of survivin levels in HepG2 as an acquired resistance mechanism, whereas treatment with a survivin antisense oligonucleotide causes S-phase arrest and significantly enhances TRAIL-induced apoptosis [28]. Previously, TRAIL was shown to induce apoptotic cell death in pancreatic cancer cells, and gemcitabine synergistically increased the antitumor effect of TRAIL [29]. Although the synergistic antitumor effect of TRAIL-expressing oncolytic adenovirus with gemcitabine was recently reported in bladder cancer cells [30], the effect has not been established for pancreatic cancer cells. As indicated, in spite of much of data published regarding survivin knockdown plus gemcitabine or survivin knockdown plus TRAIL [13, 31, 32], the triple combination of survivin knockdown with gemcitabine and TRAIL as well as oncolytic adenovirus was attempted for the first time in pancreatic cancer cells. In this study, oncolytic adenovirus treatment inducing both survivin silencing and TRAIL expression significantly increased the anti-tumorigenic activity in gemcitabine-resistant pancreatic cancer cells, which indicates greater clinical feasibility in pancreatic cancer patients.
Material and methods
Cell culture
The cancer-cell lines MiaPaCa-2, HPAC (human pancreatic cancer-cell lines), and Panc02 (murine pancreatic cancer-cell line), in addition to 293A, a subclone of the human embryonic kidney 293 cell (Invitrogen, Carsbad, CA, USA), were cultured in Dulbecco’s modified Eagle’s medium with 10 % fetal bovine serum (HyClone, Logan, UT, USA). Human normal pancreatic cells (Cat. No. T0199, Applied Biological Materials, Richmond, BC, Canada) was cultured in Prigrow II medium (Cat. No. TM002, Applied Biological Materials) with 10 % fetal bovine serum. All cells were maintained at 37 °C in a humidified atmosphere containing 5 % CO2.
Reagents
Antibody to PARP was purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies for survivin, TRAIL, and actin were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The antibody for hexon was purchased from Abcam (Cambridge, MA, USA). For immunohistochemistry, TRAIL was detected using a different antibody made from BD Biosciences (Franklin Lakes, NJ, USA). All other chemicals were purchased from Sigma-Aldrich (St Louis, MO, USA).
Construction of survivin shRNA
To construct human survivin shRNA, we screened five candidate sequences. Target selection was performed using an algorithm developed by Genolution Pharmaceuticals Inc. (Seoul, South Korea). We screened five candidate sequences after transfection of survivin shRNA into HeLa and validated them after 24 h of transfection using real-time RT-PCR. The forward primer for the real-time RT-PCR of survivin was 5′-CCTGGCAGCCCTTTCTCAA-3′, and the reverse primer was 5′- AGCAGAAGAAACACTGGGCC-3′. The selected target sequence was 5′-GGCCCCTTAGCAATGTCTTAGGAAA-3′, and the loop sequence was 5′-TCTC-3′. To express human survivin shRNA in the adenovirus, the top strand sequence (5′-GATCCGGCCCCTTAGCAATGTCTTAGGAAATCTCTTTCCTAAGACATTGCTAAGGGGCCTTTTA-3′) and the bottom strand sequence (5′-AGCTTAAAA GGCCCCTTAGCAATGTCTTAGGAAAGAGATTTCCTAAGACATTGCTAAGGGGCG-3′) were annealed and subcloned into the pSP72ΔE3-U6, an E3 shuttle vector, and then digested with BamH I and Hind III. The resulting adenoviral shuttle vector, pSP72ΔE3-U6-shsurvivin, was linearized by Xmn I digestion. The adenoviral vector dl324-IX was linearized by Spe I digestion, and the two linearized vectors were co-transformed into E. coli BJ5183 cells in order to undergo homologous recombination. The recombined adenoviral plasmids, dl324-IX-ΔE3-U6-NC (Ad-NC) and dl324-IX-ΔE3-U6-shsurvivin (Ad-shSurvivin), were then digested with Pac I and transfected into HEK-293 cells to generate the replication-incompetent adenovirus. The infectious titer of the adenovirus was determined by a limiting dilution assay in 293A cells.
Generation of oncolytic adenovirus expressing TRAIL and survivin shRNA
First, the adenoviral vector dl324-BstB I-ΔE3-U6-shSurvivin was constructed through the homologous recombination of dl324-BstB I and pSP72ΔE3-U6-shSurvivin. Then, the resulting plasmid, dl324-BstB I-ΔE3-U6-shSurvivin, was linearized with Bsp1191, while pCA14-3484-CMV-ΔE1B55-TRAIL was linearized with Drd I for a second round of homologous recombination in E. coli BJ5183 cells. To construct the shuttle vector for the oncolytic adenoviral plasmid expressing TRAIL, pEGFP-TRAIL (Addgene #10953) was cleaved by EcoR I and Sma I, and the resulting full-length TRAIL gene was subcloned into pCA14, which was digested with Hind III and blunted before EcoR I digestion (pCA14-TRAIL). Then, the TRAIL gene from pCA14-TRAIL was inserted into pCA14-3484-CMV-ΔE1B55KDa, which was previously described in detail [33]. Briefly, PCR was performed using a sense primer flanked at the 5′ end with an MfeI site (5′-CCGCAATTGCTAATTCCCTGGCATTATGCCC-3′) and an antisense primer flanked at the 5′ end with a Bgl II site (5′-GGAAGATCTTCGATGCTAGACG-3′). The resulting TRAIL PCR product was digested with Mfe I and Bgl II, subcloned into pCA14-3484-CMV- ΔE1B55KDa, and digested with EcoRI and Bgl II (pCA14-3484-CMV-ΔE1B55KDa-TRAIL). Finally, homologous recombination of dl324-BstB I-ΔE3-U6-shSurvivin, the adenoviral vector, pCA14-3484-CMV-ΔE1B55KDa-TRAIL, and the shuttle vector was performed to produce dl324-3484-CMV-ΔE1B55KDa-TRAIL-ΔE3-U6-shsurvivin. For the production of survivin shRNA-expressing oncolytic adenovirus and scrambled shRNA (negative control [NC])-expressing oncolytic adenovirus, dl324-BstB I-ΔE3-U6-shSurvivin or dl324-BstB I-ΔE3-U6-shNC was recombined with pCA14-3484-CMV-ΔE1B55. All viruses were amplified for purification according to standard methods. Titration was performed in 293A cells using a limiting dilution assay to estimate the number of infectious viral particles produced.
MTS viability assay
The CellTiter 96® Aqueous Assay kit (Promega, Madison, WI, USA) was composed of solutions containing a novel tetrazolium compound (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt; MTS) and an electron coupling reagent (phenazine ethosulfate; PES). MTS was bioreduced by cells into a formazan product that was soluble in tissue culture media. MiaPaCa-2 and HPAC cells were treated in a 6-well format with either gemcitabine treatment (10 μM) for 48 h or adenoviral infection (dl324-IX-ΔE3-U6-shNC or dl324-IX-ΔE3-U6-shsurvivin) at an MOI of 50 for 1 day, followed either by gemcitabine treatment (10 μM) for 48 h or TRAIL treatment (200 ng/ml) for 4 h. After treatment, 50 μl of supernatant from each well was transferred into a new well of a 96-well flat-bottom plate. Absorbance of formazan at 490 nm was measured directly from the 96-well assay plates without additional processing. Formazan concentration was directly proportional to the number of living cells in each culture.
Production of recombinant TRAIL
A human TRAIL cDNA fragment (amino acids 114–281) was cloned into pET-23d [34] (Novagen, Madison, WI). His-tagged TRAIL protein was expressed and purified using the Qiaexpress protein purification system (Qiagen, Valencia, CA).
Protein extracts and polyacrylamide gel electrophoresis
Cells were lysed with 1× Laemmli lysis buffer (62.5 mM Tris, pH 6.8, 2 % sodium dodecyl sulfate (SDS), 10 % glycerol, 0.002 % bromophenol blue) and boiled for 10 min. Protein concentration was measured using BCA Protein Assay Reagent (Pierce, Rockford, IL, USA). Samples were diluted with 1× lysis buffer, and β-mercaptoethanol was added to a final concentration of 350 mM. Equivalent amounts of protein were loaded into each well of 10 % SDS polyacrylamide gels. SDS–polyacrylamide gel electrophoresis (PAGE) was performed using a Hoefer gel apparatus.
Immunoblot analysis
After SDS-PAGE, proteins were electrophoretically transferred to nitrocellulose membranes. Each nitrocellulose membrane was blocked with 5 % nonfat dry milk in PBS-Tween-20 (0.1 %, v/v) at room temperature for 1 h. The membrane was then incubated with primary antibody (diluted according to the manufacturer’s instructions) for 2 h. Horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG was used as a secondary antibody. Immunoreactive proteins were visualized with a chemiluminescent detection kit (ELPIS biotech, Taejon, Korea). For imaging, a ChemiDoc system (Syngene, Frederick, MD, USA) was used.
FACS analysis
An FITC Annexin V Apoptosis Detection Kit II was purchased from BD Pharmingen™ (BD Biosciences, Franklin Lakes, NJ, USA) to conduct Annexin V-PI staining. After (1) adenoviral infection (dl324-IX-ΔE3-U6-shNC or dl324-IX-ΔE3-U6-shSurvivin) at an MOI of 50 or TRAIL treatment (200 ng/ml) for 4 h, (2) adenoviral infection (dl324-IX-ΔE3-U6-shNC or dl324-IX-ΔE3-U6-shSurvivin) at an MOI of 50 for 1 day followed by gemcitabine treatment (10 μM) for an additional 1 day, or (3) adenoviral infection (dl324-IX-ΔE3-U6-shSurvivin) at an MOI of 50 for 1 day followed by gemcitabine treatment (10 μM) for an additional 1 day and TRAIL treatment (200 ng/ml) for 4 h, trypsinized MiaPaCa-2 or HPAC cells were washed with cold PBS and then resuspended in 1× Binding Buffer at a concentration of 1 × 106 cells/ml. After transferring100 μl of the solution (1 × 105 cells) to a 5-ml culture tube with 5 μl FITC Annexin V and 5 μl PI, gently vortexed cells were incubated for 15 min at room temperature in the dark. Then, cells were analyzed by flow cytometry after adding 400 μl of 1× Binding Buffer to each tube.
Animal studies
The animal protocol (2013-0357) used in this study was reviewed and approved by the Institutional Animal Care and Use Committee in Yonsei University Health System. To generate a xenograft tumor model, 8 × 106 MiaPaCa-2 tumor cells were injected into the subcutaneous abdominal region of male BALB/c athymic nude mice. When the tumors reached an average size of 60–80 mm3, the nude mice received intratumoral injections of 1 × 109 plaque-forming units (pfu) of one of three oncolytic adenoviruses diluted in 50 μl PBS or PBS alone. The oncolytic adenoviruses used were oncolytic control adenovirus (Ad-3484-shNC), survivin shRNA-expressing oncolytic adenovirus (Ad-3484-shSurvivin), and survivin-shRNA- and TRAIL-expressing oncolytic adenovirus (Ad-3484-TRAIL-shSurvivin). Intratumoral injection was repeated every other day for a total of three injections. gemcitabine (10 mg/kg) was co-injected intratumorally on the last day of viral injection (Ad-3484-shNC, Ad-3484-shSurvivin, Ad-3484-TRAIL-shSurvivin). Intratumoral injection of gemcitabine was done rather than intravenous injection for the purpose of proof of the concept. Tumor growth was measured using a caliper every 2 days for 29 days, and tumor volume (V) was calculated using the following formula: V (mm3) = 0.52 × length (mm) × width (mm)2.
Immunohistochemistry
After 7 days of subcutaneous injection of MiaPaCa-2 cells (8 × 106) into the abdominal region of male nude mice, one of three oncolytic adenoviruses (Ad-3484-shNC, Ad-3484-shSurvivin, or Ad-3484-TRAIL-shSurvivin, 1 × 109 pfu/50 μl) was infected intratumorally every other day for a total of three injections, and gemcitabine (10 mg/kg) was intratumorally co-injected on the last day of viral injection (Ad-3484-shSurvivin, Ad-3484-TRAIL-shSurvivin). Seven days after the last viral injection, tumor tissues were extracted, fixed for 24 h in 10 % formaldehyde, and paraffin embedded for immunohistochemical (IHC) staining. IHC was performed as described below. Tissue section slides were deparaffinized twice with xylene for 10 min each, and slides were then rehydrated using a graded alcohol series. After removing endogenous peroxidases using 0.1 % H2O2, slides were washed three times with PBS. Antigen retrieval was performed using 10 mM citrate buffer (pH 6.0; DAKO, Glostrup, Denmark) and a microwave oven. Tissues were permeabilized with 0.5 % PBX (0.5 % Triton X-100 in PBS) for 30 min. After blocking for 1 h with 5 % BSA, the primary antibody was added and incubated overnight at 4 °C. For detection of specific proteins, antibodies of hexon, TRAIL (human CD253), or survivin were used respectively. Additionally, both Primary Antibody Enhancer (Thermo Fisher Scientific, Waltham, MA, USA) and HRP Polymer (Thermo Scientific) were used for signal amplification. To develop the colored product, a mixture of DAB (3,3′-diaminobenzidine) Plus Chromogen and DAB Plus Substrate (Thermo Fisher Scientific) was added for 5 min. After washing with PBS, 20 % hematoxylin counterstain was added for 2–5 min to stain the nuclei. Finally, tissue slides were dehydrated in a graded alcohol series. After clearing twice in xylene, tissues slides were coverslipped with mounting media (xylene:mount = 1:1) for microscopy.
TUNEL assay
To measure in situ apoptosis, a terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) assay was performed using tumor tissue sections prepared as described for immunohistochemistry. The TUNEL assay was carried out according to the manufacturer’s instructions (Promega, Madison, WI, USA).
Statistical analyses
Data are presented as the mean ± standard error of mean (S.E.M.). Differences between groups were examined using unpaired two-tailed t tests. P values were calculated using GraphPad Prism version 6.0. P < 0.05 was considered statistically significant. All experiments were performed three times independently.
Results
Pancreatic cancer cells were resistant to gemcitabine
Similar to previous results of gemcitabine-induced resistance, over 40–60 % cell viability was maintained in pancreatic cancer-cell lines tested in the presence of gemcitabine, even up to 50 μM for 48 h, whereas Panc02 was sensitive to gemcitabine treatment with a broad range of concentrations (Fig. 1a, b).
Pancreatic cancer cell viability after gemcitabine treatment. a Cell viability was tested using MTS viability assays in three different pancreatic cancer cell lines after gemcitabine treatment (10 μM) for 48 h. Error bars represent the standard error from three independent experiments calculated using GraphPad Prism version 6.0. b After the treatment described in (a), morphological changes were examined by microscopic analysis
Gemcitabine increased survivin expression, and survivin shRNA-expressing adenovirus effectively downregulated survivin expression in pancreatic cancer cells
As previous studies indicate that survivin downregulation enhances gemcitabine chemosensitivity in pancreatic cancer [12, 13], we designed a survivin shRNA-expressing oncolytic adenovirus as a potential therapeutic agent for pancreatic cancer. First, we confirmed that survivin, a negative regulator of apoptosis, was upregulated by gemcitabine treatment in pancreatic cancer cells (Fig. 2a). Next, we used real-time RT-PCR to analyze five survivin shRNAs, thereby identifying the best target sequence for suppression of human survivin. We found that survivin shRNA 5 corresponding to target 5 showed the greatest reduction in survivin mRNA levels (over 91 %) when expressed in HeLa cells (Supplementary Fig. 1). Survivin levels were also specifically decreased by survivin shRNA-expressing adenovirus (Fig. 2b).
Induction of survivin after gemcitabine treatment and construction of adenovirus expressing-survivin shRNA or scrambled shRNA. a Two pancreatic cancer-cell lines (MiaPaCa-2, HPAC) were treated with gemcitabine (10 μM) for 48 h, and the remaining cells were examined for survivin levels by immunoblotting. b After two pancreatic cancer-cell lines (MiaPaCa-2, HPAC) were infected with Ad-NC or Ad-shSurvivin adenovirus at 50 MOI, cellular survivin levels were examined by immunoblotting. c Schematic of adenoviral vectors expressing either survivin shRNA or both TRAIL and survivin shRNA. dl324-ΔE1A-ΔE1B-ΔE3-IX-U6-NC (Ad-NC) adenovirus: a replication-incompetent adenovirus used as the negative control. It contains the scrambled DNA sequence for shRNA, which is under the control of the U6 promoter. Oncolytic-E1A-ΔE1B-IX-ΔE3-U6-NC adenovirus: a replication-competent adenovirus used as a control. It contains E1A controlled by the CMV promoter, although it lacks the E1B region, and the remaining structure is identical to the dl324-ΔE1A-ΔE1B-ΔE3-U6-NC adenovirus. dl324-ΔE1A-ΔE1B-ΔE3-IX-U6-shSurvivin (Ad-shSurvivin) is a replication-incompetent adenovirus expressing human survivin shRNA. Oncolytic-E1A-ΔE1B-IX-ΔE3-U6-shSurvivin is a replication-competent adenovirus expressing human survivin shRNA. dl324-∆E1AΔE1B-TRAIL-IX-ΔE3-U6-shSurvivin is a replication-incompetent adenovirus expressing TRAIL and human survivin shRNA. Oncolytic-E1A-ΔE1B-TRAIL-IX-ΔE3-U6-shSurvivin is a replication-competent adenovirus expressing TRAIL and human survivin shRNA. NC, negative control
A defective adenovirus expressing survivin shRNA 5 (diagrammed in Fig. 2c along with other constructs used in this study) was delivered to pancreatic cancer cells, and the cellular levels of survivin were determined.
Survivin shRNA-expressing adenovirus in combination with gemcitabine, TRAIL, or gemcitabine/TRAIL increased apoptotic/necrotic cell death
Next, we investigated the effect of survivin downregulation on cellular apoptosis in both MiaPaCa-2 and HPAC pancreatic cancer cells. However, combined treatment with survivin shRNA-expressing adenovirus and gemcitabine or gemcitabine plus TRAIL did not increased apoptotic cell death compared to single TRAIL treatment in both cancer-cell lines (Fig. 3a). To directly determine whether cell death was really increased by survivin downregulation with gemcitabine or TRAIL, an MTS viability test was performed. Indeed, survivin downregulation combined with gemcitabine or TRAIL more effectively decreased cell viability than gemcitabine with TRAIL alone. Furthermore, the greatest cell death occurred with survivin downregulation was combined with gemcitabine plus TRAIL (Fig. 3b). This phenomenon was also confirmed by annexin V-PI analysis to demonstrate the involvement of necrotic cell death. Briefly, survivin downregulation greatly induced apoptotic/necrotic cell death with a combination of gemcitabine or gemcitabine plus TRAIL (Fig. 4a, b). Taken together, these results also imply that survivin downregulation functions as a common mediator of gemcitabine and TRAIL-induced cell death.
Enhanced cell death and decreased cell viability of gemcitabine-treated pancreatic cancer cells with survivin downregulation and subsequent TRAIL treatment. a Two pancreatic cancer cell lines (MiaPaCa-2, HPAC) were either treated with gemcitabine (10 μM) for 48 h or infected with defective adenovirus expressing survivin shRNA or TRAIL treatment (200 ng/ml) for 4 h or survivin shRNA-expressed adenoviral infection followed by gemcitabine treatment (10 μM) for 48 h either with or without subsequent TRAIL treatment (200 ng/ml) for 4 h. After treatment, cell lysates were immunoblotted for PARP. b Cell viability was quantified using MTS viability assays in MiaPaCa-2 or HPAC cells after infection with adenovirus expressing survivin shRNA (50 MOI) or scrambled shRNA as a negative control (50 MOI), followed by either gemcitabine treatment (10 μM) for 48 h, TRAIL treatment (200 ng/ml) for 4 h, or treatment with both. Error bars represent the standard error from the three independent experiments. P values lower than 0.01 indicate a very significant difference of gemcitabine and/or TRAIL-combined Ad-shSurvivin compared to the control (Ad-shSurvivin) in all given conditions. P values were calculated using the program GraphPad Prism version 6.0. NC, negative control
Enhanced cell death and decreased cell viability of gemcitabine-treated pancreatic cancer cells with survivin downregulation and subsequent TRAIL treatment. FACS analysis by Annexin V/PI staining in MiaPaCa-2 (a) or HPAC cells (b) was performed for the determination of apoptosis after (1) adenoviral infection (Ad-NC or Ad-shSurvivin) at an MOI of 50 for 2 days or TRAIL treatment (200 ng/ml) for 4 h, (2) adenoviral infection (Ad-NC or Ad-shSurvivin) at an MOI of 50 for 1 day followed by gemcitabine treatment (10 μM) for an additional 1 day, or (3) adenoviral infection (Ad-shSurvivin) at an MOI of 50 for 1 day followed by gemcitabine treatment (10 μM) for an additional 1 day and TRAIL treatment (200 ng/ml) for 4 h
Oncolytic adenovirus expressing TRAIL and survivin shRNA increased tumor regression with gemcitabine in a mouse model
To examine the potential of a survivin-shRNA- and TRAIL-expressing oncolytic adenovirus as a new therapeutic in combination with gemcitabine, we treated nude mice with MiaPaCa-2 cell grafts. After confirming TRAIL expression and survivin downregulation following treatment with an oncolytic adenovirus expressing both survivin shRNA and TRAIL in vitro (Fig. 5a), and insignificant toxicity of various oncolytic adenoviruses in normal pancreatic cell (Fig. 5b), various oncolytic adenoviruses (expressing survivin shRNA, survivin shRNA and TRAIL, or negative control [NC]) were tested in nude mice with or without gemcitabine for suppression of tumor growth and compared to PBS or gemcitabine alone or shRNA survivin-expressed adenoviral infection only. As expected, tumor suppression was greatest when treated with oncolytic shSurvivin/TRAIL plus gemcitabine. Other treatments suppressed tumor growth in the following order: oncolytic shSurvivin plus gemcitabine and oncolytic shSurvivin plus TRAIL (Fig. 5c). But, most groups except of oncolytic shSurvivin/TRAIL plus gemcitabine were expired due to the tumor burden (Fig. 5d). Expression of adenovirus, survivin, and TRAIL in tumor tissue after infection followed by gemcitabine treatment was determined by immunostaining, and levels were as expected (Fig. 6a). Apoptotic including necrotic events were also increased by combined treatment with oncolytic adenovirus armed with survivin shRNA and TRAIL plus gemcitabine, oncolytic survivin shRNA and TRAIL, or oncolytic survivin shRNA plus gemcitabine compared to gemcitabine or oncolytic survivin shRNA only when using TUNEL assay (Fig. 6b, c).
Antitumor effect of oncolytic adenovirus in MiaPaCa-2 tumors grown in male athymic nude mice. a After infection of MiaPaCa-2 cells with oncolytic shSurvivin/TRAIL adenovirus, immunoblotting was performed on cell lysates using either a TRAIL antibody or a survivin antibody. TRAIL expression and survivin downregulation occurred upon infection with an oncolytic adenovirus armed with shSurvivin and TRAIL. b Cell viability was quantified using MTS viability assays in pancreatic primary cell after infection with adenovirus expressing survivin shRNA or survivin shRNA and TRAIL or scrambled shRNA as a negative control (50 MOI) (left, upper panel). Cell viability was also quantified using MTS viability assays in pancreatic primary cell (right, upper panel), MiaPaCa-2 (left, bottom panel) or HPAC (right, bottom panel) after infection with adenovirus expressing survivin shRNA and TRAIL followed by gemcitabine treatment (10 μM) for 48 h, or gemcitabine treatment only (10 μM) for 48 h. Error bars represent the standard error from the three independent experiments. c Tumors were established and various viral injections with or without gemcitabine were performed according to the Animal studies of “Material and methods”. Tumor growth was measured every 2 days for almost 30 days. Error bars represent the standard error from the five mice in each experimental group. The asterisk indicates a significant difference between the group of oncolytic adenovirus expressing survivin shRNA plus gemcitabine and oncolytic adenovirus expressing survivin shRNA and TRAIL plus gemcitabine (P < 0.05). d Kaplan–Meier survival curves was generated by the same experimental condition described in the Animal studies of “Material and methods”
Immunohistochemical analysis after oncolytic adenovirul infection expressing various combinations of survivin shRNA, TRAIL with or without gemcitabine or single gemcitabine treatment. Experimental conditions are described in detail in the “Material and methods” section. a After established tumors were subjected to infection with oncolytic adenovirus expressing survivin shRNA or survivin shRNA and TRAIL plus gemcitabine treatment, tumor tissue sections were prepared. b The TUNEL assay was performed on tissue sections to quantify apoptotic cell death as described in the “Material and methods” section. c The percentage of TUNEL-positive cells were determined by counting the TUNEL-positive cells under 10 non-continuous low-power fields (magnification ×100)
Discussion
In this study, we examined whether virotherapy with armed oncolytic adenovirus synergistically potentiated the antitumor effect of gemcitabine in gemcitabine-resistant pancreatic cancer cells. Survivin expression was downregulated, greatly increasing gemcitabine and TRAIL-induced cell death. As expected, gemcitabine alone, or in combination with survivin shRNA, was not sufficient to make a meaningful cytotoxicity in pancreatic cancer cells. Instead, combining TRAIL expression with survivin shRNA and gemcitabine effectively killed pancreatic cancer cells (Figs. 1, 3, 4, 5). However, the underlying mechanism of synergistic pancreatic cancer-cell death by gemcitabine and TRAIL is not yet fully understood, although DR5 induction by gemcitabine may be enough to implicate TRAIL involvement [35, 36]. Furthermore, core signaling molecules related to innate and acquired resistance induced by gemcitabine are also related to TRAIL-induced resistance (e.g., survivin, XIAP, and Bcl-xL) [25, 26, 37, 38]. Our present findings also suggest that survivin functions as a mediator of gemcitabine and TRAIL-induced acquired resistance, as downregulation of cellular survivin levels synergistically increased gemcitabine plus TRAIL-induced cell death in vitro and in vivo (Figs. 3, 4, 5, 6). While these results are promising, the xenograft nude mouse model used here does not accurately mimic pancreatic cancer. Thus, we are currently developing a syngenic pancreatic-cancer animal model to accurately evaluate the potential of an oncolytic adenovirus expressing both survivin shRNA and TRAIL in combination with gemcitabine treatment, as well as targeted delivery of oncolytic adenovirus [39–41]. In fact, we have already established a mouse melanoma model system for the oncolytic adenovirus [42].
Abbreviations
- RT-PCR:
-
Reverse-transcription polymerase chain reaction
- TRAIL:
-
Tumor necrosis factor-related apoptosis-inducing ligand
- MTS:
-
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
- FLIP:
-
FLICE-inhibitory protein
- IAP:
-
Inhibitor of apoptosis protein
- BIR:
-
Baculovirus IAP repeat
- FACS:
-
Fluorescence-activated cell sorter
- FITC:
-
Fluorescein isothiocyanate
- PI:
-
Propidium iodide
- XIAP:
-
X-linked inhibitor of apoptosis protein
- HEK-293:
-
Human embryonic kidney-293
- PARP:
-
Poly (ADP-ribose) polymerase
References
Kim MP, Gallick GE (2008) Gemcitabine resistance in pancreatic cancer: picking the key players. Clin Cancer Res: Off J Am Assoc Cancer Res 14:1284–1285
Hilbig A, Oettle H (2008) Gemcitabine in the treatment of metastatic pancreatic cancer. Expert Rev Anticancer Ther 8:511–523
Hidalgo M (2010) Pancreatic cancer. N Engl J Med 362:1605–1617
Long J, Zhang Y, Yu X, Yang J, LeBrun DG, Chen C et al (2011) Overcoming drug resistance in pancreatic cancer. Expert Opin Ther Targets 15:817–828
Goldstein D, El-Maraghi RH, Hammel P, Heinemann V, Kunzmann V, Sastre J et al (2015) nab-Paclitaxel plus gemcitabine for metastatic pancreatic cancer: long-term survival from a phase III trial. J Nat Cancer Inst 107:dju413
Lo Re G, Santeufemia DA, Foltran L, Bidoli E, Basso SM, Lumachi F (2015) Prognostic factors of survival in patients treated with nab-paclitaxel plus gemcitabine regimen for advanced or metastatic pancreatic cancer: a single institutional experience. Oncotarget 6:8255–8260
Ambrosini G, Adida C, Altieri DC (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 3:917–921
Uchida H, Tanaka T, Sasaki K, Kato K, Dehari H, Ito Y et al (2004) Adenovirus-mediated transfer of siRNA against survivin induced apoptosis and attenuated tumor cell growth in vitro and in vivo. Mol Ther: J Am Soc Gene Ther 10:162–171
Altieri DC (2008) New wirings in the survivin networks. Oncogene 27:6276–6284
Azuhata T, Scott D, Griffith TS, Miller M, Sandler AD (2006) Survivin inhibits apoptosis induced by TRAIL, and the ratio between survivin and TRAIL receptors is predictive of recurrent disease in neuroblastoma. J Pediatr Surg 41:1431–1440
Kanwar JR, Kamalapuram SK, Kanwar RK (2013) Survivin signaling in clinical oncology: a multifaceted dragon. Med Res Rev 33:765–789
Liu WS, Yan HJ, Qin RY, Tian R, Wang M, Jiang JX et al (2009) siRNA directed against survivin enhances pancreatic cancer cell gemcitabine chemosensitivity. Dig Dis Sci 54:89–96
Yoon DH, Shin JS, Jin DH, Hong SW, Jung KA, Kim SM et al (2012) The survivin suppressant YM155 potentiates chemosensitivity to gemcitabine in the human pancreatic cancer cell line MiaPaCa-2. Anticancer Res 32:1681–1688
Altieri DC (2008) Survivin, cancer networks and pathway-directed drug discovery. Nat Rev Cancer 8:61–70
Zhang R, Ma L, Zheng M, Ren J, Wang T, Meng Y et al (2010) Survivin knockdown by short hairpin RNA abrogates the growth of human hepatocellular carcinoma xenografts in nude mice. Cancer Gene Ther 17:275–288
Li QX, Zhao J, Liu JY, Jia LT, Huang HY, Xu YM et al (2006) Survivin stable knockdown by siRNA inhibits tumor cell growth and angiogenesis in breast and cervical cancers. Cancer Biol Ther 5:860–866
Ambrosini G, Adida C, Sirugo G, Altieri DC (1998) Induction of apoptosis and inhibition of cell proliferation by survivin gene targeting. J Biol Chem 273:11177–11182
Olie RA, Simoes-Wust AP, Baumann B, Leech SH, Fabbro D, Stahel RA et al (2000) A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res 60:2805–2809
Pennati M, Colella G, Folini M, Citti L, Daidone MG, Zaffaroni N (2002) Ribozyme-mediated attenuation of survivin expression sensitizes human melanoma cells to cisplatin-induced apoptosis. J Clin Investig 109:285–286
Shen W, Wang CY, Wang XH, Fu ZX (2009) Oncolytic adenovirus mediated Survivin knockdown by RNA interference suppresses human colorectal carcinoma growth in vitro and in vivo. J Exp Clin Cancer Res 28:81
Kunnumakkara AB, Guha S, Krishnan S, Diagaradjane P, Gelovani J, Aggarwal BB (2007) Curcumin potentiates antitumor activity of gemcitabine in an orthotopic model of pancreatic cancer through suppression of proliferation, angiogenesis, and inhibition of nuclear factor-kappaB-regulated gene products. Cancer Res 67:3853–3861
Wang S, El-Deiry WS (2003) TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 22:8628–8633
Kauh J, Fan S, Xia M, Yue P, Yang L, Khuri FR et al (2010) c-FLIP degradation mediates sensitization of pancreatic cancer cells to TRAIL-induced apoptosis by the histone deacetylase inhibitor LBH589. PLoS ONE 5:e10376
Fulda S, Meyer E, Debatin KM (2002) Inhibition of TRAIL-induced apoptosis by Bcl-2 overexpression. Oncogene 21:2283–2294
Zhu H, Guo W, Zhang L, Davis JJ, Wu S, Teraishi F et al (2005) Enhancing TRAIL-induced apoptosis by Bcl-X(L) siRNA. Cancer Biol Ther 4:393–397
Zhang L, Fang B (2005) Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther 12:228–237
Premkumar DR, Jane EP, Foster KA, Pollack IF (2013) Survivin inhibitor YM-155 sensitizes tumor necrosis factor- related apoptosis-inducing ligand-resistant glioma cells to apoptosis through Mcl-1 downregulation and by engaging the mitochondrial death pathway. J Pharmacol Exp Ther 346:201–210
He SQ, Rehman H, Gong MG, Zhao YZ, Huang ZY, Li CH et al (2007) Inhibiting survivin expression enhances TRAIL-induced tumoricidal activity in human hepatocellular carcinoma via cell cycle arrest. Cancer Biol Ther 6:1247–1257
Xu ZW, Kleeff J, Friess H, Buchler MW, Solioz M (2003) Synergistic cytotoxic effect of TRAIL and gemcitabine in pancreatic cancer cells. Anticancer Res 23:251–258
Mao L, Yang C, Li L, Nai L, Fan L, Wang J et al (2014) Replication-competent adenovirus expressing TRAIL synergistically potentiates the antitumor effect of gemcitabine in bladder cancer cells. Tumour Biol: J Int Soc Oncodev Biol Med 35:5937–5944
Yang J, Ouyang J, Ouyang L, Ouyang L, Chen Y (2013) Inhibition of cell proliferation and increase of chemosensitivity by simultaneous knockdown of XIAP and survivin in pancreatic carcinoma cells. Oncol Res 21:43–50
Retzer-Lidl M, Schmid RM, Schneider G (2007) Inhibition of CDK4 impairs proliferation of pancreatic cancer cells and sensitizes towards TRAIL-induced apoptosis via downregulation of survivin. Int J Cancer 121:66–75
Kim SY, Kang S, Song JJ, Kim JH (2013) The effectiveness of the oncolytic activity induced by Ad5/F35 adenoviral vector is dependent on the cumulative cellular conditions of survival and autophagy. Int J Oncol 42:1337–1348
Seol DW, Billiar TR (1999) A caspase-9 variant missing the catalytic site is an endogenous inhibitor of apoptosis. J Biol Chem 274:2072–2076
Seol JW, Chaudhari AA, Lee YJ, Kang HS, Kim IS, Kim NS et al (2007) Regulation of DR-5 protein and mitochondrial transmembrane potential by gemcitabine, a possible mechanism of gemcitabine-enhanced TRAIL-induced apoptosis. Oncol Rep 18:523–529
Rajeshkumar NV, Rasheed ZA, Garcia-Garcia E, Lopez-Rios F, Fujiwara K, Matsui WH et al (2010) A combination of DR5 agonistic monoclonal antibody with gemcitabine targets pancreatic cancer stem cells and results in long-term disease control in human pancreatic cancer model. Mol Cancer Ther 9:2582–2592
Arlt A, Muerkoster SS, Schafer H (2013) Targeting apoptosis pathways in pancreatic cancer. Cancer Lett 332:346–358
Ibrahim SM, Ringel J, Schmidt C, Ringel B, Muller P, Koczan D et al (2001) Pancreatic adenocarcinoma cell lines show variable susceptibility to TRAIL-mediated cell death. Pancreas 23:72–79
Alemany R (2012) Chapter four-design of improved oncolytic adenoviruses. Adv Cancer Res 115:93–114
Alemany R (2013) Viruses in cancer treatment. Clin Trans Oncol: Offl Publ Fed Span Oncol Soc Nat Cancer Inst Mexico 15:182–188
Sharma A, Tandon M, Bangari DS, Mittal SK (2009) Adenoviral vector-based strategies for cancer therapy. Curr Drug Ther 4:117–138
Kang S, Kim JH, Kim SY, Kang D, Je S, Song JJ (2014) Establishment of a mouse melanoma model system for the efficient infection and replication of human adenovirus type 5-based oncolytic virus. Biochem Biophys Res Commun 453:480–485
Acknowledgments
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) and funded by the Ministry of Education, Science, and Technology (NRF-2013R1A1A2A100005494) and a faculty research grant of Yonsei University College of Medicine for 2014 (6-2014-0138). Zhezhu Han, Seungha Lee and Suyeon Je were supported by the Brain Korea 21 Plus project for Medical Science (Yonsei University, College of Medicine, Seoul, Republic of Korea).
Authors’ contributions
ZZH and SHL carried out overall research, experimental studies and data acquisition. SYJ participated in the animal study and helped to western blots. CYE participated in the immunohistochemistry and data acquisition. HJC carried out the data acquisition, and helped to draft the manuscript. JJS participated in the overall study design and drafted and revised the manuscript. JHK proposed the study and participated in its design and helped to draft, and assisted writing the manuscript. All authors had already read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Zhezhu Han and Seungha Lee contributed equally to this study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
10495_2015_1208_MOESM1_ESM.tif
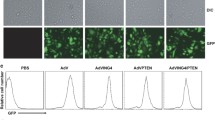
Supplementary figure 1. Screening of human survivin short hairpin RNAs (shRNAs), with sequences of five shRNA oligomers targeting survivin. The selected target sequence is indicated in bold (bottom of top panel). Five oligomers of the target and the positive control shRNA were transfected into HeLa cells. The knockdown efficiency of each oligomer was measured using quantitative real-time PCR to amplify survivin. Relative expression levels of survivin were plotted after normalization to the scrambled shRNA as a negative control (bottom panel) (TIFF 1082 kb)
10495_2015_1208_MOESM2_ESM.tif
Supplementary figure 2. The number of infectious viral particles was determined as a measure of oncolytic adenoviral replication. MiaPaCa-2 cells were infected with Ad-3484-NC or Ad-3484-shSurvivin or Ad-3484-TRAIL-shSurvivin adenovirus at an MOI of 50 for various times. After infection, the supernatants were examined for virus production. Error bars represent standard errors from three independent experiments (TIFF 419 kb)
Rights and permissions
About this article
Cite this article
Han, Z., Lee, S., Je, S. et al. Survivin silencing and TRAIL expression using oncolytic adenovirus increase anti-tumorigenic activity in gemcitabine-resistant pancreatic cancer cells. Apoptosis 21, 351–364 (2016). https://doi.org/10.1007/s10495-015-1208-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-015-1208-z