Abstract
While apoptosis regulation has been studied extensively in Drosophila melanogaster, similar studies in other insects, including disease vectors, lag far behind. In D. melanogaster, the inhibitor of apoptosis (IAP) protein DIAP1 is the major negative regulator of caspases, while IAP antagonists induce apoptosis, in part, by binding to DIAP1 and inhibiting its ability to regulate caspases. In this study, we characterized the roles of two IAP antagonists, Michelob_x (Mx) and IMP, in apoptosis in the yellow fever mosquito Aedes aegypti. Overexpression of Mx or IMP caused apoptosis in A. aegypti Aag2 cells, while silencing expression of mx or imp attenuated apoptosis. Addition of recombinant Mx or IMP, but not cytochrome c, to Aag2 cytosolic extract caused caspase activation. Consistent with this finding, AeIAP1 bound and inhibited both initiator and effector caspases from A. aegypti, and Mx and IMP competed with caspases for binding to AeIAP1. However, a difference was observed in the BIR domains responsible for Dronc binding by AeIAP1 versus DIAP1. These findings demonstrate that the mechanisms by which IAP antagonists regulate apoptosis are largely conserved between A. aegypti and D. melanogaster, although subtle differences exist.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis is a genetically controlled mechanism of cell death which is important in development, tissue homeostasis, and innate immunity. Apoptotic cells are characterized by nuclear chromosomal condensation and fragmentation, cell membrane blebbing, and formation of apoptotic bodies. These features of apoptosis are directly caused by caspases, a family of cysteine proteases which are activated following an apoptotic stimulus. A core apoptosis pathway exists in nematodes, insects, and vertebrates (referred to as the intrinsic pathway in vertebrates) which regulates apoptosis by controlling the activation of caspases. In broad terms, the core apoptotic pathway controls the activation of initiator caspases, which occurs by formation of the apoptosome, a complex that promotes initiator caspase dimerization. Activated initiator caspases cleave and activate effector caspases, which cleave key cellular substrates, leading to the stereotypical morphological changes associated with apoptosis.
The major components of the core apoptotic pathway are largely conserved among metazoans, but there are some differences in how caspase activation is regulated between phyla [1, 2]. At this point, our knowledge of apoptosis regulation in insects comes almost entirely from studies done in a single insect species, Drosophila melanogaster. Given the immense diversity which exists among insects, and the varied evolutionary pressures experienced by different insect groups, it is important to examine the process of apoptosis regulation in other insects. For example, even fruitflies and mosquitoes, which are both members of the order Diptera, are separated by ~250 million years of evolution [3], and these two groups of insects have been exposed to very different evolutionary pressures, with mosquitoes having an aquatic larval stage and relying on vertebrate blood feeding for reproduction. In addition, the sequencing of 11 additional Drosophila genomes has revealed that there are differences in the number of caspase genes even within this genus [4].
In D. melanogaster, there are three initiator caspases (Dronc, Dredd, and Strica), and four effector caspases (Drice, DCP-1, Damm and Decay). Among these, Dronc and Drice are the most important in carrying out apoptosis, with DCP-1 playing a minor role that is only evident in certain cell types in the absence of Drice [5, 6]. Dronc is recruited by the oligomerizing factor Ark into an apoptosome complex, resulting in Dronc activation through dimerization followed by auto-cleavage [7–10]. Activated Dronc cleaves and activates Drice, leading to apoptosis [7].
In vertebrates, the release of cytochrome c from mitochondria in response to an intrinsic-type apoptotic stimulus plays an essential role in apoptosome formation because of the role of cytochrome c as an essential cofactor for Apaf-1 (reviewed in [9]). Conversely, in most Drosophila cell types, apoptosome formation and Dronc activation occurs constitutively, without a requirement for cytochrome c [11–14]. However, cytochrome c is required for apoptosis in the fly retina [15] and for caspase activation during sperm development [16]. In addition there appears to be a role for mitochondria in Drosophila apoptosis, although not necessarily in caspase activation [17, 18]. A recent report also indicates that cytochrome c is released from mitochondria and plays an important role in caspase activation in lepidopteran cells [19]. Thus more research is still required to define the role of cytochrome c in apoptosis in Drosophila and other insects.
In most types of unstimulated D. melanogaster cells, apoptosome formation is constitutive and apoptosis is avoided only because the IAP protein DIAP1 is able to bind Dronc and Drice and cause their ubiquitination [20, 21]. Thus, interruption of the expression of DIAP1 protein or the ability of DIAP1 to bind to Dronc or Drice leads to rapid apoptosis, even in the absence of an exogenous apoptotic signal [11, 12, 22, 23]. DIAP1 contains two baculovirus IAP repeat (BIR) domains and a RING domain; the BIR domains are responsible for the physical interaction with caspases, while the RING domain confers E3 ubiquitin ligase activity.
The BIR2 domain of DIAP1 interacts with Dronc by binding to a twelve residue motif located between the prodomain and the large catalytic subunit of Dronc [24]. Effector caspase (Drice and DCP-1) binding is accomplished by the BIR1 domain of DIAP1, which binds to a motif that is revealed following cleavage of the caspases, and the resulting binding blocks enzymatic activity through steric occlusion [25, 26]. After binding, the N-terminal 20 amino acids of DIAP1 are removed by active Drice, which relieves auto inhibition of DIAP1 by the N-terminal region [26, 27]. However, the physical interaction between DIAP1 and Drice or Dronc cannot completely inhibit these caspases. The RING domain of DIAP1 is also required because of its ability to promote caspase ubiquitination, which can result in caspase degradation via the proteosome [20, 24]. In addition, non-degradative polyubiquitination of Drice and DCP-1 by DIAP1 reduces the activation of these caspases by steric interference with binding of substrate [21].
IAP antagonists are a group of proteins which share little sequence similarity other than a highly conserved N-terminal motif called the IAP binding motif (IBM), which allows binding to BIR domains. Elevated levels of IAP antagonists induce apoptosis, in part by competing with caspases for the binding sites of DIAP1. IAP antagonists in D. melanogaster include the cytoplasmic proteins Reaper, Hid, Grim and Sickle. These IAP antagonists have different binding affinities for DIAP1 BIR domains, with Reaper and Grim showing equal preference for binding to the BIR1 and BIR2 domains, while Hid and Sickle have higher affinity for BIR2 than BIR1 [28]. In addition to competing for IAP binding with caspases, IAP antagonists also induce apoptosis through other mechanisms, including stimulation of DIAP1 auto-ubiquitylation [20, 29, 30] and global inhibition of protein translation, a property shared by Reaper and Grim [31]. Reaper, Grim and Sickle also contain a GH3 domain, which can stimulate cell death in the absence of an IBM [32].
Aedes aegypti is an important disease vector, being the principal vector for dengue and yellow fever viruses. The availability of mosquito genome sequences has allowed the initiation of the study of apoptosis in mosquitoes. A number of genes have been identified in A. aegypti and other mosquitoes that share sequence homology with apoptosis regulatory genes in D. melanogaster [33–35]. Expression of Aedes albopictus IAP1 protects mammalian cells from bluetongue virus-induced apoptosis and rescues insect Sf9 cells from apoptosis induced by overexpression of Hid [36]. In addition, silencing of A. aegypti IAP1 (AeIAP1) in adult females caused significant death in mosquitoes, but the mechanism involved was not explored [37]. The core apoptosis pathway appears to be largely conserved in A. aegypti, as silencing of iap1 causes spontaneous apoptosis in A. aegypti Aag2 cells, and apoptosis is dependent on Dronc and Ark, while silencing the effector caspases casps7 or casps8 partially inhibits apoptosis [38]. In addition to AeIAP1, which is an ortholog of DIAP1, the A. aegypti genome also contains orthologs of the IAP-related genes DIAP2, survivin, and Bruce [33, 34].
Two IAP antagonist genes have been identified in A. aegypti, Michelob_x (Mx) and IMP. Like other IAP antagonists, Mx and IMP share little overall homology with each other or other IAP antagonists outside of the IBM, suggesting rapid evolution of these genes [39]. Although Mx was recently shown to contain a partial GH3 domain, it is not capable of inducing apoptosis without the IBM [40]. Expression of these genes in A. albopictus C6/36 cells induces apoptosis, which is dependent on the N-terminal IBM [34, 39, 41]. However, it is not clear whether the A. aegypti IAP antagonists normally function in apoptosis, and if they do, whether the mechanisms involved are similar to those used by IAP antagonists found in D. melanogaster.
In this study, we have examined the proapoptotic functions of Mx and IMP in A. aegypti cells. By silencing expression of mx and imp, we found that both genes are involved in apoptosis. We further characterized the ability of IAP antagonists and A. aegypti caspases to bind IAP1, and examined the competition between IAP antagonists and caspases for binding to IAP1. This work is the first to characterize the interactions between IAP1, IAP antagonists, and caspases in mosquitoes.
Results
Overexpression of IAP antagonists induces apoptosis in mosquito cells
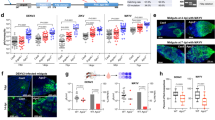
Four IAP antagonist genes, A. aegypti michelob_x (mx), A. albopictus michelob_x (Almx), A. aegypti imp (imp), and D. melanogaster reaper (rpr) have been shown to induce apoptosis when overexpressed in A. albopictus C6/36 cells [34, 39, 41]. To determine whether overexpression of these IAP antagonists also induces apoptosis in the A. aegypti cell line Aag2, we expressed the proteins using a recombinant Sindbis virus (SINV) expression system due to low transfection efficiency in existing A. aegypti cell lines. We previously constructed a series of recombinant SINVs which express Mx and Rpr by inserting the coding regions of these genes into the TE5′2J(TE) SINV infectious clone in either sense or antisense (as a control) orientations [41]. For this study, additional clones were constructed containing AlMx and Imp (sense and antisense). When Aag2 cells were infected at high multiplicity of infection (100 pfu/cell) with TE/Mx, TE/AlMx, TE/IMP or TE/Rpr, the cells underwent apoptosis within the first 24 h p.i., while cells infected with the viruses containing antisense inserts or the parental TE virus did not die, and instead exhibited typical signs of persistent infection. At 24 h p.i., Aag2 cells expressing the IAP antagonists exhibited extensive plasma membrane blebbing and formation of apoptotic bodies (Fig. 1a). Similar results were obtained when C6/36 cells were infected with the viruses expressing IAP antagonists ([41] and data not shown). Thus, overexpression of IAP antagonists induced apoptosis in Aag2 cells, although induction of apoptosis required infecting at a higher multiplicity of infection than in C6/36 cells ([41] and data not shown). This was probably due to the fact that Aag2 cells do not support SINV replication to the same degree as C6/36 cells, which exhibit extremely high levels of SINV replication and lack an antiviral RNA interference response [42, 43].
SINVs expressing Mx, IMP, or Rpr cause apoptosis in Aag2 cells. a–d Cells were mock-infected or infected with the indicated viruses and analysis was conducted at 24-h post infection. Virus names ending in “as” indicate control constructs where the insert is in antisense orientation. a Morphology of infected cells. Aag2 cells were photographed at 400× magnification. b Aag2 cell viability was determined by MTT assay. c, d At 24 h p.i., cell lysate was prepared and caspase activity was determined using Ac-IETD-AFC or Ac-DEVD-AFC as a substrate. Data are shown as mean ± SEM of three independent experiments (*** P < 0.0001; *P < 0.05 by Student’s t test)
To quantify the death of Aag2 cells infected with viruses expressing IAP antagonists, cell viability was quantified by MTT assay, which measures metabolic activity [41]. Although the MTT assay does not specifically measure apoptotic death, apoptotic cells lose metabolic activity within a short time, and the MTT assay is a convenient assay to determine cell viability. Aag2 cells infected with TE/Mx, TE/AlMx, TE/IMP or TE/Rpr had significantly lower viability than mock-infected cells or cells infected with the control viruses containing antisense inserts (Fig. 1b).
We further examined initiator and effector caspase activity in Aag2 cells using the caspase substrates Ac-IETD-AFC and Ac-DEVD-AFC, respectively. Consistent with the extensive morphological signs of apoptosis seen in Fig. 1a, Aag2 cells expressing IAP antagonists exhibited dramatically increased caspase activity compared to uninfected cells and cells infected with control viruses (Fig. 1c, d). These data, along with the morphological characteristics of the dying cells (Fig. 1a), confirmed that cell death induced by overexpression of IAP antagonists was due to apoptosis.
Silencing of IAP antagonists attenuates apoptosis in Aag2 cells
Although overexpression of IAP antagonists induced apoptosis in Aag2 cells, this does not necessarily mean that these proteins are normally involved in carrying out apoptosis. To study the role of A. aegypti IAP antagonists in apoptosis, we used RNA interference (RNAi) to silence expression of mx and imp in Aag2 cells. Transcript levels of mx and imp were dramatically decreased by 12 h after dsRNA treatment and remained low through at least 48 h, as compared with control dsRNA treatment and mock-treated cells (Fig. S1).
Next, we tested whether silencing mx or imp could protect cells from apoptosis stimulated by actinomycin D (Act D) or iap1 dsRNA. Both of these stimuli have been shown to induce apoptosis in many insect cell lines, including Aag2 [12, 22, 23, 38, 44]. After cells were treated with mx or imp dsRNA for 24 h, cells were treated with 50 ng ml−1 of Act D or 5 μg ml−1 iap1 dsRNA for 12 h. Cell viability was then measured using MTT assay (Fig. 2), and RT-PCR was used to confirm that the genes were being silenced (Fig. S2). A pan-caspase inhibitor, Z-VAD-FMK, inhibited the majority of Act D- or iap1 RNAi-stimulated cell death, indicating that the cell death was caspase-dependent. Silencing of mx or imp increased the viability of Act D- or Aeiap1 dsRNA-treated cells, while silencing mx and imp together had an additive effect (Fig. 2). These data indicate that Mx and IMP function in the apoptotic pathway or pathways stimulated by Act D and iap1 dsRNA.
Silencing expression of mx or imp protects Aag2 cells from apoptotic stimuli. Aag2 cells were treated with the indicated dsRNAs for 24 h, and then 50 ng ml−1 of Act D (a) or 5 μg ml−1 of Aeiap1 dsRNA (b) were added. Twelve hours later, cell viability was determined by MTT assay. Data are shown as mean ± SEM of three independent experiments (***P < 0.0001; **P < 0.001 by Student’s t test)
IAP antagonists physically interact with AeIAP1
To study the biochemical interactions between IAP antagonists and IAP1, several recombinant proteins were expressed in and purified from E. coli (Fig. S3). Recombinant Mx, IMP, and Rpr proteins were C-terminally His-tagged, and either included or lacked the N-terminal IBM. In addition, a series of N-terminally GST-tagged AeIAP1 proteins were expressed and purified, including full length AeIAP1, BIR1 alone, BIR2 alone, or BIR1 and 2 together (Fig. S4).
To test whether Mx or IMP interact with AeIAP1, we used pull down assays. Initially, equal amounts of His-tagged recombinant IAP antagonist proteins were incubated with in vitro translated, 35S-labeled AeIAP1, and the resulting protein complexes were purified using the His-tag. Mx, IMP and Rpr were each able to pull down AeIAP1, and this interaction was dependent on the presence of the IBM in the IAP antagonists (Fig. 3a). Mx and IMP appeared to have a higher affinity for AeIAP1 than Rpr, which is consistent with the species of origin of the proteins, and could potentially be due to a single amino acid difference in the IBM sequence of Rpr versus Mx and IMP (Fig. S3a).
A. aegypti IAP antagonists directly bind to AeIAP1, and binding is dependent on the IBM. a The indicated His-tagged recombinant proteins were incubated with 35S-labeled AeIAP1, after which protein complexes were purified using Talon resin and examined by autoradiography. GB is a control protein, the B1 domain of streptococcal protein G [50]. b Streptavidin-agarose beads were incubated with biotinylated peptides containing amino acids 2–6 of Mx and IMP, or 1–11(Met-Hid) of Hid, or buffer. Recombinant proteins (GST-BIR1, GST-BIR2, or GST) were added, and the protein that bound was eluted and detected by immunoblotting with anti-GST antibody. Input represents 10% of the amount of the relevant protein added to the beads
To further define the sites of interaction between AeIAP1 and the IAP antagonists, GST-tagged BIR1 and BIR2 of AeIAP1 were pulled down with a biotin-labeled IBM peptide representing amino acids 2-6 of both Mx and IMP (AIAFYK-biotin). We found that the IBM peptide bound equally well to both BIR1 and BIR2, suggesting that Mx and IMP can bind to either BIR domain (Fig. 3b). Consistent with previous results using IAP antagonist proteins [28, 45–47], the interaction required the first Ala residue of the IBM to be exposed at the N terminus, since a control peptide containing an N-terminal Met residue (Met-Hid) had greatly reduced binding (Fig. 3b).
AeIAP1 inhibits IAP antagonist-induced caspase activation
To test whether AeIAP1 can counteract the proapoptotic activity of IAP antagonists, we examined caspase activity in C6/36 cells and lepidopteran SF-21 cells expressing IAP antagonists with or without AeIAP1 or DIAP1. Expression of Mx, IMP or Rpr caused caspase activation in these two cell lines, while co-expression of AeIAP1 or DIAP1 decreased the level of caspase activity induced by the IAP antagonists (Fig. 4) and also inhibited IAP antagonist-induced apoptosis (data not shown).
AeIAP1 and DIAP1 inhibit IAP antagonist-induced caspase activation. C6/36 cells (a) or SF-21 cells (b) were co-transfected with IAP antagonist constructs and plasmids expressing AeIAP1 or DIAP1, or a control irrelevant plasmid. Cell lysates were prepared at 23 h post-transfection and caspase activity was determined using Ac-DEVD-AFC as a substrate. Data are shown as mean ± SEM of three independent experiments (***P < 0.0001; *P < 0.01; # P < 0.05 by Student’s t test)
AeIAP1 physically interacts with caspases
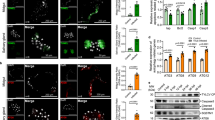
To test whether AeIAP1 physically interacts with caspases, we first examined the interaction between GST-tagged AeIAP1 or its BIR domains and in vitro translated caspase proteins by pull down assays using glutathione beads (Fig. 5a). The effector caspases CASPS7 and CASPS8 mainly interacted with BIR1; however, CASPS7 also interacted to a lesser degree with BIR2 and appeared to have a higher affinity for AeIAP1 than CASPS8, based on the ratio of bound versus input for each protein. Initiator caspase AeDronc bound more strongly to BIR1 although weaker binding to BIR2 was observed (BIR1 and BIR2 bound 70.9% and 45.2% of input, respectively) while AeDredd and Drosophila Dronc only interacted with BIR1 (Fig. 5a). In contrast, Drosophila Dronc bound only to BIR2 of DIAP1, not BIR1 (Fig. S5), as has been previously reported [24].
Interactions between AeIAP1 and initiator or effector caspases. a In vitro translated caspases were incubated with recombinant proteins (GST-BIR1, GST-BIR2, GST-BIR1+2, GST-AeIAP1, or GST) or buffer alone, and protein complexes were purified using glutathione-agarose beads and examined by autoradiography. b Coommassie Blue staining of recombinant proteins used in the pull down assay. c Full-length in vitro translated AeDronc was incubated with active recombinant AeDronc to obtain labeled, processed AeDronc, which was then incubated with GST-BIR1, GST-BIR2, GST-AeIAP1 or GST and pulled down with glutathione-agarose beads and examined by autoradiography. Putative processing sites are predicted based on homology with D. melanogaster Dronc. d Active His-tagged CASPS7 or CASPS8 recombinant protein was incubated with GST-tagged recombinant protein (GST-BIR1, GST-BIR2, GST-AeIAP1 or GST) and pulled down with glutathione-agarose beads. Proteins that bound were eluted and detected by immunoblotting with anti-His antibody
The assay in Fig. 5a used full length (unprocessed) caspase protein. In vitro translated caspase proteins are normally full length, while recombinant caspases expressed in bacteria often undergo auto-activating cleavage due to the effect of concentration during the purification process [8, 48]. To test whether there was a difference in the interaction between AeIAP1 and activated (cleaved) caspases versus full length caspases, we activated in vitro translated AeDronc by incubation with a small amount of recombinant AeDronc, and then tested the interaction between AeIAP1 and activated AeDronc by pull down assay. In vitro translated, full length AeDronc was partially processed by active recombinant AeDronc into 4 fragments, plus some remaining full-length protein (Fig. 5c). Based on the size of the fragments and homology with D. melanogaster Dronc, the 40 KD fragment most likely consisted of the prodomain and large (p20) subunit, the 23 KD fragment represented the large subunit, the 17 KD fragment was the prodomain, and the 11.8 KD fragment was the small (p10) subunit. In D. melanogaster, DIAP1 binds to a motif (residues 114-125) located between the prodomain and the p20 subunit of Dronc [24]. Similarly, binding was only observed between AeIAP1 and full length AeDronc or fragments of AeDronc containing residues 1-147 (Fig. 5c). Binding was observed with both BIR1 and BIR2 but was stronger with BIR1 (BIR1 and BIR2 bound 79.9 and 50.2% of input, respectively), similar to the results observed with full-length AeDronc (Fig. 5a).
To test the interaction between AeIAP1 and processed CASPS7 and CASPS8, purified CASPS7 or CASPS8 from E. coli were incubated with AeIAP1 and interacting caspase proteins were visualized by western blotting with anti-His antibody (Fig. 5d). The interaction between active CASPS7 and BIR1 was stronger than between CASPS8 and BIR1. Interaction was observed between the p10 subunit of CASPS7 and BIR1, while a weak interaction between CASPS8 and BIR1 was observed for the partially processed p20 + p10 intermediate. CASPS7 interacted only with BIR1 (Fig. 5d), consistent with the results obtained using unprocessed CASPS7 and CASPS8 (Fig. 5a). Interestingly, active CASPS7 was not pulled down by full length GST-AeIAP1. Since D. melanogaster Drice cleaves DIAP1 after residue 18 [26], a possible explanation for the lack of interaction with GST-AeIAP1 is that active CASPS7 may cleave AeIAP1 near its N terminus, removing the GST domain and not allowing purification of AeIAP1-associated proteins. A similar cleavage site (DFTD18) is conserved in AeIAP1. The GST-BIR1 construct used in this experiment contains residues 24–118, and so did not include this putative cleavage site (Fig. S4).
AeIAP1 inhibits the activity of initiator and effector caspases
Having shown that AeIAP1 binds to AeDronc, CASPS7, and CASPS8, we next investigated whether AeIAP1 is able to inhibit the activity of these caspases, both in cells and in vitro. After C6/36 cells were transfected with CASPS7, CASPS8 or AeDronc, cells exhibited higher activity against the effector caspase substrate Ac-DEVD-AFC than mock-transfected cells, which received only transfection reagent (Fig. 6a). Cotransfection of AeIAP1 or DIAP1 with each of the caspases decreased the caspase activity (Fig. 6a). However, since endogenous caspases were probably activated by overexpressed CASPS7, CASPS8, or AeDronc, it was not possible to conclude which caspase(s) were directly inhibited by AeIAP1 based on these results.
AeIAP1 and DIAP1 inhibit the activity of AeDronc and effector caspases CASPS7 and CASPS8. a C6/36 cells were transfected with CASPS7 or CASPS8 constructs, with or without a plasmid expressing AeIAP1 or DIAP1 or a control plasmid. Cell lysates were prepared at 23 h post transfection and caspase activity was determined using Ac-DEVD-AFC as a substrate. b–d Recombinant protein (AeIAP1, DIAP1, P35 or GST) (10 μM) or Z-VAD-FMK (100 μM) was incubated with 0.5 μM active caspases (AeDronc, CASPS7 or CASPS8) at 30°C for 1 h. Caspase activity was determined using Ac-IETD-AFC or Ac-DEVD-AFC as a substrate. Data are shown as mean ± SEM of three independent experiments (***P < 0.0001; **P < 0.001; NS non-significant by Student’s t test)
To examine the ability of AeIAP1 to inhibit caspases directly, we purified recombinant IAP and caspase proteins from E. coli and performed in vitro caspase assays. AeIAP1 and DIAP1 inhibited AeDronc, while the baculovirus P35 protein did not (Fig. 6b), which is consistent with the known specificity of P35 for effector caspases [48, 49]. AeIAP1 and DIAP1 also inhibited the activity of CASPS7 and CASPS8, which were also inhibited by P35 and Z-VAD-FMK (Fig. 6c, d). These data demonstrate that AeIAP1 functions as a caspase inhibitor through direct physical interaction with AeDronc, CASPS7, and CASPS8.
IAP antagonists release initiator and effector caspases from inhibition by AeIAP1
In D. melanogaster, it has been shown that IAP antagonists are able to compete with caspases for binding to DIAP1 [24, 26]. To test whether this can also occur in A. aegypti, we performed an in vitro competition assay. The ability of AeIAP1 to inhibit AeDronc was reduced by the addition of IAP antagonists Mx, IMP or Rpr, as compared with the addition of IAP antagonist IBM mutants or control GB protein [50] (Fig. 7a). Interestingly, Mx exhibited stronger ability to replace AeDronc from AeIAP1 than IMP, as demonstrated in the in vitro caspase assay and pull down assay (Fig. 7a, b). The ability of AeIAP1 to inhibit CASPS7 was also reduced upon addition of recombinant Mx, IMP, or Rpr (Fig. 7c). These results indicated that IAP antagonists can compete with caspases for binding to the BIR domains of AeIAP1, and thus release caspases from inhibition by AeIAP1.
IAP antagonists release AeDronc from inhibition by AeIAP1. a Recombinant GST-AeIAP1 (10 μM) was incubated with 0.5 μM of active AeDronc, followed by addition of recombinant IAP antagonists (Mx, IMP or Rpr) (10 μM) or control GB protein (10 μM). After 1 h incubation, caspase activity was determined using Ac-IETD-AFC as a substrate. Data are shown as mean ± SEM of three independent experiments (***P < 0.0001; *P = 0.0255 by Student’s t test). b In vitro translated AeDronc was incubated with recombinant GST-AeIAP1 (2.5 μM) for 1 h, after which 2.5 μM IAP antagonists (Mx, Imp or Rpr) or corresponding mutants or GB control protein were added and incubated for 1 h. Radioactive labeled AeDronc bound to GST-AeIAP1 was pulled down with glutathione agarose beads and examined by autoradiography. c The same experiment in a was performed using CASPS7 instead of AeDronc
The role of cytochrome c in caspase activation
The role of cytochrome c in apoptosis in D. melanogaster and other insects remains controversial. To test whether cytochrome c is involved in caspase activation in Aag2 cells, we homogenized Aag2 cells in the presence of a high concentration of sucrose to prevent the rupture of mitochondria, and separated the cell lysate into P10 (heavy membrane) and S100 (cytosolic) fractions. Cytochrome c was only detected in the P10 fraction (Fig. S6), consistent with its mitochondrial localization and with previous reports using D. melanogaster cells [13, 14, 51, 52]. As expected, the S100 fraction from unstimulated Aag2, C6/36 or S2 cells did not exhibit significant caspase activity (Figs. 8, S7). However, incubation with recombinant Mx or IMP protein induced caspase activation in S100 from Aag2 or C6/36 cells (Figs. 8a, S7), similar to previous results obtained using S2 cells [14]. These results indicated that an excess of IAP antagonist protein was able to activate caspases in S100, presumably by relieving caspase inhibition by AeIAP1.
IAP antagonists induce caspase activation in mosquito cell cytoplasmic lysate, and cytochrome C does not. a Fifty μg S100 lysate isolated from Aag2 cells was incubated with recombinant protein (Mx or IMP or control GB) (10 μM) prior to determining caspase activity using Ac-IETD-AFC as a substrate. b Fifty μg S100 lysate was incubated with cytochrome C (2 μg), ATP (2 mM) and MgCl2 (1 mM), and caspase activity was determined using Ac-DEVD-AFC as a substrate. Data are shown as mean ± SEM of three independent experiments (***P < 0.0001; NS non-significant by Student’s t test)
We next asked whether addition of cytochrome c was capable of activating caspases in cytosolic extract. Addition of purified cytochrome c and dATP to S100 from mammalian cells causes apoptosome formation and caspase activation, but exogenous cytochrome c does not cause caspase activation in S100 from D. melanogaster embryos or S2 cells [13, 14, 51]. Mixing S100 from Aag2 cells or S2 cells with purified bovine cytochrome c protein and dATP did not show increased caspase activity as compared to mock treatment (Fig. 8b), which indicated that cytochrome c was not able to stimulate caspase activation in S100 from Aag2 cells. It is possible that the source of the cytochrome c used in the assay (bovine) could be the cause of this negative result. However, we consider this unlikely since bovine cytochrome c can activate caspases in lepidopteran cell extract [19], and cytochrome c from D. melanogaster is able to cause caspase activation in extracts from human cells [14].
Discussion
The molecular pathways that regulate apoptosis are largely conserved among metazoans; however, there are significant differences in how apoptosis is regulated between nematodes, insects, and mammals. To date, apoptosis has only been studied in detail in a single insect, D. melanogaster. Although initial characterizations of a few genes involved in mosquito apoptosis have been reported, we are still in the early stages of understanding apoptosis regulation in mosquitoes. In this study we explored the mechanisms of apoptosis regulation in a cell line from the mosquito A. aegypti.
Overexpression of the IAP antagonists Rpr, Mx, AlMx and IMP in Aag2 cells induced apoptosis, demonstrating these proteins are sufficient to induce apoptosis. Furthermore, they are also involved in apoptosis, since silencing expression of IAP antagonists Mx or IMP in Aag2 cells reduced apoptosis induced by Act D or Aeiap1 RNAi. Silencing Mx and IMP together had an additive effect, but still did not provide complete protection. This could be due to incomplete silencing of these genes at the protein level; however, it is also quite possible that additional IAP antagonists exist in the A. aegypti genome which have not yet been discovered due to the low level of sequence similarity between most IAP antagonists. Also consistent with this is the observation that silencing of Mx or IMP provided only temporary (up to around 12 h) protection from Act D- and Aeiap1 silencing-induced death, even when both Mx and IMP were silenced. By 24 h, the protective effect was no longer evident, which is not the case when other genes such as Aedronc are silenced ([38] and data not shown).
IAP antagonists are thought to act upstream of IAPs, which raises the question why we observed partial protection against apoptosis induced by silencing Aeiap1 when we co-silenced Mx or IMP (Fig. 2b). One possibility is that the silencing of Aeiap1 is incomplete, such that reducing the levels of Mx and/or IMP allows lower levels of AeIAP1 to still be able to function and prevent caspase activation. However, it is also possible that Mx and IMP function both upstream and downstream of AeIAP1.
The IBM sequence is indispensable for the interaction between IAP antagonists and IAPs, with the first 4 amino acids being especially critical [2]. The IBM sequences of Mx and IMP are identical over the first 5 amino acids, raising the question of whether Mx and IMP can bind equally to AeIAP1. We found that D. melanogaster Rpr, whose IBM differs from that of Mx and IMP in the second residue, had less affinity for AeIAP1 than A. aegypti Mx and IMP (Fig. 3a). Even though IMP and Mx showed similar binding affinity to AeIAP1 and similar ability to displace CASPS7 from AeIAP1, Mx displaced AeDronc from AeIAP1 better than IMP (Fig 7a; b). However, further experiments are necessary to determine whether Mx and IMP are redundant, or whether they play different roles in apoptosis, as seen for Drosophila IAP antagonists.
Our results indicate that there is a difference in the BIRs responsible for binding to Dronc in AeIAP1 versus DIAP1. Effector caspases CASPS7 and CASPS8 only interacted with BIR1 but not BIR2 of AeIAP1, similar to the way that DIAP1 interacts with Drice and DCP-1. Binding was observed between AeDronc and both BIR1 and BIR2, but binding was stronger to BIR1. However, Drosophila Dronc only binds to BIR2 of DIAP1 ([24]; Fig. S5). Interestingly, Drosophila Dronc also bound only to the BIR1 domain of AeIAP1 in our assays (Fig. 5a). These results suggest that the detailed mechanism of Dronc binding may be slightly different between AeIAP1 and DIAP1, involving mainly BIR1 in AeIAP1 instead of BIR2. However, both IAPs appear to bind to a similar region in AeDronc and Drosophila Dronc, which is a motif located between the prodomain and the large subunit [24]. While binding of AeIAP1 was able to inhibit AeDronc, CASPS7 and CASPS8 in vitro, whether or not ubiquitination is involved in caspase inhibition by AeIAP1 requires further study.
The role of cytochrome c in insect apoptosis is still not clear. While most Drosophila cell types do not appear to require cytochrome c for apoptosis, it is required in the retina and during spermatogenesis [15, 16]. Furthermore, a recent study suggest that cytochrome c plays a role in apoptosis in a lepidopteran cell line [19], suggesting that this aspect of apoptosis regulation may be different in other insects. Our results suggest that cytochrome c is not required for caspase activation in Aag2 cells, since addition of purified cytochrome c did not induce caspase activation in cytoplasmic extract, while caspases were activated by addition of recombinant Mx or IMP. However, additional research will need to be done before a firm conclusion can be drawn about the role of cytochrome c in A. aegypti.
Despite the immense and rich diversity that exists among insects, prior to this and another recent related study [38], our understanding of the molecular mechanisms involved in apoptosis regulation in insects was mostly based on a single species, D. melanogaster. A more detailed understanding of the mechanisms of apoptosis regulation in disease vectors such as mosquitoes will not only enhance our knowledge of apoptosis regulation in insects, but also help us to better understand the interactions between pathogens and mosquitoes and allow for enhanced disease control methods targeting the apoptosis pathway in disease vectors.
Materials and methods
Cell culture
Aedes aegypti Aag2 cells (obtained from Carol Blair, Colorado State University) and D. melanogaster S2 cells (Invitrogen) were maintained in Schneider’s medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Atlanta Biologicals). A. albopictus C6/36 cells were cultured in Leibovitz’s medium (Invitrogen) containing 10% FBS. Spodoptera frugiperda SF-21 cells were propagated in TC-100 medium (Invitrogen) with 10% FBS. BHK-21 cells were propagated in Dulbecco’s modified Eagle’s medium (DMEM) (Mediatech) supplemented with 10% FBS. Aag2, C6/36, S2 and SF-21 cells were maintained at 27°C, and BHK21 cells were cultured at 37°C in 8% CO2.
Recombinant viruses
The coding regions of A. aegypti imp and A. albopictus mx cDNAs were amplified by PCR and cloned into the SINV DNA infectious clone pTE5′2J (TE) in the sense and antisense orientation. Construction of TE/Rpr, TE/Mx and TE viruses have been described previously [41]. Viruses were generated using BHK-21 cells and virus titres were determined by tissue culture infectious dose (TCID50) assay in BHK-21 cells as previously described [41]. Cells were infected as previously described [41] except that infections were done at an moi of 100/pfu/cell.
Caspase activity assay
Caspase activity was measured using Ac-DEVD-AFC or Ac-IETD-AFC (Enzyme Systems Products) as a substrate. Cell lysate was prepared and assays were performed as previously described [41].
RNAi procedure
Full-length ORFs were PCR-amplified such that T7 polymerase promoter sites were incorporated onto both ends. The PCR products were used as template to synthesize dsRNA using AmpliScribe T7 High Yield Transcription kit (EPICENTRE Biotechnologies). One million Aag2 cells were plated in Schneider’s medium without serum and the dsRNA was added directly to the medium at a concentration of 10 μg ml−1. After the cells were incubated with dsRNA for 1–2 h, FBS was added to a final concentration of 10%. The cells were incubated for 24 h before proceeding with the rest of the experiments.
RT-PCR
Aag2 cells were treated with dsRNA and at the indicated time points after treatment, total RNA was isolated using Trizol reagent (Invitrogen). One μg total RNA was used as a template to generate cDNA in a 10 μl reverse transcriptase reaction (Promega) with poly(dT) primer. Two microliters of cDNA was then used as template for PCR with gene-specific primers.
Transfection assay
The coding regions of Aeiap1, CASPS7, CASPS8, Aedronc, mx, imp, and Drosophila rpr and diap1 were inserted under control of a hsp70 promoter in the pHSP70PLVI+ vector [53]. One million SF-21 or C6/36 cells were transfected with 5 μg of each plasmid and 5 μl lipofectamine in medium without serum. After 6 h, cells were washed 3 times and replaced with medium plus 10% FBS. Twenty hours later, cells were incubated at 42°C for 30 min and harvested 3 h after the heat shock.
Recombinant protein and peptide preparation
N-terminally tagged GST-AeIAP1, GST-AeIAP1 BIR1 (containing amino acid residues 24-118 of AeIAP1), GST-AeIAP1 BIR2 (residues 188-278) and GST-AeIAP1 BIR1+2 (residues 17-349) were cloned into the pGEX-3x vector. GST-DIAP1 was inserted in the pGEX-4T-1 vector. C-terminally tagged CASPS7-His6, CASPS8-His6, Rpr-His6, Rpr-IBM-His6, IMP-His6, IMP-IBM-His6, Mx-His6 and Mx-IBM-His6 were cloned into pET23b. His6-AeDronc (N-terminally tagged) was inserted into pET32. GB-His6/pET30a(+) was a gift from Katsura Asano (Kansas State University). These vectors were transformed into BL21pLysS(DE)3 E. coli (Stratagene). Bacterial cultures were grown at room temperature to OD600 = 0.6, at which time they were induced with 0.1 M IPTG for 3 h. Proteins were purified using either glutathione-agarose beads (Sigma) for GST-tagged proteins or with Talon Metal Affinity Resin (Clontech) for the His6-tagged proteins, according to the instructions of the manufacturer. The IBM peptide (AIAFYK-biotin) was synthesized by New England Peptide and the Met-Hid peptide (MAVPFYLPEGGK-biotin) was previously described [14, 47].
Pull down assay
To examine the interaction of IAP antagonists (Mx, IMP and Rpr) with full length AeIAP1, 3.5 μM of recombinant His-tagged IAP antagonists or corresponding mutants were incubated with 10 μl of in vitro translation reaction containing AeIAP1 in a 100 μl final reaction with NP-40 lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 1 mM dithiothreitol, 1 mM phenylmethyl sulfonyl fluoride) at 30°C for 1 h. Full length AeIAP1 was made using the TNT T7/SP6 Coupled Reticulocyte Lysate System (Promega). After incubation, the reaction was mixed with 40 μl of Talon Metal Affinity Resin (Clontech) and rocked at 4°C for 2 h. The beads were washed three times with NP-40 lysis buffer. The bound proteins were pulled down with beads and dissolved in Laemmli buffer by heating the samples at 100°C for 5 min. Proteins were separated by 15% SDS-PAGE. After electrophoresis, gels were treated with fixing solution (30% methanol and 10% acetic acid) for 1 h and soaked in 16% salicylic acid for 5 min. After drying, gels were exposed to film at −80°C.
To examine IAP binding motif (IBM) interaction with BIR domains of AeIAP1, 5 μg of IBM peptide (AIAFYK-biotin) was added to 40 μl of streptavidin-conjugated beads (Sigma) with NP-40 lysis buffer up to 500 μl as a final volume and rocked for 1 h at 4°C to allow the peptide to bind to the beads. The beads were washed three times with NP-40 lysis buffer and incubated with 0.5 μM of GST-AeIAP1 BIR1 or BIR2 recombinant protein in 500 μl of NP-40 lysis buffer for 1 h at 4°C. After incubation, the beads were washed three times with NP-40 lysis buffer and bound protein was dissolved in Laemmli buffer by heating beads at 100°C for 5 min. Proteins were analyzed by immunoblotting using anti-GST monoclonal antibody conjugated to horse radish peroxidase (Santa Cruz Biotechnology) at 1:1000.
To test caspase interaction with AeIAP1, 10 μl of 35S-labeled in vitro translation reaction for each caspase were incubated with 2.5 μM of a GST-tagged protein in a 100 μl reaction with NP-40 lysis buffer at 30°C for 1 h. After incubation, the reaction was added to 40 μl of glutathione-agarose beads (Sigma) and rocked overnight at 4°C. The beads were washed three times with NP-40 lysis buffer and the bound proteins were separated by SDS-PAGE. The gels were dried and exposed to film at −80°C.
To study the interaction of active caspases and AeIAP1, cleaved 35S labeled AeDronc or active CASPS7 or CASPS8 recombinant proteins were used. To obtain cleaved 35S labeled AeDronc, 10 μl of full length in vitro translated AeDronc was incubated with 0.5 μM active recombinant AeDronc at 30°C for 1 h. The active 35S labeled AeDronc and the active CASPS7 or CASPS8 recombinant proteins were incubated with 0.5 μM GST-tagged protein (GST-BIR1, GST-BIR2, GST-AeIAP1 or GST) at 30°C for 1 h, and further incubated with 40 μl of glutathione-agarose beads for 1 h at 4°C, followed by washing and visualization as described above.
Viability assays
The viability of Aag2 cells was measured by MTT assay as previously described [41].
Cell fractionation
Cell fractionation was performed as previously described [14]. Cells were harvested by centrifugation at 500g for 5 min. Cell pellets were re-suspended in 1 ml of Caspase Buffer A [14] (20 mM Hepes KOH, pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 1 mM DTT in 250 mM sucrose supplemented with protease inhibitor cocktail). Cells were lysed using 50 strokes of a Dounce homogenizer and cells were further centrifuged at 500g for 10 min at 4°C. After the supernatant was centrifuged at 10,000g for 15 min at 4°C, the pellet (P10) was re-suspended in 1 ml of Caspase Buffer A with 10% glycerol and stored at −80°C. After the supernatant was further centrifuged at 100,000g for 30 min at 4°C, 10% glycerol was added to the resulting supernatant (S100) and stored at −80°C. The ability of cytochrome c purified from bovine heart to cause caspase activation was assayed as previously described [14].
Immunoblotting
Protein or cell lysate were mixed with SDS-PAGE loading buffer, heated at 100°C for 5 min and resolved by 15% SDS-PAGE, and then transferred to PVDF membrane. The GST-tagged or His-tagged recombinant proteins were detected with 1:1000 anti-GST-HRP or anti-His-HRP antibody (Santa Cruz Biotechnology). The P10 and S100 fractions from cells were immunoblotted for cytochrome c using 1:1000 anti-cytochrome c antibody ab28137 (Abcam) and 1:5000 anti-mouse IgG-HRP.
References
Hay BA, Guo M (2006) Caspase-dependent cell death in Drosophila. Annu Rev Cell Dev Biol 22:623–650
Yan N, Shi Y (2005) Mechanisms of apoptosis through structural biology. Annu Rev Cell Dev Biol 21:35–56
Grimaldi D, Engel MS (2005) Evolution of the insects. Cambridge University Press, Cambridge
Bryant B, Ungerer MC, Liu Q, Waterhouse RM, Clem RJ (2010) A caspase-like decoy molecule enhances the activity of a paralogous caspase in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol 40:516–523
Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, Guo M, Baehrecke EH, Hay BA (2006) The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development 133:3305–3315
Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A (2006) The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ 13:1697–1706
Dorstyn L, Kumar S (2008) A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death Differ 15:461–470
Muro I, Monser K, Clem RJ (2004) Mechanism of Dronc activation in Drosophila cells. J Cell Sci 117:5035–5041
Shi Y (2008) Apoptosome assembly. Methods Enzymol 442:141–156
Snipas SJ, Drag M, Stennicke HR, Salvesen GS (2008) Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ 15:938–945
Wang SL, Hawkins CJ, Yoo SJ, Muller H-AJ, Hay BA (1999) The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell 98:453–463
Muro I, Hay BA, Clem RJ (2002) The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem 277:49644–49650
Dorstyn L, Mills K, Lazebnik Y, Kumar S (2004) The two cytochrome c species, DC3 and DC4, are not required for caspase activation and apoptosis in Drosophila cells. J Cell Biol 167:405–410
Means JC, Muro I, Clem RJ (2006) Lack of involvement of mitochondrial factors in caspase activation in a Drosophila cell-free system. Cell Death Diffeer 13:1222–1234
Mendes CS, Arama E, Brown S, Scherr H, Srivastava M, Bergmann A, Steller H, Mollereau B (2006) Cytochrome c-d regulates developmental apoptosis in the Drosophila retina. EMBO Rep 7:933–939
Arama E, Agapite J, Steller H (2003) Caspase activity and a specific cytochrome C are required for sperm differentiation in Drosophila. Dev. Cell 4:687–697
Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K (2007) Mitochondrial disruption in Drosophila apoptosis. Dev Cell 12:793–806
Goyal G, Fell B, Sarin A, Youle RJ, Sriram V (2007) Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell 12:807–816
Kumarswamy R, Seth RK, Dwarakanath BS, Chadna S (2009) Mitochondrial regulation of insect cell apoptosis: Evidence for permeability transition pore-independent cytochrome-c release in the Lepidopteran Sf9 cells. Int J Biochem Cell Biol 41:1430–1440
Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, Steller H, Meier P (2002) The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol 4:445–450
Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KT, Elliott R, Zvelebil M, Blagoev B, Bergmann A, Meier P (2008) Inactivation of effector caspases through nondegradative polyubiquitylation. Mol Cell 32:540–553
Igaki T, Yamamoto-Goto Y, Tokushige N, Kanda H, Miura M (2002) Downregulation of DIAP1 triggers a novel Drosophila cell death pathway mediated by Dark and Dronc. J Biol Chem. 277:23103–23106
Zimmermann KC, Ricci J-E, Droin NM, Green DR (2002) The role of ARK in stress-induced apoptosis in Drosophila cells. J Cell Biol 156:1077–1087
Chai J, Yan N, Huh JR, Wu J-W, Li W, Hay BA, Shi Y (2003) Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol 10:892–898
Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P (2005) IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 7:70–77
Yan SJ, Wu JW, Chai J, Li W, Shi Y (2004) Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim. Nat Struct Mol Biol 11:420–428
Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, Meier P (2003) Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol 5:373–376
Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P (2003) IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J 22:6642–6652
Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H (2002) Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol 4:432–438
Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, Feldman RMR, Clem RJ, Müller H-AJ, Hay BA (2002) Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol 4:416–424
Colon-Ramos DA, Shenvi CL, Weitzel DH, Gan EC, Matts R, Cate J, Kornbluth S (2006) Direct ribosomal binding by a cellular inhibitor of translation. Nat Struct Mol Biol 13:103–111
Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M (2002) GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. EMBO J 21:3327–3336
Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, Blandin S, Christensen BM, Dong Y, Jiang H, Kanost MR, Koutsos AC, Levashina EA, Li J, Ligoxygakis P, Maccallum RM, Mayhew GF, Mendes A, Michel K, Osta MA, Paskewitz S, Shin SW, Vlachou D, Wang L, Wei W, Zheng L, Zou Z, Severson DW, Raikhel AS, Kafatos FC, Dimopoulos G, Zdobnov EM, Christophides GK (2007) Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316:1738–1743
Bryant B, Blair CD, Olson KE, Clem RJ (2008) Annotation and expression profiling of apoptosis-related genes in the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol 38:331–345
Bartholomay LC, Waterhouse RM, Mayhew GF, Campbell CL, Michel K, Zou Z, Ramirez JL, Das S, Alvarez K, Arensburger P, Bryant B, Chapman SB, Dong Y, Erickson SM, Karunaratne SH, Kokoza V, Kodira CD, Pignatelli P, Shin SW, Vanlandingham DL, Atkinson PW, Birren B, Christophides GK, Clem RJ, Hemingway J, Higgs S, Megy K, Ranson H, Zdobnov EM, Raikhel AS, Christensen BM, Dimopoulos G, Muskavitch MA (2010) Pathogenomics of Culex quinquefasciatus and meta-analysis of infection responses to diverse pathogens. Science 330:88–90
Li Q, Li H, Blitvich BJ, Zhang J (2007) The Aedes albopictus inhibitor of apoptosis 1 gene protects vertebrate cells from bluetongue virus-induced apoptosis. Insect Mol Biol 16:93–105
Pridgeon JW, Zhao L, Becnel JJ, Clark GG, Linthicum KJ (2008) Developmental and environmental regulation of AaeIAP1 transcript in Aedes aegypti. J Med Entomol 45:1071–1079
Liu Q, Clem RJ (2010) Defining the core apoptosis pathway in the mosquito disease vector Aedes aegypti: the roles of iap1, ark, dronc, and effector caspases. Apoptosis. doi:10.1007/s10495-010-0558-9
Zhou L, Jiang G, Chan G, Santos CP, Severson DW, Xiao L (2005) Michelob_x is the missing inhibitor of apoptosis protein antagonist in mosquito genomes. EMBO Rep 6:769–774
Bryant B, Zhang Y, Zhang C, Santos CP, Clem RJ, Zhou L (2010) A lepidopteran orthologue of reaper reveals functional conservation and evolution of IAP antagonists. Insect Mol Biol 18:341–351
Wang H, Blair CD, Olson KE, Clem RJ (2008) Effects of inducing or inhibiting apoptosis on Sindbis virus replication in mosquito cells. J Gen Virol 89:2651–2661
Brackney DE, Scott JC, Sagawa F, Woodward JE, Miller NA, Schilkey FD, Mudge J, Wilusz J, Olson KE, Blair CD, Ebel GD (2010) C6/36 Aedes albopictus cells have a dysfunctional antiviral RNA interference response. PLoS Negl Trop Dis 4:e856
Scott JC, Brackney DE, Campbell CL, Bondu-Hawkins V, Hjelle B, Ebel GD, Olson KE, Blair CD (2010) Comparison of dengue virus type 2-specific small RNAs from RNA interference-competent and -incompetent mosquito cells. PLoS Negl Trop Dis 26:e848
Crook NE, Clem RJ, Miller LK (1993) An apoptosis-inhibiting baculovirus gene with a zinc finger-like motif. J Virol 67:2168–2174
Liu Z, Sun C, Olejniczak ET, Meadows RP, Betz SF, Oost T, Herrmann J, Wu JC, Fesik SW (2000) Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 408:1004–1008
Wu G, Chai J, Suber TL, Wu J-W, Du C, Wang X, Shi Y (2000) Structural basis of IAP recognition by Smac/DIABLO. Nature 408:1008–1012
Wright CW, Clem RJ (2002) Sequence requirements for Hid binding and apoptosis regulation in the baculovirus inhibitor of apoptosis Op-IAP: Hid binds Op-IAP in a manner similar to Smac binding of XIAP. J Biol Chem 277:2454–2462
Hawkins CJ, Yoo SJ, Peterson EP, Wang SL, Vernooy SY, Hay BA (2000) The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem 275:27084–27093
Meier P, Silke J, Leevers SJ, Evan GI (2000) The Drosophila caspase DRONC is regulated by DIAP1. EMBO J 19:598–611
Reibarkh M, Yamamoto Y, Singh CR, del Rio F, Fahmy A, Lee B, Luna RE, Ii M, Wagner G, Asano K (2008) Eukaryotic initiation factor (eIF) 1 carries two distinct eIF5-binding faces important for multifactor assembly and AUG selection. J Biol Chem 283:1094–1103
Varkey J, Chen P, Jemmerson R, Abrams JM (1999) Altered cytochrome c display precedes apoptotic cell death in Drosophila. J Cell Biol 144:701–710
Dorstyn L, Read S, Cakouros D, Huh JR, Hay BA, Kumar S (2002) The role of cytochrome c in caspase activation in Drosophila melanogaster cells. J Cell Biol 156:1089–1098
Clem RJ, Miller LK (1994) Control of programmed cell death by the baculovirus genes p35 and iap. Mol Cell Biol 14:5212–5222
Acknowledgments
We thank Katsura Asano (Kansas State University) for providing the GB expression plasmid and Taryn Penabaz for culturing cells. This work was supported by NIH grants R21 AI067642 (to R.J.C.) and P20 RR16475 from the BRIN program of the National Center for Research Resources, by the Terry C. Johnson Center for Basic Cancer Research, and by the Kansas Agricultural Experiment Station. This is contribution number 10-343-J from the Kansas Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, H., Clem, R.J. The role of IAP antagonist proteins in the core apoptosis pathway of the mosquito disease vector Aedes aegypti . Apoptosis 16, 235–248 (2011). https://doi.org/10.1007/s10495-011-0575-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-011-0575-3