Abstract
Whether viral pathogens that induce ER stress responses benefit the host or the virus remains controversial. In this study we show that betanodavirus induced ER stress responses up-regulate GRP78, which regulates the viral replication and host cellular mitochondrial-mediated cell death. Betanodavirus (redspotted grouper nervous necrosis virus, RGNNV) infection resulted in the following increased ER stress responses in fish GF-1 grouper fin cells: (1) IRE-1 and ATF-6 sensors at 48 h post-infection (p.i.) that up-regulated chaperone protein GRP78; (2) activation of caspase-12; and (3) PERK phosphorylation and down-regulation of Bcl-2. Analyses of GRP78 functions during viral replication using either loss-of-function or gain-of-function approaches showed that GRP78 over-expression also enhanced viral replication and induced cell death. Then, we found that zfGRP78 localization gradually increased in mitochondria after RGNNV infection by EGFP tagging approach. Furthermore, zfGRP78 can interact with viral RNA-dependent RNA polymerase (RdRp) by using immunofluorescent and immunoprecipitation assays. Finally, we found that blocking GRP78-mediated ER signals can reduce the viral death factors protein α and protein B2 expression and decrease the Bcl-2 down-regulation mediated mitochondria-dependent cell death, which also enhances host cellular viability. Taken together, our results suggest that RGNNV infection and expression can trigger ER stress responses, which up-regulate the chaperone GRP78 at early replication stage. Then, GRP78 can interact with RdRp that may enhance the viral replication for increasing viral death factors’ expressions at middle-late replication stage, which can enhance mitochondrial-mediated cell death pathway and viral spreading. These results may provide new insights into the mechanism of ER stress-mediated cell death in RNA viruses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Betanodaviruses cause viral nervous necrosis (VNN), an infectious neuropathological condition of fish that is characterized by necrosis in the central nervous system, including the brain and retina. VNN necrosis is accompanied by clinical signs, such as abnormal swimming behavior and development of a darker body color [1]. This disease can cause mass mortality in the larval and juvenile populations of several teleost species and is of global economic importance [2]. Despite their potentially severe impact on the aquaculture industry, betanodaviruses have not been well studied. Characterizing virally-mediated ER stress regulation processes would aid in deciphering the mechanism(s) of viral pathogenesis and infection.
The family Nodaviridae is comprised of the genera Alphanodavirus and Betanodavirus. Alphanodaviruses predominantly infect insects, while Betanodaviruses predominantly infect fish [1, 3, 4]. Nodaviruses are small, non-enveloped, spherical viruses with bipartite positive-sense RNA genomes (RNA1 and RNA2) that are capped, but not polyadenylated [1]. RNA1 encodes an approximately 110 kDa non-structural protein that has been designated RNA-dependent RNA polymerase (RdRp) or protein A. This protein is vital for viral genome replication. RNA2 encodes a 42 kDa capsid protein, so-called protein α [4, 5], which may also function in inducing cell death [6, 7]. Nodaviruses also synthesize RNA3, a sub-genomic RNA species, from the 3′ terminus of RNA1. RNA3 contains two putative open reading frames that potentially encode an 111 amino acid, protein B1, and a 75 amino acid, protein B2 [1, 8]. It was recently shown for betanodavirus that B1 has an anti-necrotic death function during early replication stages [8]. It has been speculated that B2 could function in two different roles: as a potent suppressor of host siRNA silencing [9, 10] or a necrotic death factor [11].
The endoplasmic reticulum (ER) is a eukaryotic organelle that plays a vital role in a variety of cellular functions, including post-translational modifications, synthesis and folding of membrane and secretary proteins, metabolism, cell calcium storage and apoptosis [12–15]. Several stimuli can disrupt ER homeostasis and induce ER stress, such as the accumulation of unfolded or misfolded proteins, oxidative stress, perturbation of calcium homeostasis and viral infection [16, 17]. GRP78 is currently regarded as the master regulator of the unfolding protein response (UPR) pathway and is known to regulate the activation of PKR-like ER kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1(IRE1) [18–20].
Some studies have shown that viruses can induce ER stress [21, 22]. ER stress, in turn, regulates viral replication and pathogenesis that decide between cell survival and cell death [12, 23]. The Tula virus, hepatitis C virus (HCV), Japanese encephalitis virus (JEV) and mouse retrovirus induce an ER stress response via up-regulation of the ER chaperone protein GRP78 [12, 15, 24, 25]. Respiratory syncytial virus induces apoptotic cell death via caspase-12 activation [26]. However, not much is known regarding ER stress responses to aquatic viral infection. The mechanism(s) by which cells undergo apoptosis or are rescued from ER stress is also not well understood. Studies of these mechanisms could be useful for elucidating the pathways involved in host responses and viral pathogenesis.
The Bcl-2 family of proteins, comprised of both anti- and pro-apoptotic molecules, constitutes a critical, intracellular decision point regulating a common death pathway [27]. The ratio of antagonist (Bcl-2, Bcl-xL, Mcl-1, and A1) to agonist (Bax, Bak, Bcl-xs and Bad) molecules dictates whether a cell responds to a proximal apoptotic stimulus [27, 28]. These proteins also interact with mitochondria to control the balance of mitochondrial membrane potential or MMP [29]. On the other hand, Bcl-2 this anti-apoptotic protein can down-regulate by CHOP protein in some cellular contexts through IRE1 sensor triggered signaling [30].
In previous studies we demonstrated that the redspotted grouper nervous necrosis virus (RGNNV) TN1 strain induced apoptosis and post-apoptotic necrotic cell death in a grouper liver cell line GL-av, which was sub-cloned from spontaneous immortalization GL-a cell line [31]. We showed that RGNNV infection induced loss of the mitochondrial membrane potential (MMP) in GL-av cells. MMP loss was blocked by the mitochondrial membrane permeability (MMP) transition pore inhibitor BKA [31], as well as the Bcl-2 family member protein zfBcl-xL [11].
The RGNNV genome encodes two viral death inducers either protein α or B2. Protein α (42 kDa) activates caspase-3 [6] and triggers mitochondrial-mediated cell death, which may use the newly synthesized protein pathway [32]. Protein α activity can be blocked by zfBcl-xL [7]. The second death inducer encoded by the viral genome is the B2 gene, which has been shown to induce cell death via a Bax-mediated pathway [33]. On the other hand, a novel anti-necrotic death gene, B1, is an early expression gene that contributes to anti-necrotic cell earth during early replication stages and has been proposed to regulate cell death [8].
In the present study, we first demonstrate that RGNNV can induce ER stress responses for GRP78 upregulation. We show an interaction between GRP78 and RdRp that enhances viral replication on mitochondria. Then, this GRP 78-mediated ER stress for enhancing viral replication can downregulate the Bcl-2 protein, which triggers mitochondrial-mediated cell death. Our results provide new insights into the GRP78-mediated ER stress-induced cell death of RNA viruses.
Materials and methods
Cell culture and virus
The GF-1 grouper fin cell line was provided by Dr Chi (Institute of Zoology and Development of Life Science, Taiwan, ROC). Cells were grown at 28°C in Leibovitz L-15 medium (GibcoBRL, Gaithersburg, MD) supplemented with 5% fetal bovine serum and 25 μg/ml gentamycin. Naturally infected red grouper larvae were collected in 2002 in Tainan prefecture and used as the source of red spotted grouper nervous necrosis virus (RGNNV Tainan No. 1; RGNNV TN1). The virus was used to infect grouper liver-a subcloned (GL-av) cells that were grown using the same conditions as grouper fin (GF-1) cells. The virus was purified as described by Mori et al. [5], and stored at −80°C. Viral titer was determined using a TCID50 assay according to Nicholson and Dunn [34].
Western blot analysis
GF-1 cells were seeded in 60 mm diameter Petri dishes with 3 ml of medium (105 cells/ml) for 20 h. The monolayers were rinsed twice with phosphate buffered saline (PBS) and infected with RGNNV-TN1 (MOI = 5; propagated and titrated in GF-1 cells with a TCID50 of 108/0.1 ml) for 0, 24, 48 and 72 h. In a vomitoxin group (VT; GRP78 inhibitor), cells were pre-treated with VT (1 μg/ml; Sigma, St. Louis, MO, USA) for 2 h prior to infection with RGNNV for 0, 24, 48 and 72 h p.i. [35]. At the end of each incubation, the culture medium was aspirated, the cells were washed with PBS and then lysed in 0.3 ml of lysis buffer (10 mM Tris base, 20% glycerol, 10 mM sodium dodecyl sulfate [SDS], 2% ß-mercaptoethanol [ß-ME], pH 6.8). An aliquot of each lysate with 30 μg protein per sample was electrophoresed on an SDS polyacrylamide gel [36] to resolve the proteins. The gels were immunoblotted [37] with either (1) primary antibodies (1:3000 dilutions) of anti-cytochrome c (Imgenex, Suite E, San Diego, CA92121, USA), actin (UPSTATE, Charlottesville VA, USA), ATF6 (Imgenex), and EGFP (BD Biosciences, Palo Alto, CA 94303-4230, USA) monoclonal antibodies followed by secondary antibody (1:10,000 dilution) of peroxidase-labeled goat anti-mouse conjugate (Amersham Biosciences, USA), (2) primary antibodies (1:1000 dilutions) of anti-RGNNV B2, B1, protein A, protein α (home-made), cytochrome c oxidase I (Imgenex), XBP1 (BioLegend, San Diego, California, USA), PERK (BioLegend), PERK-P (BioLegend), Bcl-2 (BD Biosciences) and Hsp60 (UPSTATE) polyclonal antibodies followed by a secondary antibody (1:5,000 dilution) of peroxidase-labeled goat anti-rabbit conjugate or (3) primary antibody (1:1000 dilution) of anti-GRP78 polyclonal antibody (BD Biosciences) followed by a secondary antibody (1:5,000 dilution) of peroxidase-labeled goat anti-rat conjugate. Chemiluminescence indicative of antibody binding was captured on Kodak XAR-5 films (Eastman Kodak, USA).
In situ staining for caspase-12 activation
GF-1 cells were infected with RGNNV as described above at 28°C for 0, 24, 48 and 72 h. Culture medium was aspirated, 300 μl of staining solution was added (FITC-ATAD-FMK in L-15 medium; CaspGLOW Fluorescein Caspase-12 Staining kit; BioVision, Mountain View, CA, USA) and the dishes were incubated at 28°C for 60 min. Staining solution was aspirated, cells were washed with wash buffer, and then viewed under a fluorescence microscope using band-pass filters.
siRNA for knockdown of grouper GRP78 (gGRP78) protein expression
The sequences for gGRP78-specific siRNA (5′-GACTCTGACCTGAAGAAGT-3′) and a non-silencing control siRNA (5′-UUCUCCGAACGUGUCACGUTT-3′) (MDBio, Taipei, Taiwan) were used for gGRP78 knockdown. The siRNA specific for gGRP78 was designed using the siRNA Target Designer on the Promega website and synthesized by the MDBio Company. The gGRP78 siRNA was labeled with Fluorescein (FAM) at the 3′-end so that the efficiency of gGRP78 siRNA transfection could be tested.
To transfect the gGRP78 siRNA, 100 pmol of siRNA and 5 μl of lipofectamine 2000 (Invitrogen) were separately diluted in 100 μl of L15 medium without serum and incubated at room temperature for 5 min. The two solutions were mixed and incubated at room temperature for 15 min, after which the mixture was added to GF-1 cells that had been pre-seeded in 35-mm Petri dishes with 0.8 ml of L15-5% FBS. GF-1 cells were transfected either with 100 nM gGRP78 siRNA or 100 nM non-silencing control siRNA (siRNA-negative) as a negative control. At 24 h post-incubation, the gGRP78 siRNA-transfected GF-1 cells were infected with RGNNV. At 24, 36 and 48 h pi, cells were analyzed by fluorescence microscopy using 488 nm excitation and a 515 nm long-pass filter [31]. At 24, 48 and 72 h pi, infected cells were washed with PBS and harvested. Total protein was extracted using 100 μl of lysis buffer for Western blot analysis and culture media were harvested for virus titer assays at 24, 48 and 72 h pi.
Cloning of zfGRP78 and construction of EGFP-zfGRP78 fused-genes
Synthesis and amplification of cDNA (total mRNA from 14 day old zebra fish) used the SuperScript One-Step™ reverse transcriptase-polymerase chain reaction (RT-PCR) system kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Zebra fish GRP78 primers P1 and P2 were each added for a final concentration of 0.2 μM. PCR cycling conditions were 54°C for 30 min, 2 min at 94°C (to inactivate the reverse transcriptase), 95°C for 30 s (DNA denaturation), 57°C for 30 s (annealing), and 72°C for 45 s (extension) for a total of 35–40 cycles. The RT-PCR primers zfGRP78 P1 (5′-GGGGTACCATGCGGTTGCTTTGCCTG-3′; KpnI cutting site underlined) and zfGRP78 P2 (5′-CGGGATCCCAGCTCGTCCTTCTCTTC-3′; BamHI cutting site underlined) were used to amplify a fragment covering the coding sequence of zfGRP78. The purity and size of the amplified product was verified by 1.2% agarose gel electrophoresis and staining with ethidium bromide [38]. The cloned PCR products were sequenced by the dye termination method using an ABI PRISM 477 DNA sequencer (Applied Biosystems, Foster City, CA) and scanned against the GenBank database BLAST (www4.ncbi.nlm.nih.gov/) and PROSITE (psort.ims.u-tokyo.ac.jp/) programs. Then, PCR productions were inserted into a pEGFP-C1 vector (Clontech), which contained the human cytomegalovirus (CMV) promoter as a control, to obtain pzfGRP78-EGFP using the KpnI and BamHI enzyme cutting sites.
Assays of zfGRP78 enhanced RGNNV replication in fish cells
GF-1 cells were seeded in 60 mm diameter culture dishes (3 × 105/dish) 1 day before transfection. Cells were transfected with 2 μg of pEGFP and pzfGRP78-EGFP [33] using Lipofectamine-Plus (Life Technologies, USA) according to the manufacturer’s instructions. Then, the cells were incubated for 24 h with RGNNV infection (MOI = 5). At 24, 36 and 48 h pi, cells were analyzed by fluorescence microscopy using 488 nm excitation and a 515 nm long-pass filter [31] or at 24, 48 and 72 h pi, the medium was aspirated, the monolayers were washed with PBS and lysed in 0.3 ml of lysis buffer (10 mM Tris, 20% glycerol, 10 mM sodium dodecyl sulfate [SDS], 2% ß-mercaptoethanol, pH 6.8). Fusion proteins’ expressions were analyzed by Western blot at 0, 24, 48 and 72 h pi. Supernatants were also collected at 24, 48 and 72 h pi for additional titer assays.
Tracking mitochondrial zfGRP78 localization in fish cells
To track the localization of zfGRP78 fused with EGFP, GF-1 cells were cultured on 35-mm Petri dishes. Cells were transfected with 2 μg of pEGFP and pzfGRP78-EGFP [7, 33] using Lipofectamine-Plus (Life Technologies, USA) according to the manufacturer’s instructions. Then, the cells were incubated for 24 h with RGNNV (MOI = 5). At 24, 36 and 48 h pi, cells were stained with MitoTracker Red CM-H2XRos (Invitrogen), for which live cells were labeled with the mitochondrion-specific dye in accordance with the manufacturer’s instructions, after which cells were analyzed by fluorescence microscopy using 488 nm excitation and a 515 nm long-pass filter [31].
Preparation of mitochondria from RGNNV-infected cells
GF-1 cells were seeded in 60 mm diameter Petri dishes with 3 ml of medium (105 cells/ml) for 20 h. Cells were either treated with VT (1 μg/ml) or left untreated and infected with RGNNV (MOI = 5) for 48 h. At each time point subsequent to a change of the culture medium, 1 ml was removed. Mitochondria were isolated by a modification of a previously described protocol [7]. Briefly, GF-1 cells (2 × 106) were washed with PBS and homogenized in 0.3 ml of mitochondria isolation buffer (0.35 M mannitol, 10 mM HEPES, 0.1% bovine serum albumin, pH 7.2) using a glass homogenizer. Unbroken cells and nuclei were pelleted by centrifugation (600×g for 5 min at 4°C) for yielding a crude fraction enriched with mitochondrias. The mitochondria pellet was isolated from centrifuged supernatant (10,000×g for 10 min at 4°C) and the supernatant was collected and mixed with 25 μl of 10× sodium dodecyl sulfate sample buffer. Samples (50 μl) were boiled and subjected to Western blot analysis [37].
Tracking protein A localization with an immunofluorescence assay
GF-1 cells were cultured on 35-mm Petri dishes. Cells were transfected with 2 μg of pEGFP and pzfGRP78-EGFP using Lipofectamine-Plus (Life Technologies, USA) according to the manufacturer’s instructions [7, 33]. Then, the cells were incubated for 24 h with RGNNV (MOI = 5). At 48 h pi, cells were rinsed once with PBS, fixed with 4% paraformaldehyde for 15 min at room temperature, and then permeabilized for 10 min in 0.2% Triton X-100 in PBS at room temperature for immunofluorescence assay (IFA) [39] with 2% BSA blocking for 30 min. IFA was performed by incubating cells with primary antibodies for 1 h at room temperature, washing with PBS with 0.05% Tween 20 (PBST), incubating with secondary antibodies conjugated with tetramethyl rhodamine isocyanate for 40 min at room temperature, and washing three times with PBST. Then, cells were analyzed by fluorescence microscopy using 488 nm excitation and a 515 nm long-pass filter [31].
Immunoprecipitation assay
GF-1 cells seeded in 100 mm culture dishes were transfected with p3XFLAG-myc-CMV (Sigma) or p3XFLAG-myc-CMV-RGNNV protein A plasmids using Lipofectamine, as described above. The cells were then infected with RGNNV for 48 h. Immunoprecipitation was done as described previously [38]. Briefly, culture medium was aspirated and cells were rinsed twice with PBS (10 mM phosphate, 2.7 mM potassium chloride, 137 mM NaCl, pH 7.4). Cells in each 100 mm dish were lysed in 1 ml lysis buffer (50 mM Tris HCl, pH7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM PMSF) containing protease inhibitor (Roche, USA). Cells were incubated for 30 min on a shaker at 4°C. Soluble protein (1 mg/ml) was incubated with 50 μl of ANTI-FLAG M2 affinity gel (Sigma Chemicals, MO, USA) overnight at 4°C. Immunoprecipitates were washed three times with wash buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA and 0.5 mM PMSF), resuspended in 70 μl of 2× sample buffer (125 mM Tris HCl, pH 6.8, 4% SDS, 20% glycerol), boiled for 10 min and subjected to Western blot analysis.
Evaluation of mitochondrial membrane potential with a lipophilic cationic dye
To assess mitochondrial membrane potential (ΔΨm), RGNNV-infected GF-1 cells were stained with the fluorescent lipophilic cationic reagent Mito-Capture (provided in the MitoCapture Mitochondrial Apoptosis Detection kit). This dye is trapped in mitochondria with normal ΔΨm and is released into the cytosol by mitochondria with abnormal ΔΨm, resulting in loss of fluorescence intensity. Cell preparation and virus infection conditions are described above. GF-1 cells were infected with RGNNV and then either treated with vomitoxin (1 μg/ml; GRP78 inhibitor) at 28°C for 0, 24, 48 and 72 h or not treated. Medium was aspirated and MitoCapture reagent (500 μl) was added to each 60-mm Petri dish. Cells were incubated at 28°C for 60 min and then viewed under a fluorescence microscope using band-pass filters to detect fluorescein and rhodamine [31].
Quantification of cell viability
To assay reduced cell death for knockdown of gGRP78 expression, approximately 105 cells/ml of GF-1 cells were cultured in a 60 mm diameter Petri dish for 20 h. The GF-1 cells were transfected either with 100 nM gGRP78 siRNA or 100 nM non-silencing control siRNA (siRNA-negative). At 24 h post-incubation, the gGRP78 siRNA -transfected GF-1 cells were infected with RGNNV (MOI = 5). These cells and uninfected cells were then incubated at 28°C for 24, 36 or 48 h. In contrast, to assay for enhanced cell death for over-expression of zfGRP78, GF-1 cells were seeded in 60 mm diameter culture dishes (3 × 105/dish) 1 day before transfection. Cells were transfected with 2 μg of pEGFP and pzfGRP78-EGFP [32] using Lipofectamine-Plus (Life Technologies, USA) according to the manufacturer’s instructions. Then, the cells were incubated for 24 h with RGNNV (MOI = 5). At each time point, sets of Petri dishes were washed with PBS and treated with 0.5 ml of 0.1% trypsin-EDTA (Gibco, Grand Island, NY) for 1−2 min. Cell viability was determined in triplicate using a Trypan blue dye exclusion assay [40]. Each data point represents the mean viability of three independent experiments ± SEM. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. A P-value < 0.05 was taken as a statistically significant difference between group mean values.
Results
RGNNV induces ER stress responses in GF-1 fish cells via XBP1 expression, ATF6 cleavage, PERK phosphorylation and GRP78 up-regulation
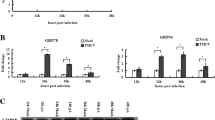
We examined if ER stress responses could be triggered by RGNNV infection in order to investigate the role of betanodavirus infection on ER stress in fish cells. We found the following results for RGNNV-infected GF-1 cells. (1) RGNNV up-regulate XBP1 expression at 24, 48 and 72 h p.i. (Fig. 1A, lanes 6–8, respectively), which may activate ER sensor IRE1 when compared with mock-infected cells (Fig. 1A, lanes 1–4 represent 0, 24, 48 and 72 h, respectively). (2) Full length ATF6 was cleaved to produce cleaved forms at 48 h and 72 h p.i. (Fig. 1A, lanes 7–8, respectively) when compared with mock-infected cells (Fig. 1A, lanes 1–4 represent 0, 24, 48 and 72 h, respectively). (3) Chaperone GRP78 was up-regulated at 48 and 72 h p.i. (Fig. 1A, lanes 7–8, respectively) when compared with mock-infected cells (Fig. 1A, lanes 1–4 represent 0, 24, 48 and 72 h, respectively). (4) PERK was significantly phosphorylated at 72 h p.i. (Fig. 1A, lane 8), which corresponded with cleaved Bcl-2 protein at 48 and 72 h p.i. (Fig. 1A, lanes 7–8) at the same times, when compared with mock-infected cells (Fig. 1A, lanes 1–4 represent 0, 24, 48 and 72 h, respectively). By comparison, RGNNV-infected GF-1 cells expressed viral proteins A and α at 24, 48 and 72 h p.i. (Fig. 1A, lanes 6–8) when compared with mock-infected cells (Fig. 1A, lanes 1–4 represent 0, 24, 48 and 72 h respectively), which showed that viral replication corresponded to ER stress responses. Actin was used as an internal loading control. On the other hand, the Brefeldin A (BFA) as an ER stress inducer can induce ER stress in fish cells. The fish GF-1 cell were treated with Brefedin A (1 μg/ml) for 0, 12, 24, 36 and 48 h. At the end points, then these cell lysates were prepared at the indicated time and analyzed by Western blotting to detect XBP1, ATF6, GRP78 and Actin at 0 h (Fig. 1B, lane 1), 12 h (Fig. 1B, lane 2), 24 h (Fig. 1B, lane 3), 36 h (Fig. 1B, lane 4) and 48 h (Fig. 1B, lane 5), respectively. Cell lysate MCF-7 as a positive control that treated with ER stress inducer is shown in Fig. 1B, lane 6. In the results, BFA also can up-regulate the XBP1 and GRP78 from 12, 24, 36 to 48 h (Fig. 1B, lane 2–5), respectively, but did not induce ATF6 cleaved form ATF6 full length form (Fig. 1B, lanes 1–5) in fish GF-1 cell, which supported RGNNV-induced ER stress signaling was significant.
RGNNV infection induces ER stress responses in fish cells. A GF-1 cell lysates were prepared at the indicated times p.i. and analyzed by Western blotting to detect XBP1, ATF6, PERK, PERK-P, GRP78, Bcl-2, RGNNV proteins (protein A, protein α, B1 and B2) and actin in mock-infected cells (lanes 1–4) and RGNNV-infected cells (lanes 5–8) at 0, 24, 36, 48 and 72 h, respectively. Cell lysate MCF-7 as a positive control that treated with ER stress inducer is shown in lane 9. B Brefeldin A (BFA) as an ER stress inducer can induce ER stress in fish cells. The fish GF-1 cell were treated with Brefedin A (1 μg/ml) for 0, 12, 24, 36 and 48 h, then these cell lysates were prepared at the indicated time and analyzed by Western blotting to detect XBP1, ATF6, GRP78 and Actin at 0 h (B, lane 1), 12 h (lane 2), 24 h (lane 3), 36 h (lane 4) and 48 h (lane 5), respectively. Cell lysate MCF-7 as a positive control that treated with ER stress inducer is shown in lane 6. C RGNNV induces caspase-12-like activation. Phase-contrast fluorescence microscopy was used to analyze staining for activated caspase-12-like in RGNNV-infected GF-1 cells at 24 and 48 h p.i. Normal control cells are shown in B-a, b, e, f, and RGNNV-infected cells are shown in B-c, d, g, h. Caspase-12-like activated cells are indicated by arrows. Scale bar = 10 μm. D The percentage of caspase-12-like activated cells at each time point is shown for control and RGNNV-infected cells. The number of positive cells per 200 cells in each sample was counted for three independent experiments. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. P < 0.01 was taken as a statistically significant difference between group mean values
Up-to-now, the caspase-12 has not yet been described in fish and that therefore the data obtained with the caspase-12 kit may not unequivocally reflect caspase-12 activity, so we refer to caspase-12-like activity throughout the paper. Furthermore, we found that caspase-12-like was gradually activated in RGNNV-infected GF-1 cells at 24 h p.i (Fig. 1C-c, d; indicated by arrows) with higher activated levels at 48 h p.i (Fig. 1C-g, h; indicated by arrows) when compared to uninfected control cells at 24 h (Fig. 1C-a, b) and 48 h (Fig. 1C-e, f). Of a total of 200 cells counted, cells positive for caspase-12-like activation in the RGNNV-infected group ranged from 12% at 24 h p.i, 72% at 48 h p.i to 95% at 72 h p.i (Fig. 1D), as compared with 1% caspase-12-like positive cells in the uninfected control cells at 24, 48 and 72 h p.i. Furthermore, we evaluated whether a caspase-12 inhibitor can block RGNNV-induced cell death and found that this treatment was just partially protective against the detrimental effect of RGNNV infection (data not shown).
Knockdown of gGRP78 reduces viral titers and increases cell viability
Based on our results shown in Fig. 1, we were interested to know why chaperone GRP78 needed to be up-regulated by betanodavirus (RGNNV). We tested a hypothesis of whether chaperone GRP78 could enhance viral replication in our system. We designed grouper GRP78 siRNA and used this to transiently transfect GF-1 cells (Fig. 2A) in order to block RGNNV-up-regulated GRP78. In the results shown in Fig. 2B, we found that gGRP78 siRNA effectively down-regulated the gGRP78 expression level by about 45% (Fig. 2B, lane 7) at 48 h pi when compared with si-negative control with 100% expression (Fig. 2B, lane 3) at 48 h pi. As shown in Fig. 2C, when compared with the siRNA negative control, knockdown of gGRP78 by siRNA could apparently enhance cell viability from 58% at 24 h, 37% at 36 h and 23% at 48 h to 87% at 24 h, 78% at 36 h and 45% at 48 h. Further, from viral titer assays done at 24, 48 and 72 h pi, all titers were reduced by about one log titer as shown in Fig. 2D, which indicated that knockdown of gGRP78 could affect viral replication.
Effects of GRP78 knockdown by siRNA on viral replication (A) Phase-contrast and green fluorescence micrographs of transiently transfected GF-1 cells with si-Negative control at 24 h (a, b), 36 h (e, f) and 48 h (i, j) and with sigGRP78 at 24 h (c, d), 36 h (g, h) and 48 h (k, l) with RGNNV (MOI = 5). Scale bar = 10 μm. B Analysis of gGRP78 expression levels by Western blot analysis in si-negative transfected (lanes 1–4) and sigGRP78 transfected GF-1 cells (lanes 5–8) at 0, 24, 48, and 72 h pi, respectively. Lane 9 is for positive cell lysate MCF-7. Actin was used as an internal loading control. C The viability of GF-1 cells infected with RGNNV with si-negative and sigGRP78 transfection was determined in triplicate at 24, 36 and 48 h pi. In this and all subsequent figures (unless otherwise noted) results are presented as the percentage of RGNNV-infected or non-infected cells at each time point determined by a dye exclusion method in triplicate; each point represents the mean of three independent experiments and error bars are SEMs. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. *P < 0.05 was taken as a statistically significant difference between group mean values. D Analysis of viral titers showed that gGRP78 knockdown could reduce viral replication with RGNNV infection, with viral titers determined at 24, 48 or 72 h. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. *P < 0.05 was taken as a statistically significant difference between group mean values
zfGRP78 over-expression enhances viral titer and reduces cell viability
We next asked if chaperone GRP78 could enhance viral replication. We cloned zebrafish GRP78 (zfGRP78; accession number. NW 001879074) from 2-week-old fish, which contained its 1953 bp and had a theoretical molecular weight of 72.1 kDa, and with the EGFP gene we constructed a fusion gene, zfGRP78-EGFP. These plasmids were transiently transfected into GF-1 cells for 24 h and then tested to determine if zfGRP78 affected viral replication. In the results shown in Fig. 3A, we found that both EGFP (Fig. 3A, lanes 1–4) and zfGRP78-EGFP (Fig. 3A, lanes 5–8) were expressed in the transfected cells at 0, 24, 48 and 72 h pi by Western blot analysis. Actin was used an internal control shown in Fig. 3A, lanes 1–8. As shown in Fig. 3B, up to 25 to 30% of the transfected cells were either EGFP-containing cells (Fig. 3B-b, f, j; indicated by arrows) or zfEGFP-EGFP-containing cells (Fig. 3B-d, h, i; indicated by arrows). As shown in Fig. 3C, over-expression of zfGRP78 with RGNNV infection apparently reduced cell viability from 66% at 24 h, 47% at 36 h and 31% at 48 h to 48% at 24 h, 34% at 36 h and 13% at 48 h, when compared with EGFP expression as a negative control. Further, cells that over-expressed zfGRP78-EGFP also had dramatically enhanced viral titers, up to two logs, compared to EGFP-containing cells at 24, 48 and 72 h pi (Fig. 3D). These results were consistent with those for knockdown of gGRP78 for reducing viral titer with RGNNV infection, as shown in Fig. 2D.
Effects of GRP78 over-expression on viral replication (A) Analysis of gGRP78 expression levels by Western blot analysis in pEGFP transfected (lanes 1–4) and pzfGRP78-EGFP transfected GF-1 cells (lanes 5–8) at 0, 24, 48 and 72 h pi, respectively. Actin was used as an internal loading control. B Phase-contrast and green fluorescence micrographs of transiently transfected GF-1 cells with EGFP control (The green fluorescent-contained cells were indicated by arrows) at 24 h (a, b), 36 h (e, f) and 48 h (i, j) and with zfGRP78-EGFP at 24 h (c, d), 36 h (g, h) and 48 h (k, l) with RGNNV (MOI = 5). Scale bar = 10 μm. C The viability of EGFP and zfGRP78-EGFP transfected GF-1 cells infected with RGNNV was determined in triplicate at 24, 36 and 48 h pi. Results are the percentage of 200 cells at each time point determined in triplicate, with each point representing the mean of three independent experiments; error bars are SEMs. D Analysis of viral titers showed that zfGRP78 over-expression could enhance viral replication with RGNNV infection, with viral titers determined at 24, 48 or 72 h. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. *P < 0.05 was taken as a statistically significant difference between group mean values. (Color figure online)
Chaperone GRP78 can interact with viral RNA-dependent RNA polymerase on mitochondria at a middle replication stage
The mechanism for gGRP78-mediated enhancement of viral replication remains unknown. Recently, nodavirus RdRp was shown to be targeted to mitochondria [41, 42] and replicated on the mitochondrial membrane [42, 43]. We asked if GRP78 played a role in regulating the function of protein A (RdRp) in mitochondria. We attempted to track the localization of GRP78 with EGFP in order to understand its localization in the fish system. With RGNNV infection, zfGRP78-EGFP was also distributed in the cytoplasm (Fig. 4A-b, f, j; as shown in green fluorescent cells) at 0, 24 and 36 h pi, respectively. These cells were also stained with a mitochondrial tracker dye to monitor mitochondrial locations (Fig. 4A-c, g, k; indicated by arrows) at 0, 24 and 36 h pi, respectively. Merged images of cells (Fig. 4A-d, h, i; as shown in orange fluorescent cells) also showed that zfGRP78-EGFP was localized in mitochondria, as shown by a more yellow color within cells, when compared with zfGRP78-EGFP absent cells that just showed a strong orange color (Fig. 4A-c, g). The enlarge cells as shown in Fig. 4A-m (from Fig. 4A-d), n (from Fig. 4A-h) and o (from Fig. 4A-l) at 0, 24 and 36 h, respectively, which apparently green and orange fluorescent overlapping cell was showed at 24 and 36 h as compared with 0 h, green fluorescent cell. This showed that GRP78 was targeted into the mitochondria. Figure 4B, lane 2 represents the membrane fraction and lane 4 represents the cytosolic fraction of RGNNV-infected GF-1 cells, which examined if GRP78 could move into mitochondria in either RGNNV-infected zfGRP78 over-expressing cells (Fig. 4A) or just RGNNV-infected cells (Fig. 4B). We also found that gGRP78 expression was nearly undetectable in uninfected cells in either the mitochondrial or cytoplasmic fractions (Fig. 4B, lanes 1 and 3). Hsp60 was used as an internal control for the mitochondrial form (Fig. 4B, lanes 1 and 2) and actin was used as an internal control for the cytosolic form (Fig. 4B, lanes 3 and 4).
Tracking zfGRP78 mitochondrial localization with RGNNV infection. A Phase-contrast and fluorescence micrographs of transiently zfGRP78-EGFP-transfected GF-1 cells. GF-1 cells were transfected with zfGRP78-EGFP plasmid for 24 h, and then infected with RGNNV and incubated for different times. Phase-contrast micrographs of cells infected with RGNNV at 0 h (a), 24 h (e), and 36 h (i); Green fluorescence micrographs of cells infected with RGNNV at 0 h (b), 24 h (i), and 36 h (j); Orange fluorescence micrographs of cells infected with RGNNV with mitochondrial tracker staining at 0 h (c), 24 h (g), and 36 h (k); and fluorescence merged micrographs of cells infected with RGNNV at 0 h (d, m), 24 h (h, n), and 36 h (l, o); carrier fluorescent cells are indicated by arrows in these figures. Scale bar = 10 μm. B Demonstration that gGRP78 was targeted to the mitochondrial membrane in RGNNV-infected GF-1 cells at 48 h p.i. Mitochondria preparation was described in Materials and methods. Lane 1 is the mitochondrial fraction from uninfected cells. Lane 2 is the mitochondrial fraction from RGNNV-infected cells. Lane 3 is the cytosolic fraction from uninfected cells and lane 4 is the cytosolic fraction from RGNNV-infected cells. Hsp60 as a mitochondrial fraction control is shown in lanes 1–2. Actin as a cytosolic fraction control is shown in lanes 3–4. (Color figure online)
Next, using immunostaining assay, we investigated potential interactions between GRP78 and RdRp on mitochondria during RGNNV infection. In the results to identify protein A localization by immunostaining, at 48 h p.i., protein A was tracked with an orange color in EGFP-transfected cells (Fig. 5A-c, g, k) and zfGRP78-EGFP-transfected cells (Fig. 5A-d, h, l) with RGNNV infection when compared with negative controls for both EGFP-transfected cells (Fig. 5A-a, e, i) and zfGRP78-EGFP-transfected cells (Fig. 5A-b, f, j), which did not have detectable protein A expressions. Then, the green fluorescence images for EGFP (Fig. 5A-a, d) and zfGRP78-EGFP-containing cells (Fig. 5A-b, c) were merged with protein A expression (Fig. 5A-h, EGFP-transfected cells; Fig. 5A-g, zfGRP78-EGFP-transfected cells and without protein A (Fig. 5A-e, EGFP-transfected cells; Fig. 5A-f, zfGRP78-EGFP-transfected cells) expressing cells, which specifically detected zfGRP78-EGFP and protein A merged cells with an orange-yellow color (Fig. 5A-k, n; as indicated by arrows), but not in the other groups (Fig. 5A-j, m).
zfGRP78 interacts with RGNNV RNA-dependent RNA polymerase at a middle replication stage. A Fluorescence micrographs of GF-1 cells transiently transfected with EGFP and zfGRP78-EGFP. GF-1 cells were transfected with EGFP or zfGRP78-EGFP plasmid and incubated for 24 h, then infected with RGNNV and incubated for 48 h pi. Fluorescence micrographs of EGFP-transfected negative control (a), zfGRP78-EGFP-tranfected negative control (b), EGFP-transfected cells with RGNNV infection (d), zfGRP78-EGFP-tranfected cells with RGNNV infection (c) were shown. Red fluorescence micrographs using an RdRp polyclonal antibody for EGFP-transfected negative control (e), zfGRP78-EGFP-tranfected negative control (f), EGFP-transfected cells with RGNNV infection (h) and zfGRP78-EGFP-tranfected cells with RGNNV infection; Yellow-orange fluorescence merged micrographs of EGFP-transfected negative control (i), zfGRP78-EGFP-tranfected negative control (j, m), EGFP-transfected cells with RGNNV infection (l), zfGRP78-EGFP-tranfected cells with RGNNV infection (k, n); Positive cells are shown in A-k, n. Scale bar = 10 μm. B Demonstration that gGRP78 interacts with RdRp (protein A) from RGNNV-infected cells. RGNNV RNA1 (encoded for protein A or designated RdRp), cloned into a Flag tagged vector (p3XFLAG-myc-CMV), was transiently transfected into GF-1 cells with lipofectamine, which were infected with RGNNV (MOI = 5) and incubated for 48 h p.i. Protein was immunoprecipitated with an anti-Flag monoclonal antibody and immunoblotted with an anti-GRP78 polyclonal antibody. Lane 1 shows protein from empty Flag vector-transfected GF-1 cells and probed with GRP78 antibody, which did not detect the GRP78 signal. Lane 2 shows protein immunoprecipitated from Flag-RNA1 vector transfected cells and immunoblotted with GRP78 polyclonal antibody, which could detect GRP78 signal. (Color figure online)
We also asked if gGRP78 could interact with protein A. Using an anti-affinity gel to immunoprecipitated Flag protein A with gGRP78 during RGNNV infection, we found that RGNNV Flag- protein A specifically co-immunoprecipiated with gGRP78 (Fig. 5B, lane 2), but did not detect gGRP78 in the without added-protein A group (Fig. 5B, lane 1). Combined with the results in Fig. 4, gGRP78 could interact with protein A on mitochondria.
gGRP78-mediated ER stress signal can regulate viral protein and Bcl-2 protein expression
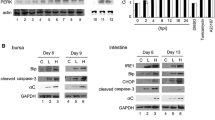
We investigated the role of GRP78 on viral replication and anti-apoptotic protein Bcl-2 expression in our system by using the either gGRP78 specific siRNA for blocking gGRP78 expression or GRP78 specific synthesis inhibitor VT to block gGRP78 synthesis [35], but may affect some proteins. In the grouper GRP78 siRNA approach for knockdown of gGRP78 was shown in Fig. 2B, lane 7 as compared with negative control Fig. 2B, lane 3 that sigGRP78 can down-regulate about 55% than siNegative control. Then, here continue to check the viral protein α and Bcl-2 expression level, in the results shown in Fig. 6a, we found that gGRP78 siRNA effectively down-regulated the gGRP78 expression that correlated to reduce 25% expression level on the viral protein α that was a reproducible effect (Fig. 6a, lane 5) at 24 h pi as compared siNegative control 100% expression (Fig. 6a, lane 7); and at 36 h pi reduces 40% (Fig. 6a, lane 6) than siNegative control 100% (Fig. 6a, lane 8). Uninfected group as a negative control was shown in Fig. 6a, lanes 1–2 (sigGRP78 group for either at 24 or 36 h) and Fig. 6a, lanes 3–4 (siNegative control group for either at 24 or 36 h). On the other hand, gGRP78 siRNA effectively down-regulated the gGRP78 expression that correlated to reduce about 27% expression level on the Bcl-2 expression level (Fig. 6a, lane 6) when compared with uninfected control with 100% expression (Fig. 6a, lane 2) at 36 h pi; and at siNegative control group, can reduce about 45% (Fig. 6a, lane 8) as compared with uninfected group with 100% expression (Fig. 6a, lane 4), which means down-regulation of gGRP78 can enhance Bcl-2 expression about 18%. Actin as an internal control was shown in Fig. 6a, lanes 1–9. Furthermore, by double check down-regulation of gGRP78 for regulating with viral protein α, B2 and Bcl-2 expression. We pre-treated GF-1 cells for 2 h with different concentrations of VT (0, 0.2, 0.5 and 1.0 μg/ml) and then infected the cells with RGNNV. We found that VT blocked GRP78 expression most efficiently at a concentration of 1 μg/ml (data not shown). Thus, we used VT at a concentration of 1 μg/ml in all subsequent experiments. As demonstrated in Fig. 6b, 1 μg/ml VT almost completely inhibited GRP78 expression in RGNNV-infected cells (Fig. 6b, lanes 5–6) as compared with RGNNV-infected group (Fig. 6b, lanes 3–4) and was correlated with reduced viral protein expressions, including protein αand B2 [Fig. 6b, lanes 5 (48 h p.i.) and 6 (72 h p.i.)], when compared to untreated cells [Fig. 6b, lanes 3 (48 h p.i.) and 4 (72 h p.i.)]. Appropriate uninfected controls were used [Fig. 6b, lanes 1 (48 h p.i.) and 2 (72 h p.i.)].
sigGRP78 and VT block GRP78 synthesis, reduce viral protein expression and reverses Bcl-2 down-regulation at a middle-late replication stage. a Western blot analysis of viral proteins and Bcl-2 protein expression in RGNNV-infected GF-1 cells with sigGRP78-transfected cells at 24 h pi (lane 5), and at 36 h pi (lane 6); RGNNV-infected GF-1 cells with siNegative-transfected cells at 24 h pi (lane 7), and at 36 h pi (lane 8); uninfected with sigGRP78-transfected cells at 24 h pi (lane 1) and 36 h (lane 2); uninfected with siNegative-transfected cells at 24 h pi (lane 3) and 36 h (lane 4). HeLa cell lysate as a positive control was shown in lane 9. Actin was used as an internal control in lanes 1–7. Quantification of viral protein αand Bcl-2 expression levels in RGNNV-infected cells at late replication stage. Protein expression level was quantified by Personal Densitometer (Molecular Dynamic). b Western blot analysis of viral proteins and Bcl-2 protein expression in RGNNV-infected GF-1 cells with VT (1 μg/ml) treatment at 48 h pi (lane 5), and at 72 h pi (lane 6); RGNNV-infected GF-1 cells at 48 h pi (lane 3), and at 72 h pi (lane 4); uninfected cells at 48 h pi (lane 1) and 72 h (lane 2). Actin was used as an internal control in lanes 1–6. Quantification of Bcl-2 expression levels in RGNNV-infected cells at late replication stage. Protein expression level was quantified by Personal Densitometer (Molecular Dynamic)
On the other hand, we found that Bcl-2 expression was apparently downregulated to 38% in RGNNV-infected cells at 72 h p.i. (Fig. 6b, lane 4). However, Bcl-2 down-regulation was reversed back to 72% by VT treatment (Fig. 6b, lane 6) when compared with uninfected control 100% expression at 72 h (Fig. 6a, lane 2), which results consistent with gRPG78 siRNA approach (Fig. 6a). This indicated that a gGRP78-mediated ER stress signal could increase viral death factors protein α and B2 expression and reduce the Bcl-2 expression level at middle-late replication stages.
Can an ER stress signal induce mitochondrial-mediated cell death?
Bcl-2 has been shown to regulate the mitochondrial membrane potential [29]. We investigated the role of ER stress-mediated Bcl-2 down-regulation in RGNNV-infected cells for MMP loss in these cells. Blocking the up-regulation of gGRP78 by its specific siRNA showed that RGNNV-induced MMP loss with siNegative-transfected group (Fig. 7A-c, g, k; as indicated by arrows) was blocked by sigGRP78 at 24 h p.i (Fig. 7A-d, h, l) when compared with siNegative-transfected cells (Fig. 7A-a, e, i) and sigGRP78-transfected cells (Fig. 7A-b, f, j) but uninfected. Mitochondrial function was evaluated using MitoCapture Reagent (Apoptosis Detection, Mitochondria BioAssayTM Kit). The Mitocapture dye aggregates in the mitochondria of healthy cells and fluoresces red. In apoptotic cells, the dye cannot accumulate in mitochondria, remains as monomers in the cytoplasm, and fluoresces green. Quantifying MMP levels (Fig. 7B) by calculating the ratios for red to green fluorescent and normalizing to control values it was observed that in sigGRP78-transfected RGNNV cells, the loss of MMP was decreased to 16% at 24 h pi and 30% at 36 h p.i as compared to 75% at 24 h, and 92% at 36 h with siNegative control. Negative control either siNegative or sigGRP78 uninfected cells all showed MMP losses of 1% (24 h) and 2% (48 h).
gGRP78-mediated ER stress signal induces mitochondrial-mediated cell death. A MMP loss was demonstrated by either strong green fluorescence or loss of red fluorescence via knockdown of gGRP78 by its siRNA. Phase-contrast and fluorescence images of uninfected with siNegative-transfected cells as a negative control (a, e, i) at 0 h; uninfected with sigGRP78-transfected cells as a negative control (b, f, j) at 0 h; RGNNV-infected with siNegative-transfected cells (c, g, k) at 24 h; RGNNV-infected with sigGRP78-transfected cells (d, h, l) at 24 h that were stained with the lipophilic cationic dye at 24 h pi; loss of red fluorescent cells (A, k and i) or strong green fluorescent cells (A, g and h) are indicated by arrows. B The percentage loss of MMP was calculated at 0, 24 and 36 h p.i. for uninfected with siNegative or sigGRP78-transfected cells, and RGNNV-infected with siNegative or sigGRP78-transfected cells. Data were analyzed using either paired or unpaired Student’s t-tests, as appropriate. *P < 0.05 was taken as a statistically significant difference between group mean values. (Color figure online)
Discussion
RGNNV-infected fish lie on their sides, float belly up or swim abnormally (e.g., swim in circles or to the right). Among the histopathological changes observed in these fish are extensive cellular vacuolation and necrotic neuronal degeneration in the central nervous system and retina [44]. The mechanisms by which betanodavirus infections induce neuronal degeneration and whether neuronal degeneration causes abnormal swimming behavior remain unknown. Previous studies showed that RGNNV infection induced post-apoptotic necrotic cell death through a disrupted mitochondrial mediated pathway [11, 31], but the upstream events before mitochondrial damage have not been defined. In this study, for the first time we have demonstrated that RGNNV-induced ER stress responses result in the up-regulation of chaperone protein GRP78 that can regulate viral replication via gGRP78 interactions with RdRp. Elucidating these regulatory pathways is essential for studying the mechanisms of pathogenesis and to develop potent therapeutic agents to combat RGNNV infections.
The effect of ER chaperone protein GRP78 on viral replication
Under normal physiological conditions, up-regulation of ER chaperone proteins (the glucose-regulated proteins, GRPs, of 54, 78 and 94 kDa) ensures the exit of properly folded proteins from the ER [19, 45, 46]. Chaperones mediate the correct assembly of a large number of proteins and are expressed in response to elevated temperatures or other cell stressors. Pathogens can also induce ER stress and the up-regulation of chaperone proteins. For example, up-regulation of the chaperone machinery has been seen in Human cytomegalovirus (DNA virus) infection of human fibroblasts where GRP78 plays a significant role in virion assembly and egress from the cytoplasm [47]. However, severe or prolonged ER stress has been shown to lead to cell death through the initiation of downstream death programs, such as activation of ER-associated caspase-12, the JNK pathway and Gadd153/CHOP [30, 48–50].
In our system, we showed that RGNNV infection triggered the activation of three markers that have been previously shown to be reflective of ER stress responses [16, 51]. In RGNNV-infected cells, we showed (1) induction of IRE1/ATF6 for the up-regulation of gGRP78, (2) activation of ER-associated caspase-12 and (3) induced PERK phosphorylation that reduced Bcl-2 expression (Fig. 1), suggesting that RGNNV infection induced an ER stress response in GF-1 cells.
In this study, we first identified that an RGNNV-induced gGRP78 signal pathway could enhance viral replication, supported by both loss-of-function (Figs. 2, 6a) and gain-of-function (Fig. 3) assays. In addition, VT treatment inhibited GRP78 synthesis (Fig. 6b). However, it is not clear if the up-regulation of GRP78 in our system had a protein refolding function.
Why gGRP78 interacts with RdRp at early-middle replication stages?
A number of cell proteins have been proposed to play roles in viral RNA-dependent RNA polymerase (RdRp) functions. Identifying host factors that are recruited to the viral replication complex is important in order to shed light on virus-cell interactions that facilitate infection and to identify the mechanistic components required for RNA replication [52]. The RNA replication complex of positive-strand RNA viruses is associated with intracellular membranes that are different from RNA viruses that target distinct membranes. It is very unique case that protein A (RdRp) contains a mitochondrial targeting sequence (MLS) [41, 42]. For alphanodavirus (FHV) and betanodavirus (GGNNV), the RNA replication complex is always associated with mitochondria [43, 53]. By comparison, heat shock chaperone protein 70 can have both positive and negative regulatory effects on flockhouse virus (alphanodavirus) RNA replication [54].
Based on these reports, we asked if gGRP78 could interact with mitochondrial RdRp (protein A) of RGNNV to enhance viral replication at early-middle replication stages that viral replication are defined as early (0–24 h pi), middle (24–48 h pi) and late (48–72 h pi) replication stages in this system. For this proposal, we first tracked GRP78 with EGFP as a fusion protein in combination with a mitochondria tracker dye to prove zfGRP78 mitochondrial targeting [Fig. 4A-n (24 h pi) and o (36 h pi)] apparently after RGNNV infection. Then, these cells were stained with RdRp antibodies conjugated with tetramethyl rhodamine isocyanate to demonstrate the interaction between GRP78 and the RGNNV RdRp enzyme on the mitochondria at 48 h pi (Fig. 5A-k, n) in RGNNV-infected fish cells, which may enhance viral replication in this special case that differs with other RNA virus replication in ER.
RGNNV-triggered ER stress signal regulates mitochondrial-mediated cell death at a middle replication stage
Recently, three major pathways that trigger apoptosis were identified: (1) the death receptor or extrinsic pathway; (2) the mitochondrial or intrinsic pathway; and (3) the ER stress mediated pathway. These pathways are activated by caspase 8, caspase 9 and caspase 12-like, respectively. The ER, like the mitochondria, is a repository for both pro-apoptotic and anti-apoptotic molecules. Bcl-2 family members, including Bcl-2, Bcl-xL, Bax, Bak and Bik, are associated with the ER to regulate calcium homeostasis and apoptosis [16, 55–58]. RGNNV infections can induce the loss of mitochondrial membrane potential (MMP). MMP loss is blocked by either the MMP transition pore inhibitor BKA or the Bcl-2 family member Bcl-xL [11, 31]. However, apoptosis in RGNNV-infected cells does not occur solely via the mitochondrial dependent pathway.
In our system, we also found that sigGRP78 blocking of gGRP78 up-regulation either conversely down-regulated Bcl-2 expression or reduced viral replication, and apparently decreased mitochondrial-mediated cell death (Fig. 7), which means ER stress could regulate viral replication and cell death through GRP78.
Figure 8 schematically depicts the process of RGNNV-induced mitochondrial-mediated cell death. RGNNV enters the cell and viral replication occurs within cells. This reaction induces the ER stress responses of ER stress sensors and markers, such as PERK phosphorylation, ATF6 cleavage, XBR1 expression and caspase-12 activation, at an early-middle replication stage. Then, chaperone GRP78 is up-regulated, which may receive ATF6 and IRE1 sensors signals. Further, GRP78 interacts with RdRp on mitochondria to enhance viral replication and synthesis of viral death factors, such as protein α [7] and B2 [33]. Finally, viral replication induces mitochondrial damage resulting in the loss of MMP, down-regulation of Bcl-2 protein and caspase-12-like activation, thereby triggering cell death via the mitochondrial-mediated pathway. Cell death via GRP78 up-regulation can be prevented by treatment with GRP78 specific siRNA or a GRP78 synthesis inhibitor (VT). These results provide new insights into RNA virus-host interactions to aid in understanding the molecular pathogenesis of RGNNV.
RGNNV induces an ER stress-mediated cell death cascade. RGNNV infection and early replication causes an ER stress response upon entry and the primary replication stage. Then, viral replication triggers an ER stress induction stage that includes: (1) activating ATF6 and IRE1 ER sensors that up-regulate chaperone protein GRP78; (2) PERK sensor is phosphorylated, which may be correlated with down-regulated Bcl-2; (3) caspase-12-like becomes activated. Further, the ER stress signal can induce GRP78 up-regulation. At the GRP78 interaction with RdRp stage, GRP78 may interact with protein A (RdRp) on the mitochondrial membrane to enhance viral replication and viral gene expression, which always includes death factors protein α and B2. Finally, viral death factors induce MMP loss and PERK-mediated downredulate Bcl-2 expression, which combined the death signals for triggering necrotic cell death at the mitochondrial dysfunction stage. Cell death could be prevented by: (a) gGRP78 siRNA to knockdown gGRP78 expression and (b) BKA treatment to block mitochondria permeability transition pore (MPTP)
References
Ball LA, Johnson KL (1999) Reverse genetics of nodaviruses. Adv Virus Res 53:229–244
Munday BL, Kwang J, Moody N (2002) Betanodavirus infections of teleost fish: a review. J Fish Dis 25:127–142
Schneemann A, Reddy V, Johson JE (1998) The structural and function of nodavirus particles: a paradigm for understanding chemical biology. Adv Virus Res 50:381–446
Toffolo V, Negrisolo E, Maltese C, Bovo G, Belvedere P, Colombo L, Valle LD (2006) Phylogeny of betanodaviruses and molecular evolution of their RNA polymerase and coat proteins. Mol Phylogenet Evol 43:298–308
Mori K, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I (1992) Properties of a new virus belonging to nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 187:368–371
Guo YX, Wei T, Dallmann K, Kwang J (2003) Induction of caspase-dependent apoptosis by betanodaviruses GGNNV and demonstration of protein alpha as an apoptosis inducer. Virology 308:74–82
Wu HC, Chiu CS, Wu JL, Gong HY, Chen MC, Lu MW, Hong JR (2008) Zebrafish anti-apoptotic protein zfBcl-xL can block betanodavirus protein alpha-induced mitochondria-mediated secondary necrosis cell death. Fish Shellfish Immunol 24:436–449
Chen LJ, Su YC, Hong JR (2009) Betanodavirus non-structural protein B1: a novel anti-necrotic death factor that modulates cell death in early replication cycle in fish cells. Virology 385:444–454
Fenner BJ, Thiagarajan R, Chua HK, Kwang J (2006) Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J Virol 80:85–94
Iwamoto T, Mise K, Takeda A, Okinaka Y, Mori K, Arimoto M, Okuno T, Nakai T (2005) Characterization of Striped jack nervous necrosis virus subgenomic RNA3 and biological activities of its encoded protein B2. J Gen Virol 86:2807–2816
Chen SP, Wu JL, Su YC, Hong JR (2007) Anti-Bcl-2 family members, zfBcl-x(L) and zfMcl-1a, prevent cytochrome c release from cells undergoing betanodavirus-induced secondary necrotic cell death. Apoptosis 12:1043–1060
Su HL, Liao CL, Lin YL (2002) Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J Virol 76:4162–4171
Harding HP, Calfon M, Urano F, Novoa I, Ron D (2002) Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol 18:575–599
Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM (2002) The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol 3:411–421
Li XD, Lankinen H, Putkuri N, Vapalahti O, Vaheri A (2005) Tula hantavirus triggers pro-apoptotic signals of ER stress in Vero E6 cells. Virology 333:180–189
Rao RV, Bredesen DE (2004) Misfolded proteins, endoplasmic reticulum stress and neurodegeneration. Curr Opin Cell Biol 16:653–662
Williams BL, Lipkin WI (2006) Endoplasmic reticulum stress and neurodegeneration in rats neonatally infected with borna disease virus. J Virol 80:8613–8626
Chen X, Shen J, Prywes R (2002) The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem 277:13045–13052
Lee K, Tirasophon W, Shen X, Prywes M, Okada R, Yoshida RH, Mori K, Kaufman RJ (2002) IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev 16:452–466
Szegezdi E, Logue SE, Gorman AM, Samali A (2006) Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Rep 7:880–885
Baltzis D, Qu LK, Papadopoulou S, Blais JD, Bell JC, Sonenberg N, Koromilas AE (2004) Resistance to vesicular stomatitis virus infection requires a functional cross talk between the eukaryotic translation initiation factor 2 alpha kinases PERK and PKR. J Virol 78:12747–12761
Netherton CL, Parsley JC, Wileman T (2004) African swine fever virus inhibits induction of the stress-induced proapoptotic transcription factor CHOP/GADD153. J Virol 78:10825–10838
He B (2006) Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ 13:393–403
Dimcheff DE, Faasse MA, McAtee FJ, Portis JL (2004) Endoplasmic reticulum (ER) stress induced by a neurovirulent mouse retrovirus is associated with prolonged BiP binding and retention of a viral protein in the ER. J Biol Chem 279:33782–33790
Tardif KD, Mori K, Siddiqui A (2002) Hepatitis C virus subgenomic replicons induce endoplasmic reticulum stress activating an intracellular signaling pathway. J Virol 76:7453–7459
Bitko V, Barik S (2001) An endoplasmic reticulum-specific stress-activated caspase (caspase-12) is implicated in the apoptosis of A549 epithelial cells by respiratory syncytial virus. J Cell Biochem 80:441–454
Farrow SN, Brown R (1996) New members of the Bcl-2 family and their protein partners. Curr Opin Genet Dev 6:45–49
Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 hetrodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 74:609–619
Zamzami N, Kroemer G (2001) The mitochondrion in apoptosis: how Pandora’s box opens. Nat Rev Mol Cell Biol 2:67–71
Oyadomari S, Mori M (2004) Roles of CHOP/GADD153 in endoplasmic reticulum stress. Cell Death Differ 11:381–389
Chen SP, Yang HL, Her GM, Lin HY, Jeng MF, Wu JL, Hong JR (2006) Betanodavirus induces phosphatidylserine exposure and loss of mitochondrial membrane potential in secondary necrotic cells, both of which are blocked by bongkrekic acid. Virology 347:379–391
Wu HC, Wu JL, Chu HL, Su YC, Hong JR (2010) RGNNV induces mitochondria-mediated cell death via newly synthesized protein dependent pathway in fish cells. Fish Shellfish Immunol 29:457–463
Su YC, Wu JL, Hong JR (2009) Betanodavirus non-structural protein B2: a novel necrotic death factor that induces mitochondria-mediated cell death in fish cells. Virology 385:143–154
Nicholson B, Dunn J (1974) Homologous viral interference in trout and Atlantic salmon cell cultures infected with infectious pancreatic necrosis virus. J Virol 14:180–182
Yang GH, Li S, Pestka JJ (2000) Down-regulation of the endoplasmic reticulum chaperone GRP78/BiP by vomitoxin (Deoxynivalenol). Toxicol Appl Pharmacol 162:207–217
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Kain SR, Mai K, Sinai P (1994) Human multiple tissue western blots: a new immunological tool for the analysis of tissue-specific protein expression. Biotechniques 17:982–987
Hong JR, Wu JL (2002) Induction of apoptotic death in cells via Bad gene expression by infectious pancreatic necrosis virus infection. Cell Death Differ 9:113–124
Chen PC, Wu JL, Her GM, Hong JR (2010) Aquatic birnavirus induces necrotic cell death via mitochondria-mediated caspases pathway that inhibited by bongkrekic acid. Fish Shellfish Immunol 28:344–353
Mullen PD, Brand RJ, Parlette GN (1975) Evaluation of dye exclusion and colony inhibition techniques for detection of polyoma-specific, cell-mediated immunity. J Natl Cancer Inst 54:229–231
Mezeth KB, Nylund S, Henrisksen H, Patel S, Nerland AH, Szilvay AM (2007) RNA-dependent RNA polymerase from Atlantic halibut nodavirus contains two signals for localization to the mitochondria. Virus Res 130:43–52
Miller DJ, Ahlquist P (2002) Flock house virus RNA polymerase is a transmembrane protein with amino-terminal sequences sufficient for mitochondrial localization and membrane insertion. J Virol 76:9856–9867
Priscilla M, Wynsberghe V, Ahlquist P (2009) 5′ cis elements direct nodavirus RNA1 recruitment to mitochondrial sites of replication complex formation. J Virol 83:2976–2988
Bovo G, Nishizawa T, Maltese C, Borghesan F, Mutinelli F, Montesi F, De Mas S (1999) Viral encephalopathy and retinopathy of farmed marine fish species in Italy. Virus Res 63:143–146
Mazzarella RA, Marcus N, Haugejorden SM, Balcarek JM, Baldassare JJ, Roy B, Li LJ, Lee AS, Green M (1994) Erp61 is GRP58, a stress-inducible luminal endoplasmic reticulum protein, but is devoid of phosphatidylinositide-specific phospholipase C activity. Arch Biochem Biophys 308:454–460
Pahl HL (1999) Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol Rev 79:683–701
Buchkovich NJ, Maquire TG, Yu Y, Paton AW, Paton JC, Alwine JC (2007) Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J Virol 82:31–39
Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103:239–252
Lamkanfi M, Kalai M, Vandenabeele P (2004) Caspase-12: an overview. Cell Death Differ 11:365–368
Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M (2001) Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem 276:13935–13940
Rutkowski DT, Kaufman RJ (2004) A trip to the ER: coping with stress. Trends Cell Biol 14:20–28
Dufresne PJ, Thivierge K, Cotton S, Beauchemin C, Ide C, Ubalijoro E, Laliberte JF, Fortin MG (2008) Heat shock 70 protein interaction with Turnip mosaic virus RNA-dependent RNA polymerase within virus-induced membrane vesicles. Virology 374:217–227
Miller DJ, Schwartz MD, Ahlquist P (2001) Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol 75:11664–11676
Weeks SA, Shield WP, Sahi C, Craig EA, Rospert S, Miller DJ (2010) A targeted analysis of cellular chaperones reveals contrasting roles for heat shock protein 70 in flock house virus RNA replication. J Virol 84:330–339
Germain M, Mathai JP, Shore GC (2002) BH-3-only BIK functions at the endoplasmic reticulum to stimulate cytochrome c release from mitochondria. J Biol Chem 277:18053–18060
Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ (2003) BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science 300:135–139
Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ (2001) Proapoptotic BAX and BAK: a requisite gateway to mitochondrial dysfunction and death. Science 292:727–730
Kim I, Xu W, Reed JC (2008) Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov 7:1013–1030
Acknowledgements
The authors are grateful to Dr. S. C. Chi (Institute of Zoology and Development of Life Science, Taiwan, ROC) for providing the grouper fin cell line GF-1. This work was supported by grant NSC 98-2313-B-006-004-MY3 from the National Science Council, Taiwan, Republic of China, awarded to Dr. Jainn-Ruey Hong.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, YC., Wu, JL. & Hong, JR. Betanodavirus up-regulates chaperone GRP78 via ER stress: roles of GRP78 in viral replication and host mitochondria-mediated cell death. Apoptosis 16, 272–287 (2011). https://doi.org/10.1007/s10495-010-0565-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-010-0565-x