Abstract
The histone deacetylase inhibitor (HDACi) LBH589 has been verified as an effective anticancer agent. The identification and characterization of new targets for LBH589 action would further enhance our understanding of the molecular mechanisms involved in HDACi therapy. The role of the tumor suppressor death-associated protein kinase (DAPK) in LBH589-induced cytotoxicity has not been investigated to date. Stable DAPK knockdown (shRNA) and DAPK overexpressing (DAPK+++) cell lines were generated from HCT116 wildtype colon cancer cells. LBH589 inhibited cell proliferation, reduced the long-term survival, and up-regulated and activated DAPK in colorectal cancer cells. Moreover, LBH589 significantly suppressed the growth of colon tumor xenografts and in accordance with the in vitro studies, increased DAPK levels were detected immunohistochemically. LBH589 induced a DAPK-dependent autophagy as assessed by punctuate accumulation of LC3-II, the formation of acidic vesicular organelles, and degradation of p62 protein. LBH589-induced autophagy seems to be predominantly caused by DAPK protein interactions than by its kinase activity. Caspase inhibitor zVAD increased autophagosome formation, decreased the cleavage of caspase 3 and PARP but didn’t rescue the cells from LBH589-induced cell death in crystal violet staining suggesting both caspase-dependent as well as caspase-independent apoptosis pathways. Pre-treatment with the autophagy inhibitor Bafilomycin A1 caused caspase 3-mediated apoptosis in a DAPK-dependent manner. Altogether our data suggest that DAPK induces autophagy in response to HDACi-treatment. In autophagy deficient cells, DAPK plays an essential role in committing cells to HDACi-induced apoptosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Histone deacetylase inhibitors (HDACi) are a relatively new class of anticancer agents and display pleiotropic activities by inhibiting deacetylation of histone and non-histone cellular proteins [1, 2]. Currently several HDAC inhibitors with anticancer activity are in clinical trials against hematologic and solid malignancies, including colon cancer [1, 3].
It has been demonstrated that HDACi such as vorinostat, SAHA and butyrate induce autophagic cell death in addition to apoptosis in HeLa cells [4, 5]. Even though the morphological and biochemical features of apoptosis and autophagic cell death are distinct [6–8], both pathways have interconnected molecular regulators. DAPK is a Ca2+/CaM serine/threonine protein kinase that induces both autophagic cell death and apoptosis [9–11]. DAPK is involved in apoptosis induced by TNF-α, interferon-γ, Fas and TGF-β [12–14]. It also mediates the induction of autophagy in response to oxidative damage [15]. The kinase activity of DAPK has been shown to be crucial for triggering a number of cell death pathways [16]. DAPK activity is known to be negatively regulated by auto-phosphorylation of serine-308 [16, 17]. However some of the biological effects of DAPK are likely mediated through protein recruitment in a phosphorylation independent manner [18, 19]. Interestingly DAPK inactivation is associated with chemotherapy resistance [20, 21].
The panHDACi panobinostat (LBH589) is currently under investigation in several preclinical studies and shows promising antitumor activity at low nanomolar concentrations [22]. Its multi-target property appears to be very attractive. LBH589 induces histone acetylation, cell cycle arrest, and apoptosis in a variety of cancer cell lines [23–25]. It has the potential to sensitize colon cancer cells to TRAIL [26] mediated apoptosis. Ellis et al. [27] reported that LBH589 induces autophagy in cells lacking effective apoptosis machinery. Moreover, Fazzone et al. [28], showed a synergistic interaction between HDACi and 5-FU in colon cancer cells. Different drug targets have been described to be responsible for its anticancer effects which include down-regulation of NFkB and IRAK1 [29], proteasomal degradation of the damage sensor CHEK1 [30], induction of the stress protein CHOP [24, 31], or degradation of c-FLIP a negative regulator of death-receptor induced cell death [32]. The role of DAPK in LBH589-induced cytotoxicity has not been investigated so far.
Because DAPK is frequently inactivated by promoter hypermethylation in human tumors [33], it is of great interest to study the role of this tumor suppressor in HDACi action especially because a few reports exist showing that DAPK is activated after HDACi treatment [9, 34].
Using HCT116 colorectal tumor cells that express different amounts of DAPK i.e. HCT116 wildtype (DAPK wt), HCT116 DAPK shRNA (hereafter referred to as DAPK shRNA cell line), and HCT116 DAPK+++ (hereafter referred to as DAPK+++ cell line), we showed that LBH589 up-regulated DAPK both in vitro and in vivo. LBH589 was found to induce DAPK-dependent autophagy but a DAPK-independent apoptosis. However, in autophagy deficient cells, DAPK plays an essential role in committing cells to apoptosis. DAPK itself appears to switch from a prosurvival to an anti-survival molecule in LBH589-treated autophagy deficient colorectal cancer cells.
Materials and methods
Drugs and chemicals
LBH589 (Panobinostat) was provided by Novartis (Basel, Switzerland). Vorinostat (SAHA) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The pan-caspase inhibitor, Z-VAD-FMK was purchased from R&D systems (Wiesbaden, Germany). The autophagy inhibitor, Bafilomycin A1, Acridine orange and MTT was purchased from Sigma-Aldrich (St. Louis, MO). The DAPK inhibitor, 2-phenyl-4-(pyridin-3-ylmethylidene)-4,5-dihydro-1,3-oxazol-5-one was purchased from MolPort (Riga, Latvia).
Tumor cell lines and cell culture
The human colon cancer cell line HCT116 was obtained from American Type Culture Collection (ATCC, Manassas, VA). The cell lines (DAPK wt, DAPK shRNA, DAPK+++) were maintained in RPMI medium and supplemented with 10 % fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml). All cell lines were maintained at 37 °C in a humidified atmosphere and 5 % CO2. Mycoplasma contamination was excluded by using the PCR based system.
After 24 h of seeding, cells were stimulated with 0.05 μM LBH589 for various time points. In case of DAPK+++ cells, cells were incubated with 25 nM tamoxifen for 6 h and subsequently stimulated with LBH589. DAPK+++ control cells were treated only with tamoxifen. For inhibitor experiments cells were pre-incubated for 1 h with corresponding inhibitors.
Generation of DAPK shRNA stable cell line
HCT DAPK shRNA stable cell line was generated using DAPK shRNA lentiviral particles according to the manufacture’s instructions (Santa Cruz). Briefly, HCT116 cells were transfected with lentiviral particles containing a set of expression constructs each encoding DAPK specific 19–25 nt shRNA to knock down DAPK expression. After transduction stably shRNA expressing cells were isolated via puromycin selection. Transduction efficiency was measured by FACS and DAPK knock down was verified by PCR and Western Blotting.
Generation of DAPK+++ overexpressing cell line
Using a recently described 4-hydroxy tamoxifen (4HT)-inducible lentiviral expression system [35, 36], we aimed to permit over-expression of DAPK in transduced HCT116 cell line in a 4-HT dose-dependent manner.
To generate cells expressing 4-HT-inducible DAPK, HCT cells were transduced with a lentivirus pF GEV16 Super PGKHygro, which expresses a Gal4 DNA binding domain fused to a mutant estrogen receptor and GEV16, and a lentivirus pF 5 UAS hs DAPK SV40 Puro, which expresses DAPK in a Gal4-dependent fashion. In the first vector, the ubiquitin promoter constitutively drives expression of a GEV16 transcription factor. In the absence of 4HT, GEV16 is retained in the cytoplasm, but in the presence of 4HT it translocates to the nucleus where the GAL4-DNA binding domain (DBD) directs DNA binding to GAL4 upstream activating sequences (UAS) expressed by the second vector whereby its VP16 transactivation domain induces gene transcription. The GEV16 and 5-UAS constructs are contained within the lentiviral 5′-long terminal repeats (LTR) and 3′-self-inactivating LTR. To generate lentiviral supernatants, 293T cells were transfected with 3 mg pMD2.G, 5 mg pMDlg/pRRE, and 2.5 mg pRSV-Rev of the lentiviral packaging vectors [37] together with the constructs described above. The supernatants were harvested 24 h post-transfection, filtered (45 mm filter, Schleicher & Schuell), and concentrated by centrifugation (19,500×g, 2 h at 121 °C). The concentrated virus was added to cells as described [36]. After 24–48 h hygromycin and puromycin are added to select cells infected with both viruses.
xCELLigence real-time cell assay
Human colon tumor cells having different endogenous DAPK levels were seeded at 7,500 cells per well in triplicate in 96X microtiter E-plates for impedance-based real-time cell analysis using the xCELLigence RTCA system (Roche Molecular Diagnostics, Mannheim, Germany). Cellular viability and proliferation was measured continuously every 15 min for 72 h as described previously [38]. Results are expressed as an arbitrary unit called Cell Index (C.I.) and are given as mean ± SD of triplicates.
Crystal violet assay
DAPK wt cells (7,500 cells/well) were seeded in 96-well tissue culture plates and were incubated for 24 h. Next day, cells were washed once with PBS and stimulated with different concentrations of LBH589 (0.005, 0.01, 0.05, 0.075 and 0.1 μM) for 24, 48, 72 h. At the end of incubation time, the cell viability of the LBH589 was evaluated by crystal violet staining. The optical density was measured at 595 nm using a microplate reader. The average absorbance values of untreated controls were taken as 100 % cell viability.
Measurement of cell viability
Cells were seeded in 96-well flat bottom microtiter plates at a density of 7,500 cells in 200 μl per well. After treatment, 10 μl of 5 mg/ml MTT (3-[4,5-dimethyl-thiazol-2-yl]-2,5-diphenyltetrazolium-bromide, Sigma) solution was added to each well and incubated in a humidified 5 % CO2 incubator at 37 °C for 2 h. The medium with MTT solution was removed and 100 μl of DMSO was added and incubated for 15 min at RT. The absorbance of the solution was read spectrophotometrically at 450 nm with a reference at 650 nm using a microtitre plate reader (VictorX3).
Immunoprecipitation
Immunoprecipitation was performed according to the manufacturer’s instructions (Invitrogen, Karlsruhe, Germany). Briefly, protein G magnetic Dynabeads were coated with DAPK antibody (1:250 dilution) for 2 h with rotation at RT. Supernatants were removed using a magnetic stand and Dynabeads–antibody complex was washed with 200 μl Antibody Binding and Washing Buffer. 900 μg of protein lysate was added to the Dynabeads–antibody complex and gently resuspended by pipetting. The Dynabeads–antibody–antigen complex was incubated overnight at 4 °C with rotation. The Dynabeads–antibody–antigen complex was washed three times with 200 µl washing buffer each. Immunoprecipitates were eluted in 20 μl elution buffer. Eluted immunoprecipitates were resuspended with SDS reducing loading buffer and incubated 5 min at 95 °C. The proteins were separated by 10 % SDS-PAGE and analysis was performed by Western Blotting.
In vitro kinase assay
In vitro kinase assay was performed as described previously with minor modifications [39]. Briefly, immunoprecipitation was performed as described above. Dynabeads–antibody–antigen complex was resuspended in washing buffer and supernatant was removed using magnetic stand. The Dynabeads–antibody–antigen complex was resuspended in kinase buffer (60 mM HEPES, pH 7.5, 3 mM Mncl2, 3 mM Mgcl2, 3 μM sodium orthovanadate, 1.2 mM DTT, 2.5 μg/μl PEG, 2 μg RB-S6P) before the addition of 7.5 μCi [γ32-P] ATP (GE Healthcare, Amersham Biosciences) and incubated at 25 °C for 30 min. Samples were boiled with SDS reducing buffer at 95 °C for 5 min and separated using 10 % SDS-poly acrylamide gel. After gel electrophoresis, the proteins were transferred to nitrocellulose membrane and autoradiographed.
Clonogenic assay
Cells (1,000 cells/well) were seeded in 6 cm tissue culture plates and incubated for 24 h after which they were washed with PBS and treated with 0.05 μM LBH589. After overnight incubation, the drug-containing medium was replaced by growth medium and cells were incubated for 12–14 days. As the colonies became visible, the cells were fixed with 70 % methanol for 20 min. Colonies were then stained with crystal violet for 20 min, washed with water and dried. Pictures of stained colonies were made and area of interest was analyzed by Image-Pro Plus software.
Western Blot analysis
Whole cell lysates were prepared from DAPK wt, DAPK shRNA, and L27 DAPK+++ tumor cells. Protein concentration of lysates was determined with Bio-Rad Dc Protein Assay (BioRad Laboratories, Hercules, CA), and 30 μg proteins were loaded onto 10 or 12 % SDS-polyacrylamide gel electrophoresis. The proteins were transferred to nitrocellulose membranes before immunodetection processing with anti-DAPK (BD Transduction Laboratories, Lexington NY), anti-phosphoDAPKSer308 (Sigma), anti-LC3 (Nanotools, Teningen, Germany), anti-pH2AX (Millipore corporation, Billerica, USA), anti-Beclin 1, anti-Atg7, anti-p62, anti-caspase 3, anti-PARP (Cell Signaling Technology Inc.), anti-histone H3 acetyl rabbit poly clonal, anti-histone H4 pan-acetyl rabbit poly clonal (Activ Motif, Rixensart, Belgium) and with secondary antibodies (anti-mouse or anti-rabbit IgG peroxidase conjugated; Thermo Scientific, Rockford, IL). Bound antibodies were detected by incubating the blots in Immobilon western chemiluminescent HRP substrate (Millipore Corporation). Immunoreactivity was measured as peak intensity using an image capture and analysis system (GeneGnome, Syngene, UK). Hybridization with anti-β-actin or GAPDH was used to control equal loading and protein quality. Image J analysis software (version 1.45s) was used to quantify band intensities.
Flow cytometric analysis of cell cycle distribution
The distribution of cells in different phases of cell cycle was determined by flow cytometric analysis of DNA content. DAPK shRNA, DAPK wt, and DAPK+++ cells were stimulated with LBH589 for 24 and 48 h. After indicated times, both floated and adherent cells were harvested, washed twice with PBS, and stained with PI solution (50 μg/ml PI, 0.1 % sodium citrate, 0.1 % Triton X-100 in PBS). Distribution of cell cycle phases with different DNA contents was determined using a flow cytometer (Becton–Dickinson, CA, USA). Analysis of cell cycle distribution and the percentage of cells in the sub-G1, G1, S, and G2/M phase of the cell cycle were determined using the software CellQuest Pro (BD).
Detection of autophagic vacuoles by acridine orange
Cells were plated at a density of 2 × 105 on glass cover slips in six-well plates and incubated for 24 h. Cells were treated with 0.05 μM LBH589 for 24 and 48 h. At the appropriate time points, cells were incubated with 1 μg/ml acridine orange (Sigma, Germany) for 15 min. Cells were treated with 3 nM bafilomycin A1 for 60 min before the addition of acridine orange to inhibit the acidification of autophagic vacuoles. The acridine orange was removed and fluorescent photographs were obtained using an inverted fluorescence microscope (Nikon).
Detection of autophagosomes by LC3 immunofluorescence staining
Cells were plated at a density of 2 × 105 on glass cover slips in six-well plates and incubated for 24 h. Cells were treated with 0.05 μM LBH589 for 24 and 48 h. At the appropriate time points, cells were fixed with 4 % (w/v) paraformaldehyde for 30 min and then made permeabilized with methanol at −20 °C for 10 min. The cells were then covered with 10 % (v/v) goat serum for 30 min at room temperature to block non-specific adsorption of antibodies to the cells. After this procedure, the cells were incubated with primary antibody against LC3 at 4 °C overnight. Cells were then probed with Alexa Fluor 488 goat anti-mouse secondary antibodies and incubated at room temperature for another 2 h. Fluorescent signals were detected using inverted fluorescence microscope (Nikon).
Real-time reverse transcription-PCR
Total cellular RNA was extracted using RNeasy Kit (Qiagen, Hilden, Germany) and cDNA was synthesized using QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. The real-time RT-PCR was performed in a final volume of 25 μl using a CFX 96 (Bio-Rad) and threshold cycle numbers were determined using the Bio-Rad CFX manager software. DAPK primer sequences were sense 5-CCTTGCAAGACTTCGAA AGGATA-3 and antisense 5-GATTCCCGAGTGGCCAAA-3. The final reaction mixture contained the forward and reverse primer at 10 pmol each, PCR was performed under the following conditions: 95 °C for 5 min, followed by 45 cycles of 95 °C for 10 s, 60 °C for 30 s using QuantiFast SYBR Green PCR Kit (Qiagen). Real-time RT-PCR was performed in duplicate, and the threshold cycle numbers were averaged. Expression of genes of interest was normalized to β2-microglobulin and is expressed as arbitrary units.
Xenograft model
In vivo experiments were performed as described previously [24]. Briefly, 5.0 × 106 DAPK wt or DAPK shRNA cells suspended in sterile physiologic NaCl solution were injected into the flank regions of the 6–8 week old male NMRI mice (Harlan Winkelmann GmbH, Germany). Seventeen to twenty seven animals were used in the xenograft studies. Tumors were measured every day using a caliper square. When tumors reached a diameter of 7 mm, animals were treated with LBH5889 (10 mg/kg) or physiologic saline solution for 30 days by daily intraperitoneal injections. Animals were sacrificed by cervical dislocation at the end of the treatment period or when abortion criteria were met. Tumor xenografts were fixed in 10 % phosphate-buffered formalin or snap-frozen in liquid nitrogen. Ethical approval was granted by the local government authority (Government of Lower Franconia, Würzburg, Germany) before the beginning of experiments.
Tissue microarray and immunohistochemistry
Tumor tissues were fixed in formalin and embedded in paraffin. The punches were transferred onto a new paraffin block to form the tissue microarrays (TMAs). Immunohistochemical studies were performed on 3 μm thick slices using EnVision (Dako) detection system. TMAs were deparaffinized in xylene for 30 min at 72 °C and rehydrated in descending concentrations of ethanol. Antigen retrieval was done using pressure cooker (120 °C, 5 min, 1 mM Tris–EDTA buffer). Endogenous peroxidases activity was inhibited by incubating the slices for 5 min with blocking solution (Dako). All slices were incubated with primary antibodies (anti-DAPK (1:200) and anti-ki67 (1:50, Dako) for 30 min at RT, followed by washing with washing buffer (Dako) and incubation with secondary antibody linked with horseradish peroxidase (goat-anti-mouse, Dako) at RT for 30 min. Positive immunohistochemical reactions were detected using DAB + (Dako) as chromogen substrate. Nuclei were counterstained with hematoxylin (Dako).
Assessment of immunohistochemical protein expression
The TMAs were investigated by two independent reviewers blinded to other data [AH, MG]. For DAPK protein expression, staining intensity (SI) and the percentage of positive cells were assessed semi quantitatively using the following system: SI was classified as 0 (no staining), 1 (weak), 2 (moderate), and 3 (strong); number of positive cells (PC): 0 (0 %), 1 (<10 %), 2 (10–50 %), 3 (51–80 %), and 4 (>80 %). For DAPK, an immunoreactive score (IRS) was calculated using the formula [SI * PC = IRS]. Ki-67 staining was estimated by counting the percentage of positive nuclei in a representative tumor area. An average IRS of triplicates was finally estimated for each group.
Statistical analysis
Statistical analysis was performed using SPSS (SPSS Inc., Chicago, IL, USA). Differences in mRNA expression, cell viability and colony forming ability were compared using the one-way analysis of variance (ANOVA) followed by Tukey’s HSD, Dunnett’s t, and Student–Newman–Keuls post hoc tests. Significance of in vivo data was calculated using the t test for independent samples. p values less than 0.05 were considered statistically significant.
Results
LBH589 decreases cell viability in HCT116 DAPK wt cells and induces acetylation of histones H3 and H4
In order to find the optimal concentration of LBH589 in DAPK wt cells, cells were treated with increasing doses of LBH589. As shown in supplementary Fig. 1, LBH589 treatment decreased the cell viability in a time-and dose-dependent manner in DAPK wt cells. 0.05 μM LBH589 reduced the cell viability by approximately 50 % at 24 h. No significant viability effects were observed with doses lower than the 0.01 μM LBH589 (Supplemental Fig. 1a). Therefore, we used 0.05 μM LBH589 for all experiments in this study, a dose that has been used also by other groups [28, 32].
Next, DAPK shRNA, DAPK wt and DAPK+++ cells were treated with 0.05 μM LBH589 for 24, 48 and 72 h and analyzed for the acetylation status of histones H3 and H4. As expected, treatment with LBH589 induced acetylation of histone H3 and H4 in all the three cells lines (Supplemental Fig. 1b).
LBH589 up-regulates DAPK in HCT116 cancer cells in vitro and in vivo
To investigate if DAPK is involved in LBH589 action in tumor cells, we assessed the expression of DAPK at the mRNA and protein levels in cell lines that have different levels of DAPK i.e. DAPK wt, DAPK shRNA, and DAPK+++. The latter cells were used as an internal control of unphysiologically high DAPK levels and to clarify the involvement of DAPK in LBH589 activity. Cells were stimulated with 0.05 μM LBH589 for 24, 48 and 72 h. Although LBH589 significantly (p < 0.05) up-regulated the expression of DAPK at the mRNA level (Fig. 1b) in all three cell lines, the overall levels were approximately 4 and 20-fold lower in DAPK shRNA cells when compared to DAPK wt and DAPK+++ cells, respectively. Although DAPK+++ cells showed a slight increase in expression of DAPK at mRNA level with tamoxifen or LBH589 treatment alone (between 2- and 3-fold) only the combination of tamoxifen and LBH589 remarkably enhanced mRNA expression (up to 100-fold) verifying that the inducible lentiviral expression system is suitable (Supplemental Fig. 3a).
LBH589 enhances DAPK in vitro and in vivo. DAPK shRNA, DAPK wt, and DAPK+++ cells were treated with 0.05 μM LBH589 for 24, 48 and 72 h. a Inactive (pDAPK) and total levels of DAPK were analyzed by Western Blotting using antibodies against pDAPK and DAPK. Results are representative of three independent experiments. The band intensities were quantified by densitometry analysis.b The mRNA expression was analyzed using real-time RT-PCR. The mRNA levels were normalized to levels of ß2-microglobulin. DAPK+++ control cells (c) were treated with tamoxifen only. Results summarized in the bar graphs are representative of two independent experiments, *p < 0.05 (when compared with untreated controls). c LBH589 up-regulated the DAPK levels in DAPK wt and DAPK shRNA mice xenograft tissues. Arrows indicate extensive necrosis after LBH589 treatment. Magnification: ×200, insets magnification: ×400
In addition, there was a gradual increase in DAPK protein levels from DAPK shRNA cells to DAPK wt to DAPK+++ cells (Fig. 1a). There was a high level of inhibitory autophosphorylated form pDAPKSer308 in all three cell line controls. Under LBH589, the pDAPK levels were decreased over time. The subsequent reduction in pDAPK/DAPK ratio reflects the activation of DAPK. Indeed there was an increase in kinase activity of DAPK after LBH589 treatment in all the three cell lines. As expected from the highest pDAPK/DAPK ratio in DAPK shRNA cells they showed the lowest basal level of kinase activity whereas DAPK wt and DAPK+++ cells did not differ in their basal kinase activity. After LBH589 treatment, the kinase activity was increased in all three cell lines reaching the same levels in DAPK shRNA and DAPK wt cells and the highest level in DAPK+++ cells (Supplemental Fig. 2). DAPK+++ cells showed a slight decrease in the pDAPK/DAPK ratio also with tamoxifen or LBH589 treatment alone (between 2- and 3-fold). Nevertheless only the combination of tamoxifen and LBH completely activated DAPK with nearly total loss of the pDAPK inactive form (Supplemental Fig. 3b).
Thus these three cell lines will be a suitable model to study the DAPK-dependent effects of LBH589 in colorectal cancer cells.
In a separate experiment, we used the chemically related hydroxamic acid derivative, vorinostat, to generalize the HDACi effect on DAPK. The three cell lines were stimulated with vorinostat for 24, 48 and 72 h. Vorinostat induced an up-regulation of DAPK in all three cell lines (Supplemental Fig. 1c) with simultaneous activation of DAPK activity which is similar to LBH589.
To evaluate the in vivo DAPK regulation in LBH589-treated mice, DAPK wt and DAPK shRNA colon tumor xenografts were generated in nude mice and treated with 10 mg/kg LBH589 for 30 days. LBH589 induced an increase of DAPK protein levels in the cytoplasm of tumor cells of both DAPK wt and DAPK shRNA xenografts but to a higher extent in DAPK wt cells (Fig. 1c). LBH589 significantly suppressed the growth of both colon xenografts irrespective of DAPK levels (Supplemental Fig. 4a). Immunohistochemical analysis shows that Ki67 score is significantly (p < 0.05) decreased in the nuclei of both colon xenografts (Supplemental Fig. 4b). In addition, there was a higher number of necrotic cells with loss of cell integrity and nuclei in both xenografts (Fig. 1c).
LBH589 reduces the cell viability and long-term survival in HCT116 cells in a DAPK-independent manner
To study if the different DAPK levels in the three cell types affect their viability, we used the impedance-based xCELLigence system to monitor dynamic cell proliferation in real-time. As shown in Fig. 2a, the three cell lines showed similar growth kinetics. To further evaluate the effect of LBH589 on cellular viability, we performed MTT tests. As shown in Fig. 2b, LBH589 significantly (p < 0.05) reduced the cell viability in all three cell lines at 24, 48 and 72 h. Reduction in cell viability was most prominent in DAPK+++ cells.
Cell viability and long term survival is independent on the DAPK status in HCT116 cancer cells. a The indicated cell types were seeded at a density of 7,500 cells per well in 96X E-plates. The attachment, spreading and proliferation of cells were monitored every 15 min until 72 h using the xCELLigence real-time cell-analyzer. The results are expressed as normalized cell index. b The indicated cell types were seeded at a density of 7,500 cells per well in 96-well plates and treated with LBH589 for 24, 48 and 72 h. Cell viability was determined by the MTT assay. Results are expressed as percentages of control cells. Each value is the mean of two separate experiments performed in triplicates, *p < 0.05 compared with untreated controls. #p < 0.05 for comparison between LBH589 treated cells. c The indicated cell types were seeded at a density of 500 cells in 5 cm dishes and incubated with LBH589. After overnight incubation, drug-free medium was applied to the samples. Cells were then allowed to form colonies for 12–14 days. Colonies were fixed in methanol, stained with crystal violet, washed with water and photographed. *p < 0.05 compared with untreated controls, #p < 0.01 for comparison between LBH589 treated cells
To further study the long-term growth inhibitory effect of LBH589, an anchorage dependent clonogenic assay was performed. Results showed that treatment with LBH589 significantly (p < 0.05) reduced colony formation of all three colon cancer cell lines (Fig. 2c). In line, DAPK+++ cells were more sensitive to LBH589.
LBH589 activates caspase 3-mediated apoptosis and cell cycle arrest in a DAPK-independent manner
Various studies have already shown that HDACi LBH589 induces apoptosis in cancer cell lines [23, 24, 28]. First, we examined potential DAPK-dependent cell cycle regulatory effects of LBH589. Each cell line was stimulated with 0.05 μM LBH589 for 24 and 48 h and DNA content was analyzed by flow cytometry after staining with propidium iodide. After 24 h of LBH589 treatment, sub-G1 population that is indicative of cell death was observed in all the three cell lines. In addition, LBH589 caused a G2/M cell cycle arrest with decreased G1/S fractions when compared to untreated controls in all three cell lines. The results suggest that cells with different DAPK levels might have similar modes of cell cycle regulation after LBH589 treatment indicating that LBH589-induced cell cycle arrest was DAPK-independent (Supplemental Fig. 5).
To further determine DAPK’s role in LBH589-induced apoptosis, we investigated the cleavage of caspase 3 and PARP, two hallmarks of apoptosis, in LBH589-treated cells. There was no difference in activation of caspase 3 and PARP cleavage in DAPK wt and DAPK shRNA cells whereas we observed a strong apoptosis induction in DAPK+++ cells (Fig. 3a).
LBH589 induces apoptosis in human colon tumor cell lines in a DAPK-independent manner. a The cells having different DAPK status were treated for 24, 48 and 72 h with or without LBH589. Western Blot analysis shows activation of caspase-3, PARP and pH2AX in all three cell types after treatment with LBH589. The data are representative of two independent experiments. b The indicated cell types were preincubated with a general caspase inhibitor, zVAD for 1 h and then stimulated with LBH589 for 24, or 48 h, respectively. Western Blotting analysis for caspase 3 and PARP was performed. The data are representative of two independent experiments. c The indicated cell types were treated with or without vorinostat for various time points. Protein levels of caspase 3 and PARP were detected by Western Blotting. The data are representative of two independent experiments
To provide further evidence that caspases are involved in LBH589-induced apoptosis, we treated the cells with the general caspase inhibitor zVAD. Cells were pre-stimulated with 40 μM zVAD for 1 h and then treated with LBH589 for the indicated time points. Pre-treatment with zVAD completely abolished LBH589-induced caspase and PARP cleavage (Fig. 3b) suggesting that LBH589-induced cell death is mediated by caspase 3. Interestingly additional caspase 3 cleaved bands were observed after combined LBH589 and zVAD treatment (Fig. 3b), suggesting that zVAD protects caspase 3 p20 from full maturation or that it is protected by effector caspase inhibitors such as XIAP [40]. Considering the fact that zVAD did not rescue DAPK wt cells from LBH589 induced cell death in crystal violet assay we hypothesize that LBH589 activates also caspase-independent mechanisms (Supplemental Fig. 6).
To verify the role of DAPK kinase activity in LBH589-induced apoptosis we used a potent and selective DAPK inhibitor, 2-phenyl-4-(pyridin-3-ylmethylidene)-4,5-dihydro-1,3-oxazol-5-one (hereafter referred as DAPK inhibitor) for inhibiting its catalytic activity [41]. First, we measured the effect of single and combination treatment in the DAPK wt cells using crystal violet assay. Pre-treatment with DAPK inhibitor did not influence the LBH589 induced caspase 3-mediated cell death at 24 and 48 h, respectively, when compared to LBH589 alone (Supplemental Fig. 7a). Furthermore, pre-treatment of DAPK wt and DAPK+++ cells with the DAPK inhibitor did not alter the LBH589-induced caspase 3 and PARP cleavage suggesting that DAPK’s kinase activity is not important for the observed caspase 3-dependent cell death (Supplemental Fig. 7b, c). Indeed, DAPK shRNA and DAPK wt cells demonstrated nearly equal levels of kinase activity after LBH589 treatment, whereas DAPK+++ cells showed the most remarkable DAPK kinase activity.
As shown in supplemental Fig. 8, LBH589 induces time-dependent increase in the number of apoptotic bodies when compared to the respective controls; however this increase was DAPK-independent. Cells with artificial overexpression of DAPK (DAPK+++) showed the most prominent apoptotic body formation at 24 h. In conclusion, there were no differences in LBH589 sensitivity between DAPK shRNA and DAPK wt cells whereas DAPK+++ cells were highly susceptible to LBH589-induced apoptosis.
We then investigated if apoptosis is caused by DAPK-mediated DNA damage. To detect DNA double strand breaks after LBH589 exposure, we studied the levels of the DNA damage marker, pH2AX, by Western Blotting. An increase in pH2AX levels was detected in all three cell lines. DNA damage was less pronounced in DAPK+++ cells at 24 h. These data suggest that in this cell line the damaged cells are more efficiently eliminated by apoptosis or that there are other mechanisms protecting the cells from excessive damage. Because DAPK is known to interact with p53 a link to p53-mediated repair cannot be excluded.
To determine if HDACi-induced apoptotic effects are generally DAPK-independent we tested caspase 3 and PARP activation in our cell system treated with vorinostat. As expected, we could not observe DAPK-dependent apoptosis induction between the three cell lines after vorinostat treatment suggesting that DAPK does not seem to play a role in apoptosis induction by panHDACi (Fig. 3c).
LBH589 induces autophagy in HCT116 cancer cells in a DAPK-dependent manner
To investigate the mechanism of LBH589-induced cell death we studied whether LBH589 induces typical hallmarks of autophagy, a process where DAPK has been shown to be a key player [10, 11].
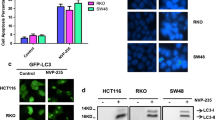
We used immunofluorescence and immune blot analysis to detect LC3-II positive punctuate staining and autophagosome formation. As shown in Fig. 4a–c, treatment with LBH589 caused the punctuate accumulation of LC3-II, representing the formation of new autophagic vacuoles in the cytoplasm of all three cell lines. Furthermore, the LC3-II positive punctuation correlated with DAPK levels, suggesting that DAPK enhances autophagosome formation. Consistent with the immunofluorescence data, Western Blot analysis with an anti-LC3 antibody revealed that LBH589 treatment caused an accumulation of autophagosomes in the three colon cancer cell lines with an increased conversion of cytosolic LC3-I to autophagosomal membrane-bound LC3-II. LC3-II levels were more pronounced in DAPK+++ cells followed by DAPK wt and DAPK shRNA cells (Fig. 4d). Overall, these results suggest that LBH589-induced autophagy is potentiated by DAPK in colon cancer cells. Based on our data on DAPK kinase activity, we suggest that LBH589-induced autophagy seems to be rather caused by DAPK protein interactions than by its kinase activity.
DAPK-dependent autophagy induction after LBH589 treatment. a–c The three cell types were seeded on glass coverslips and treated with LBH589 for various time points as indicated. Endogenous LC3 aggregation was detected using immunofluorescence with anti-LC3 antibody. The appearance of punctuated signals of LC3 is a hallmark of autophagy. The picture shows one representative experiment out of two independent experiments. d The indicated cell types were treated with or without LBH589 for various time points. Protein levels of LC3 were detected by Western Blotting. The data are representative of two independent experiments. e HCT116 cells having different DAPK status were stimulated with 2 μM Vorinostat for 24, 48 and 72 h. After the treatment, whole-cell protein lysates were prepared and Western Blotting analysis was performed against LC3
To find out whether other essential markers of autophagy are induced in response to LBH589 treatment, we analyzed protein expression of Beclin1 and Atg7. Immunoblotting analysis against Beclin1 and Atg7 revealed nearly no change in protein levels in all three cell lines (Supplemental Fig. 9).
To study if DAPK-dependent autophagy is a general effect observed with other HDACi we treated cells with vorinostat and analysed LC3-II levels. As shown in Fig. 4e, stimulation with vorinostat induced LC3-II autophagosome formation in all three cell lines in a DAPK-dependent manner. These data indicate a DAPK-mediated autophagosome formation as a general effect of HDACi in the treatment of colorectal cancer cells and reflects that DAPK seems to be involved in the immediate early stress response to HDACi-induced DNA-damage.
LBH589 increases acidification of vesicular organelles and causes loss of LC3-II and p62 protein in a DAPK-dependent manner
Previous studies have shown that autophagy is associated with acidification of vesicular organelles [42, 43]. However, it is not yet known if DAPK mediates the effects of LBH589 on acidification of vesicular organelles. To test this hypothesis, we used bafilomycin A1, an inhibitor of autophagy that acts by inhibiting the H+-ATPase responsible for acidification of the autophagolysosomal vacuoles [44]. The formation of acidic vesicular organelles was determined by acridine orange staining. As shown in Fig. 5a–c, control cells displayed green fluorescence whereas LBH589-treated cells displayed red fluorescence, indicating formation of numerous autophagolysosomal vacuoles. On the other hand, pre-treatment with bafilomycin A1 reduced LBH589-induced acidification of vesicular organelles.
LBH589 induces formation of acidic autophagic vacuoles and p62 protein degradation in a DAPK-dependent manner. a–c The three cell types were treated with LBH589 for 24 or 48 h in the presence or absence of autophagic inhibitor bafilomycin A1. The formation of acidic vesicular organelles seen by acridine orange staining was observed using inverted fluorescence microscope. d Cells were prestimulated with bafilomycin A1 for 1 h and then stimulated with LBH589 for 24 and 48 h. Western Blotting was performed against LC3 and p62 protein. The band intensities were quantified by densitometry analysis. In each cell line the control was adjusted to one after normalization to β-actin. Bafilomycin A1 increased the accumulation of autophagosomes and p62 protein. All data are representative of two independent experiments
LC3-II is known to accumulate as a result of increased autophagosome formation or impaired autophagosome-lysosome fusion [45]. To determine which of these two possibilities occur in LBH589-treated cells we investigated autophagic flux in human colon cancer cells by assessing the LC3-II turnover in the presence and absence of bafilomycin A1. As shown in Fig. 5d, treatment with LBH589 caused an increase in LC3-II levels. Moreover, the increase in LC3-II potentiated over time and correlated with DAPK levels, suggesting a DAPK-dependent formation of autophagosomes. When compared with cells treated with bafilomycin A1 alone, treatment with both, bafilomycin A1 and LBH589, caused not only further induction of LC3-I but also a significant increase in conversion of LC3-I to LC3-II suggesting that the increase in LC3-II by DAPK was not due to the blockage of autophagic degradation (Fig. 5d, upper blot).
The increased autophagic flux was further confirmed by the decrease of p62, an accepted substrate of autophagy, whose level decreases upon autophagy induction and accumulates when autophagy is inhibited [45, 46]. LBH589 treatment caused a slight decrease of p62 levels in all three cell lines and this reduction was prevented by bafilomycin A1, confirming an autophagy-mediated p62 degradation. Treatment with bafilomycin A1 alone resulted in accumulation of p62 which was most pronounced in DAPK+++ cells (Fig. 5d, lower blot). This suggests that the extent of autophagosome-lysosome fusion is dependent on endogenous DAPK level of a cell. In DAPK shRNA and DAPK wt cells LBH589 did not reverse the bafilomycin-induced increase in p62 levels. In DAPK+++ cells LBH589 decreased p62 levels to the levels seen in control cells. Thus we suggest that in DAPK+++ cells p62 might lead to tumor suppression possibly by its own forced degradation or because bafilomycin did not cause a sufficient inhibition of p62 degradation in cells having artificially high DAPK levels.
Crosstalk between LBH589-induced autophagy and apoptosis
As a next step we aimed to find out if DAPK plays a role in the crosstalk between LBH589-induced autophagy and apoptosis. At first, we inhibited caspase 3-dependent apoptosis by zVAD and studied autophagosome formation by Western Blotting for LC3-II. In general, combined treatment with zVAD reinforced the LBH589-induced increase in LC3-II levels in all three cell lines (Fig. 6a) independent of the DAPK status. This suggests that the less efficient elimination of damaged tumor cells by apoptosis caused a general increase in autophagosome formation.
Crosstalk between autophagy and apoptosis. The three cell types were treated with LBH589 in the presence or absence of either zVAD or bafilomycin A1 for various time points as indicated. a The cross inhibition and/or activation were analysed by Western Blotting against LC3, b Caspase 3 and PARP. The data are representative of two independent experiments
Next, we analyzed whether inhibition of autophagy by bafilomycin A1 influences LBH589-induced apoptosis. First, we measured the effect of single and combination treatment in the DAPK wt cells using crystal violet assay. Pre-treatment with bafilomycin A1 further increased the rate of apoptotic cells in comparison to LBH589 alone (Supplemental Fig. 10). Interestingly, pre-treatment with bafilomycin A1 alone caused an increase in active caspase 3 levels in a time-dependent manner in all three cell lines. PARP cleavage was more prominently induced in DAPK+++ cells. The enhanced caspase cleavage at 24 and 48 h was strongly DAPK-dependent (Fig. 6b). Complete PARP cleavage at 48 h was observed only in DAPK+++ cells. These data show that the induction of autophagy by DAPK partly protects cells from LBH589-induced apoptosis suggesting a pro-survival role for DAPK. A schematic model of DAPK’s action after LBH589 treatment in autophagy competent and deficient cells is given in Fig. 7.
Schematic model illustrating that DAPK commits cells to apoptosis under autophagy-deficient conditions. LBH589 treatment simultaneously induces DAPK-dependent autophagy but DAPK-independent apoptosis in human colon tumor cells. Inhibition of autophagy sensitized the tumor cells to treatment-induced apoptosis and DAPK may be responsible to initiate apoptosis if the capacity for autophagy is insufficient. This indicates that under autophagy inhibition DAPK could act to switch between autophagy and apoptosis
Discussion
Previous studies have demonstrated that the HDACi LBH589 induces apoptosis in various cancer cell lines, including colon cancer [1, 47]. In the last decade the tumor suppressor DAPK has been linked with regulation of apoptosis and autophagy [48]. DAPK is a calcium/calmodulin regulated cytoskeleton-associated serine/threonine kinase and many DAPK substrates are cytoskeleton-associated proteins [18]. DAPK interacts with different MAPKs such as ERK [49] or p38 [39] in response to inflammatory apoptotic stimuli. Otherwise DAPK phosphorylation by RSK and Src is considered to rather protect from cell death [50, 51]. Thus, DAPK can serve as a mediator between death-inducing and pro-survival signals [52].
The role of DAPK in LBH589-induced cytotoxicity has not yet been studied. For the first time we show that dephosphorylation of DAPK at ser 308 is the most important mechanism in DAPK activation by HDACi. Whereas LBH-induced apoptosis seems to be independent of DAPK, autophagy induction is predominantly caused by DAPK protein interactions than by its catalytic activity. Under autophagy deficient conditions, DAPK plays a role in sensitizing human colon tumor cells to LBH589-induced apoptosis. Autophagy is either a protective and survival mechanism in response to intracellular stress such as radiotherapy and chemotherapy or a mechanism of cell death under certain conditions especially in response to treatments that trigger caspase-independent autophagy [53]. It has been reported that DAPK phosphorylates the autophagy-associated molecule beclin 1, reducing the BH3-pocket interaction between beclin 1 and its inhibitor Bcl-2, finally promoting autophagy [54]. Down-regulation of Bcl-2 triggers autophagy, but does not promote apoptosis in leukemic cells [55]. Lin Yao et al. [18] reviewed that DAPK can act either as a pro-survival factor or promotes apoptosis in autophagy signalling pathways. Previous studies have reported that blockage of autophagy with bafilomycin A1 led to an accumulation of autophagosomal structures [44]. Here we show that bafilomycin A1 prevented the acidification of lysosomes and caused accumulation of LC3-II protein. In this regard, Harrison et al. [56] showed that DAPK interacts with microtubule-associated protein 1B and LC3 protein resulting in the stimulation of DAPK-dependent membrane blebbing and autophagy. LBH589 treatment did not change the activity of autophagy-related proteins beclin1 and Atg7. This might be due to the fact that LBH589-DAPK pathway is a non-canonical autophagy pathway. Further studies are required to investigate that interesting notion.
We also observed that LBH589 induces phosphorylation of H2AX, a DNA damage marker, in all the three cell lines in a DAPK-independent manner. This observation is in line with previous studies showing that histone deacetylase inhibitors TSA [57] and LBH589 [58] phosphorylate H2AX as an early cellular response to DNA damage. However, DNA damage was less evident in DAPK+++ cells despite pronounced apoptosis. The reason might be that under conditions of high DAPK activity, damaged cells are more efficiently eliminated by autophagy-driven cell death or that autophagy seems to protect the cells from severe damage altogether. If autophagy mediates the cytotoxicity we would expect that bafilomycin A1 co-treatment would lead to an attenuated cell death. We analysed PARP, which detects and signals downstream of DNA strand breaks and subsequently activates DNA repair programs or cell death pathways. It is activated at an intermediate stage of apoptosis and is then cleaved and inactivated at a late stage by apoptotic proteases, such as caspase 3. In our system, an increase in PARP cleavage was noted by combination treatment in all three cell lines and complete cleavage occurred in DAPK+++ cells further indicating that DAPK-driven autophagy might initially be triggered to protect the cells by sequestering and degrading damaged organelles. Thus, inhibition of autophagy sensitized the tumor cells to treatment-induced apoptosis and DAPK may be responsible to initiate apoptosis if the capacity for autophagy is insufficient. This indicates that under autophagy inhibition DAPK could act to switch between autophagy and apoptosis.
p62 is known to be a selective substrate of the autophagic machinery and impaired autophagy is accompanied by the accumulation of p62. In this study, bafilomycin A1 treatment increased the level of p62 in a DAPK-dependent manner suggesting a role for DAPK in autophagic flux induced by LBH589. Bafilomycin A1 failed to block the LBH589-induced p62 degradation in the DAPK+++ cells perhaps due to their artificially high DAPK levels. Alternatively, a proteasomal degradation of p62 after combination treatment cannot be excluded [59]. Furthermore we showed that bafilomycin A1 inhibited lysosomal hydrolases by decreasing the acidification of lysosomal compartment. These data suggest that the vacuolar type H+-ATPase is essential for the regulation of the late stage of macroautophagy after LBH589 treatment. Consistent with other studies, our reports showed that a commonly synthetic peptide inhibitor of caspases, zVAD, slightly enhanced LC3-II levels in all three cell lines irrespective of the DAPK expression and activity. The reason could be that zVAD inhibits both apoptosis and autophagic flux by preventing lysosomal degradation of autophagosome contents [60, 61]. Our data show that caspase 3 cleavage in LBH589-treated cells was recovered upon zVAD treatment, yet there was no rescue of death in crystal violet assay. This suggests both caspase-dependent and caspase-independent apoptotic mechanisms are at play after LBH589 treatment. We have recently shown in liver cancer cells that LBH589 up-regulated CHOP, a marker of the unfolded protein response and endoplasmatic reticulum (ER) stress [24]. There is evidence that ER stress can facilitate autophagosome formation and thus trigger autophagy [62]. ER-stress-induced autophagy is important for clearing polyubiquitinated protein aggregates and consequently protects against cell death. In this respect, it has been shown that treatment of neuroblastoma cells with ER stressors induced autophagosome formation and autophagy-inhibited cells demonstrated an increased vulnerability to ER stress and increased caspase activity [63]. In our study, there was a clear increase in PARP cleavage when autophagy was blocked as a sign of augmented apoptosis induction supporting the initiation of cytoprotective autophagy in a DAPK-dependent manner. Gozuacik et al. [10] have been reported that DAPK is important in integrating autophagy and apoptosis induced by ER stress. They emphasize the fact that there are always multiple executers securing redundancy in signalling pathways. This finally results in a robust and fine-tuned cellular stress response and DAPK’s regulatory role could be rather upstream of these signalling cascades.
Taken together, our data suggest that manipulation of autophagy could improve the efficacy of anticancer therapies and should be taken into account in tumor cells that overexpress DAPK, e.g. in non-methylated tumors. The current study further establishes the potential utility of LBH589 for the treatment of colorectal cancer.
References
Atadja P (2009) Development of the pan-DAC inhibitor panobinostat (LBH589): successes and challenges. Cancer Lett 280:233–241
Glozak MA, Sengupta N, Zhang X, Seto E (2005) Acetylation and deacetylation of non-histone proteins. Gene 363:15–23
Mariadason JM (2008) HDACs and HDAC inhibitors in colon cancer. Epigenetics 3:28–37
Shao Y, Gao Z, Marks PA, Jiang X (2004) Apoptotic and autophagic cell death induced by histone deacetylase inhibitors. Proc Natl Acad Sci USA 101:18030–18035
Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X (2012) Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci USA 109:6561–6565
Bursch W, Hochegger K, Torok L, Marian B, Ellinger A, Hermann RS (2000) Autophagic and apoptotic types of programmed cell death exhibit different fates of cytoskeletal filaments. J Cell Sci 113:1189–1198
Bursch W (2001) The autophagosomal-lysosomal compartment in programmed cell death. Cell Death Differ 8:569–581
Ogier-Denis E, Codogno P (2003) Autophagy: a barrier or an adaptive response to cancer. Biochim Biophys Acta 1603:113–128
Wu J, Hu CP, Gu QH, Li YP, Song M (2010) Trichostatin A sensitizes cisplatin-resistant A549 cells to apoptosis by up-regulating death-associated protein kinase. Acta Pharmacol Sin 31:93–101
Gozuacik D, Bialik S, Raveh T, Mitou G, Shohat G, Sabanay H et al (2008) DAP-kinase is a mediator of endoplasmic reticulum stress-induced caspase activation and autophagic cell death. Cell Death Differ 15:1875–1886
Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A (2002) DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol 157:455–468
Cohen O, Inbal B, Kissil JL, Raveh T, Berissi H, Spivak-Kroizaman T et al (1999) DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol 146:141–148
Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A (1995) Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev 9:15–30
Jang CW, Chen CH, Chen CC, Chen JY, Su YH, Chen RH (2002) TGF-beta induces apoptosis through Smad-mediated expression of DAP-kinase. Nat Cell Biol 4:51–58
Eisenberg-Lerner A, Kimchi A (2012) PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ 19:788–797
Jin Y, Blue EK, Gallagher PJ (2006) Control of death-associated protein kinase (DAPK) activity by phosphorylation and proteasomal degradation. J Biol Chem 281:39033–39040
Shamloo M, Soriano L, Wieloch T, Nikolich K, Urfer R, Oksenberg D (2005) Death-associated protein kinase is activated by dephosphorylation in response to cerebral ischemia. J Biol Chem 280:42290–42299
Lin Y, Hupp TR, Stevens C (2010) Death-associated protein kinase (DAPK) and signal transduction: additional roles beyond cell death. FEBS J 277:48–57
Chuang YT, Lin YC, Lin KH, Chou TF, Kuo WC, Yang KT et al (2011) Tumor suppressor death-associated protein kinase is required for full IL-1β production. Blood 117:960–970
Eisenberg-Lerner A, Kimchi A (2012) DAPk silencing by DNA methylation conveys resistance to anti EGFR drugs in lung cancer cells. Cell Cycle 11:2051
Ogawa T, Liggett TE, Melnikov AA, Monitto CL, Kusuke D, Shiga K et al (2012) Methylation of death-associated protein kinase is associated with cetuximab and erlotinib resistance. Cell Cycle 11:1656–1663
Neri P, Bahlis NJ, Lonial S (2012) Panobinostat for the treatment of multiple myeloma. Expert Opin Investig Drugs 21:733–747
LaBonte MJ, Wilson PM, Fazzone W, Groshen S, Lenz HJ, Ladner RD (2009) DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines. BMC Med Genomics 2:67
Di Fazio P, Schneider-Stock R, Neureiter D, Okamoto K, Wissniowski T, Gahr S et al (2010) The pan-deacetylase inhibitor panobinostat inhibits growth of hepatocellular carcinoma models by alternative pathways of apoptosis. Cell Oncol 32:285–300
Pettazzoni P, Pizzimenti S, Toaldo C, Sotomayor P, Tagliavacca L, Liu S et al (2011) Induction of cell cycle arrest and DNA damage by the HDAC inhibitor panobinostat (LBH589) and the lipid peroxidation end product 4-hydroxynonenal in prostate cancer cells. Free Radic Biol Med 50:313–322
Lee SC, Cheong HJ, Kim SJ, Yoon J, Kim HJ, Kim KH et al (2011) Low-dose combinations of LBH589 and TRAIL can overcome TRAIL-resistance in colon cancer cell lines. Anticancer Res 31:3385–3394
Ellis L, Bots M, Lindemann RK, Bolden JE, Newbold A, Cluse LA et al (2009) The histone deacetylase inhibitors LAQ824 and LBH589 do not require death receptor signaling or a functional apoptosome to mediate tumor cell death or therapeutic efficacy. Blood 114:380–393
Fazzone W, Wilson PM, Labonte MJ, Lenz HJ, Ladner RD (2009) Histone deacetylase inhibitors suppress thymidylate synthase gene expression and synergize with the fluoropyrimidines in colon cancer cells. Int J Cancer 125:463–473
LaBonte MJ, Wilson PM, Fazzone W, Russell J, Louie SG, El-Khoueiry A et al (2011) The dual EGFR/HER2 inhibitor lapatinib synergistically enhances the antitumor activity of the histone deacetylase inhibitor panobinostat in colorectal cancer models. Cancer Res 71:3635–3648
Brazelle W, Kreahling JM, Gemmer J, Ma Y, Cress WD, Haura E et al (2010) Histone deacetylase inhibitors downregulate checkpoint kinase 1 expression to induce cell death in non-small cell lung cancer cells. PLoS ONE 5:e14335
Rao R, Nalluri S, Fiskus W, Savoie A, Buckley KM, Ha K et al (2010) Role of CAAT/enhancer binding protein homologous protein in panobinostat-mediated potentiation of bortezomib-induced lethal endoplasmic reticulum stress in mantle cell lymphoma cells. Clin Cancer Res 16:4742–4754
Kauh J, Fan S, Xia M, Yue P, Yang L, Khuri FR et al (2010) c-FLIP degradation mediates sensitization of pancreatic cancer cells to TRAIL-induced apoptosis by the histone deacetylase inhibitor LBH589. PLoS ONE 5:e10376
Esteller M, Herman JG (2002) Cancer as an epigenetic disease: DNA methylation and chromatin alterations in human tumours. J Pathol 196:1–7
Zhang X, Yashiro M, Ren J, Hirakawa K (2006) Histone deacetylase inhibitor, trichostatin A, increases the chemosensitivity of anticancer drugs in gastric cancer cell lines. Oncol Rep 16:563–568
Vince JE, Wong WW, Khan N, Feltham R, Chau D, Ahmed AU et al (2007) IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131:682–693
Diessenbacher P, Hupe M, Sprick MR, Kerstan A, Geserick P, Haas TL et al (2008) NF-kappaB inhibition reveals differential mechanisms of TNF versus TRAIL-induced apoptosis upstream or at the level of caspase-8 activation independent of cIAP2. J Invest Dermatol 128:1134–1147
Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J et al (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33:401–406
Gloesenkamp CR, Nitzsche B, Ocker M, Di Fazio P, Quint K, Hoffmann B et al (2012) AKT inhibition by triciribine alone or as combination therapy for growth control of gastroenteropancreatic neuroendocrine tumors. Int J Oncol 40:876–888
Bajbouj K, Poehlmann A, Kuester D, Drewes T, Haase K, Hartig R et al (2009) Identification of phosphorylated p38 as a novel DAPK-interacting partner during TNFalpha-induced apoptosis in colorectal tumor cells. Am J Pathol 175:557–570
Leverkus M, Sprick MR, Wachter T, Mengling T, Baumann B, Serfling E et al (2003) Proteasome inhibition results in TRAIL sensitization of primary keratinocytes by removing the resistance-mediating block of effector caspase maturation. Mol Cell Biol 23:777–790
Okamoto M, Takayama K, Shimizu T, Muroya A, Furuya T (2010) Structure-activity relationship of novel DAPK inhibitors identified by structure-based virtual screening. Bioorg Med Chem 18:2728–2734
Paglin S, Hollister T, Delohery T, Hackett N, McMahill M, Sphicas E et al (2001) A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res 61:439–444
Newman RA, Kondo Y, Yokoyama T, Dixon S, Cartwright C, Chan D et al (2007) Autophagic cell death of human pancreatic tumor cells mediated by oleandrin, a lipid-soluble cardiac glycoside. Integr Cancer Ther 6:354–364
Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct 23:33–42
Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS et al (2008) Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 4:151–175
Li J, Hou N, Faried A, Tsutsumi S, Kuwano H (2010) Inhibition of autophagy augments 5-fluorouracil chemotherapy in human colon cancer in vitro and in vivo model. Eur J Cancer 46:1900–1909
Hague A, Manning AM, Hanlon KA, Huschtscha LI, Hart D, Paraskeva C (1993) Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int J Cancer 55:498–505
Bialik S, Kimchi A (2006) The death-associated protein kinases: structure, function, and beyond. Annu Rev Biochem 75:189–210
Chen CH, Wang WJ, Kuo JC, Tsai HC, Lin JR, Chang ZF et al (2005) Bidirectional signals transduced by DAPK-ERK interaction promote the apoptotic effect of DAPK. EMBO J 24:294–304
Anjum R, Roux PP, Ballif BA, Gygi SP, Blenis J (2005) The tumor suppressor DAP kinase is a target of RSK-mediated survival signaling. Curr Biol 15:1762–1767
Wang WJ, Kuo JC, Ku W, Lee YR, Lin FC, Chang YL et al (2007) The tumor suppressor DAPK is reciprocally regulated by tyrosine kinase Src and phosphatase LAR. Mol Cell 27:701–716
Michie AM, McCaig AM, Nakagawa R, Vukovic M (2010) Death-associated protein kinase (DAPK) and signal transduction: regulation in cancer. FEBS J 277:74–80
Gozuacik D, Kimchi A (2007) Autophagy and cell death. Curr Top Dev Biol 78:217–245
Zalckvar E, Berissi H, Eisenstein M, Kimchi A (2009) Phosphorylation of Beclin 1 by DAP-kinase promotes autophagy by weakening ist interactions with Bcl-2 and Bcl-XL. Autophagy 5:720–722
Saeki K, Yuo A, Okuma E, Yazaki Y, Susin SA, Kroemer G et al (2000) Bcl-2 down-regulation causes autophagy in a caspase-independent manner in human leukemic HL60 cells. Cell Death Differ 7:1263–1269
Harrison B, Kraus M, Burch L, Stevens C, Craig A, Gordon-Weeks P et al (2008) DAPK-1 binding to a linear peptide motif in MAP1B stimulates autophagy and membrane blebbing. J Biol Chem 283:9999–10014
Zhang F, Zhang T, Teng ZH, Zhang R, Wang JB, Mei QB (2009) Sensitization to gamma-irradiation-induced cell cycle arrest and apoptosis by the histone deacetylase inhibitor trichostatin A in non-small cell lung cancer (NSCLC) cells. Cancer Biol Ther 8:823–831
Scuto A, Kirschbaum M, Kowolik C, Kretzner L, Juhasz A, Atadja P et al (2008) The novel histone deacetylase inhibitor, LBH589, induces expression of DNA damage response genes and apoptosis in Ph-acute lymphoblastic leukemia cells. Blood 111:5093–5100
Myeku N, Figueiredo-Pereira ME (2011) Dynamics of the degradation of ubiquitinated proteins by proteasomes and autophagy: association with sequestosome 1/p62. J Biol Chem 286:22426–22440
Wu YT, Tan HL, Huang Q, Kim YS, Pan N, Ong WY et al (2008) Autophagy plays a protective role during zVAD-induced necrotic cell death. Autophagy 4:457–466
Yu L, Alva A, Su H, Dutt P, Freundt E, Welsh S et al (2004) Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 304:1500–1502
Cheng Y, Yang JM (2011) Survival and death of endoplasmic-reticulum-stressed cells: role of autophagy. World J Biol Chem 2:226–231
Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S et al (2006) Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol 26:9220–9231
Acknowledgments
The authors would like to thank Mr. Rudolf Jung and Mrs. Maria Leidenberger for excellent technical assistance and Mrs. Gabriele Krumholz for assisting in conducting animal experiments.
Author information
Authors and Affiliations
Corresponding author
Additional information
An erratum to this article is available at http://dx.doi.org/10.1007/s10495-016-1224-7.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gandesiri, M., Chakilam, S., Ivanovska, J. et al. DAPK plays an important role in panobinostat-induced autophagy and commits cells to apoptosis under autophagy deficient conditions. Apoptosis 17, 1300–1315 (2012). https://doi.org/10.1007/s10495-012-0757-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-012-0757-7