Abstract
A label-free method for detecting the attachment of human cancer cells to a biosensor surface for rapid screening for biological activity is described, in which attachment of a cell results in highly localized increase of the resonant reflected wavelength of a photonic crystal narrowband reflectance filter incorporated into a standard 96-well microplate. An imaging detection instrument is used to determine the spatial distribution of attached cells by mapping the shift in reflected resonant wavelength as a function of position. The method enables monitoring of cancer cell attachment, cell proliferation, and cell detachment that is induced by exposure of the cells to drug compounds. We demonstrate the efficacy of this method as an early screening technique for the rapid quantification of the rate of cancer cell proliferation on the sensor surface, and subsequently as a means for quantifying cell detachment resulting from apoptosis that is induced by exposure of the cells to cytotoxic chemicals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cell-based assays are becoming an increasingly important part of the preclinical pharmaceutical discovery and validation process, as researchers directly study the effects of chemical compounds upon a wide variety of cell types. The ability to rapidly quantify the toxicity and selectivity of potential drug treatments to both targeted cell populations and to non-targeted cells would provide a means for efficiently selecting compounds for further study that are likely to perform a desired function without undesired side effects. A variety of assays for measuring necrosis, apoptosis, and cell proliferation are currently in widespread usage that are capable of providing information from cell populations or individual cells.
The most common necrosis assay measures the increased plasma membrane permeability of dying cells, as leaky membranes permit perfusion of a dye or stain [1]. Conversely, colorimetric assays for necrosis can measure the metabolic activity of mitochondria in live cells by their ability to metabolize dyes [1], [2]. However, because the early phases of apoptosis do not affect membrane permeability or alter mitochondrial activity, necrosis assays are not suitable for characterizing apoptosis [2].
Methods for determining apoptosis typically rely upon detection of fragmented genomic DNA [3], [4], labeling of the protein phosphatidylserine (PS) translocated to the outer surface of the plasma membrane [2], [5], detection of released proteins such as cytochrome C and apoptosis inducing factor (AIF) [6], or detection of activated caspases [7], [8]. These methods measure biochemical consequences resulting from the apoptosis process, but each requires substantial manipulation of the cells to extract the required analyte, label it with a dye, and to perform the measurement. Regardless of whether the protocol calls for ELISA, flow cytometry, a fluorescence microplate reader, or fluorescence microscopy, each method results in removal of the cells from their culture environment, the destruction of the cells being measured, special reagents, and multiple washing/centrifugation steps. These methods are either capable of measuring the aggregate properties of large cell populations or small populations of individual cells, but generally not both simultaneously, particularly with high assay throughput.
In cell proliferation assays, a defined number of cells are typically plated onto an appropriate matrix and the number of colonies that are formed after a period of growth are enumerated [9]. Because this method is time-consuming and laborious, it is not practical for large numbers of samples, particularly for establishing growth curves for cell populations. Therefore, indirect analysis of cell proliferation through measurement of DNA synthesis rate through the incorporation of labeled DNA precursors during cell division [9]. As with apoptosis assays, each proliferation assay requires staining of the cells with proprietary reagents (resulting in cell death), removal of cells from their culture environment, a multi-step assay protocol, and detection of either the aggregate properties of cell populations (radiometry or fluorescence microplate scanner) or the individual properties of a small number of cells with low throughput (flow cytometry, fluorescence microscopy, or light microscopy).
In this work, a label-free detection system based upon the unique properties of photonic crystal biosensors and a high resolution imaging detection instrument is described that is capable of quantifying cytotoxicity and proliferation. The system is capable of detecting individual cells, but also measures large populations of cells with high throughput. The sensor is incorporated within standard 96-well microplates, and allows the same cells to be measured many times without removal from their liquid environment. The assay protocols for measuring cytotoxicity and proliferation require no additional reagents, centrifugation steps, wash steps, and are not subject to quenching. Here, we demonstrate the first use of an imaging instrument to detect the attachment of cells to the surface of a photonic crystal biosensor. The imaging instrument enables spatial maps of the attached cell density in the bottom of the microplate wells to be measured by detecting the local changes in the reflection spectrum provided by the photonic crystal. The capability for image-based label-free detection of cell attachment to the sensor is used to visualize and quantify the rates of human breast cancer cell proliferation and subsequent cell apoptosis induced by the introduction of a cytotoxic chemical compound.
Materials and methods
Photonic crystal biosensors
Optical biosensors offer a means for detecting the attachment of cells to the surface of a transducer through their increased dielectric permittivity with respect to their liquid media [10]. While optical biosensors based upon surface plasmon resonance have been used for many years to characterize macromolecular affinity interactions [11], they have not found widespread usage for detecting cells for several reasons. The detection system is generally arranged only to interrogate a small (<1 mm2) region of the sensor surface within a narrow (∼50 μm wide by 50 μm tall) flow channel under a constant flow of buffer. Blockage of the flow channel by cells, low assay throughput, and assay conditions not amenable to maintaining viable cells (such as lack of incubation) have prevented biosensor-based methods from becoming widely adopted.
A new class of optical biosensors based on the unique properties of optical device structures known as photonic crystals have been recently developed [12], [13]. A photonic crystal is composed of a periodic arrangement of dielectric material in two or three dimensions [14], [15]. A photonic crystal structure geometry can be designed to concentrate light into extremely small volumes and to obtain very high local electromagnetic field intensities. As shown in previous publications, the photonic crystal biosensor behaves as a narrowband wavelength reflectance filter, where the sensor reflects ∼100% of incident light at the resonant wavelength, while all other wavelengths are transmitted through the sensor structure.
Recently, we demonstrated a photonic crystal optical biosensor that can be incorporated into standard format 96-well microplates for the purpose of detecting the attachment of cells to the biosensor surface without the use of fluorescent labels or any other type of marker, and that the attachment of cells to the biosensor can be modulated by the immobilization of cognizant ligands to the sensor surface that selectively recognize expressed outer membrane proteins [16]. In our previous work, cells were detected with an instrument that was capable of quantifying the attached cell density on the surface of the photonic crystal biosensor within the microplate wells, but that did not provide images of the attached cell density distribution.
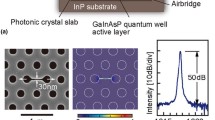
(a) Schematic diagram of the imaging detection instrument. (b) Cross-section schematic of photonic crystal biosensor. (c) Reflected resonant spectrum measured from a single ∼22.3 × 22.3 μm2 pixel of the photonic crystal with water on its surface, where “PWV” represents the Peak Wavelength Value of the reflected peak
The sensor operates by measuring changes in the wavelength of reflected light as biochemical or cellular binding events take place within an evanescent field region adjacent to the surface. The resonant reflected wavelength of the sensor can be measured by illuminating the photonic crystal at normal incidence with white light, and collecting the reflected light with a spectrometer. Illumination and collection may be performed through optical fibers to gather single measurements over a large illuminated spot, or free space optics can be used to generate a spatial image of the resonant reflected wavelength. Image-based detection with high spatial resolution is enabled by the photonic crystal structure, which is designed to cut off lateral propagation of the resonant wavelength, thus eliminating pixel-to-pixel optical cross-talk. The sensors are inexpensively fabricated on sheets of plastic film, and incorporated into standard microplates that are compatible with cell culture methods and standard microplate handling procedures.
The photonic crystal is comprised of a 1-dimensional periodic grating surface structure (period = 550 nm) formed within a UV-cured polymer on a transparent polyester sheet using a room-temperature replica molding process. The low refractive index grating is subsequently coated with a film of high refractive index TiO2 by sputtering to achieve the final photonic crystal cross section shown in Fig. 1. The biosensors are cut from the polyester sheet and attached with adhesive to form the bottom surface of standard 96-well format microplates [17]. The detection instrument, shown in Fig. 1, illuminates the sensor surface at normal incidence with a white light lamp (Oriel), and a line from the sensor surface is imaged into the entrance slit of an imaging spectrometer (Acton Research). The reflected spectrum of each pixel across the image line is gathered (Fig. 1(b)), and the Peak Wavelength Value (PWV) of each resonant reflection spectrum is mathematically determined. A PWV image is constructed by sequential scanning of the sensor across the imaged line region in 22.3 μm increments. The detection instrument measures changes in the resonant reflected PWV of the biosensor surface as the detected output on a pixel-by-pixel basis that can generate images of PWV with a spatial resolution as low as 4×4 μm2. An imaging resolution of 22.3×22.3 μm2 was used for all the images reported in this work, as higher resolution scans require time and computer memory in proportion to number of image pixels. Detection of cell attachment to the sensor requires measuring a shift in PWV, so the sensor surface is scanned twice: once before and once after cells have been immobilized on the surface. The two images are aligned and subtracted mathematically to determine the difference in PWV as detected by the sensor. Because the PWV is only increased for pixels in which a cell has attached to the sensor, the imaging instrument measures the density of cell binding as a function of position within the microplate well.
MCF-7 cell line
The human breast cancer cell line MCF-7 was purchased from American Type Culture Collection (ATCC). The cells were cultured in minimum essential medium with 10% fetal bovine serum and antibiotic-antimycotic. The cells were maintained in an incubator at 37°C with 5% carbon dioxide.
Incubation with known drugs and plant extracts
The anticancer agents doxorubicin hydrochloride and curcumin were purchased from Sigma Aldrich. Plant extracts from the species Protium serratum and Sapindus mukurossi were provided by Professor R Chowdhury from University of Dhaka, Bangladesh. Stock solutions of the plant extracts were prepared by dissolving them in ethanol at 25 mg/ml. The plant extracts were dissolved in ethanol and diluted with cell culture media to a concentration of 100 μg/ml. Doxorubicin was dissolved in sterile cell culture grade water and curcumin was dissolved in ethanol. Both drugs were diluted with cell culture media to a concentration of 100 μm. The final concentration of ethanol was less than 1% in sample solution.
Doxorubicin IC50 measurement
The IC50 value of doxorubicin exposed to MCF-7 breast cancer cells was measured using the photonic crystal biosensor and compared to a standard colorimetric assay (MTT). Eight concentrations of doxorubicin were introduced to triplicate wells of MCF-7 cells at 0.5, 1.0, 2.5, 5.0, 10, 25, 50, and 100 μM. The cells were allowed to stabilize and scanned after 24 h of incubation and scanned again after 24 h of doxorubicin incubation.
MCF-7 cancer cell detection protocol
The assay protocol is outlined in Fig. 3. The biosensor microplate wells are initially filled with 100 μL cell culture media, and a baseline PWV image is scanned (Scan “A”). Next, cells are introduced to the wells by pipetting ∼500–1000 cells per well and allowing them to attach to the sensor surface for ∼24 h in the CO2 incubator. After the cells have attached to the sensor surface, a second PWV image is scanned (Scan “B”). Subtraction of Scan “A” from Scan “B” generates a PWV shift image that measures the distribution of attached cells. Next, chemical compounds are introduced to the microplate wells by pipette, and the microplate is returned to the incubator for an additional 24 h before a third PWV image is scanned (Scan “C”). Subtraction of Scan “A” from Scan “C” generates a new PWV shift image of the cell distribution. The PWV shift images of the original cell distribution may be compared with the final cell distribution after chemical exposure to determine whether the cells proliferated or detached from the sensor during the final 24-h period. All conditions were measured in triplicate wells, and wells without chemical compound introduction were utilized as experimental control.
(a) PWV shift image of a single 6 mm diameter well of a microplate with MCF-7 cells immobilized on the surface and (b) the PWV shift as a function of lateral position for one horizontal line from the PWV image, selected to intersect single cells and cell clusters, where the threshold PWV shift value selected to indicate cell attachment is shown. (c) Histogram of PWV shift values for the pixels within the image, showing (inset) the distribution of pixels above the PWV shift threshold for cell attachment
(a) PWV shift images of a single 6 mm diameter microplate well after MCF-7 cell attachment (upper), and the same cells after 24 h of incubation (lower) without chemical exposure, where proliferation results in ∼2× increase in the number of attached cells. (b) PWV shift images of attached MCF-7 cells before and after 24 h incubation in the presence of extract from Sapindus mukurossi and (c) Protium serratum, which display accelerated proliferation with respect to the control and the formation of cell clusters. (d) PWV shift images of attached MCF-7 cells before and after 24 h incubation in the presence of doxorubicin and (e) curcumin, in which ∼90% of the cells undergo apoptosis, and release from the biosensor surface
Results
Figure 2(a) shows a typical PWV shift image for the attachment of MCF-7 cells to the biosensor (Scan “B”—Scan “A”). The PWV shift as a function position is represented as a false color image, and the PWV shift through a horizontal line that intersects multiple cells is shown (Fig. 2(b)). Each pixel location with an attached cell results in a locally elevated PWV that does not extend to neighboring pixels, thus demonstrating negligible pixel-to-pixel optical cross-talk. The cells are found distributed randomly across the biosensor area, but are often found to cluster in groups. Cell clusters were observed visually by microscope corresponding to the locations of cells clusters observed by the PWV shift image. Each pixel with a single attached cell results in a PWV shift of ∼0.6 nm, and pixels within a cell cluster result in PWV shifts up to ∼2.0 nm, possibly due to the presence of multiple cells (∼10 μm diameter) within a single pixel.
In order to estimate the number of immobilized cells, we apply a PWV shift threshold to each pixel in the PWV shift image to determine whether cell attachment has resulted in a locally higher PWV. The horizontal bar shown in Fig. 2(b) represents a PWV shift threshold of 0.62 nm. For each well, a histogram can be used to visualize the proportion of pixels with a PWV shift above the selected threshold, as shown in Fig. 2(c). For this work, all pixels above the threshold are counted as a single cell, thus it is possible that this simple algorithm would undercount cells for pixels with large PWV shift due to multiple cells/pixel.
An example of MCF-7 cell proliferation is shown in Fig. 3(a), where the PWV shift image is shown after cell attachment after subsequent proliferation in the CO2 incubator for 24 h. Without introduction of chemical compounds, the cell count shows an average of ∼240% increase.
Having established that the sensor and detection instrument are capable of measuring cell attachment and cell proliferation, we studied how the process of cell proliferation may be modified by the addition of a chemical compound during the proliferation phase of the assay. Curcumin and doxorubicin are both known to induce apoptosis in MCF-7 cells [18]–[21], while the plant extracts, Protium serratum and Sapindus mukurossi [22], have unknown effect on the breast cancer cells. However, Protium serratum is a medicinal plant that has been explored in earlier studies of allelopathic activity [23]. Sapindus mukurossi is also a medicinal plant that is known to possess potent spermicidal activity and hence can be used as an aid in contraceptive methods [24], and monodesmosides extracted from pericarps of Sapindus mukurossi have been reported to enhance the rectal absorption of antibiotics in rats [25].
Figure 3(b) and (c) shows the PWV shift images before and after the MCF-7 proliferation in the presence of Sapindus mukurossi and Protium serratum, respectively. In each case, the cells are observed to grow rapidly, as quantified by the cell count, which increased by ∼200% for Sapindus mukurossi and 283% for Protium serratum. Figure 3(d) and (e) shows that the curcumin and doxorubicin wells have undergone termination of cell growth and loss of attachment to the biosensor. Since curcumin and doxorubicin are known to induce apoptosis in MCF-7 cells, the cells should be released from the surface of the sensor, thus returning the PWV image to its original “blank” baseline. The curcumin wells show an average of 93% decrease in cell count and the doxorubicin-induced wells show an average of 89% decrease, which closely correspond to the experimental results published previously [20].
In order to validate the biosensor cell viability measurement against a conventional apoptosis assay, the dose-response characteristic was obtained by exposure of the MCF-7 cells to range of concentrations of doxorubicin and compared to published results using the same cell line and drug using an MTT assay [26]. Figure 4 shows the cell viability as a function of doxorubicin concentration plotted on the same graph as the MTT assay (reference) for doxorubicin concentration ranging from 0.0001 to 100 μM. The MTT assay results in an IC50 value of approximately 1 μM while the biosensor assay results in an IC50 value of ∼2 μM. The correlation between the methods, plotted in the inset of Fig. 4, reveals excellent agreement between the two assays.
The dose-response characteristic for cell viability of MCF-7 cells exposed to doxorubicin results in an IC50 determination of ∼2 μM by the photonic crystal biosensor assay (solid line) and 1 μM by the MTT assay (dotted line). The correlation is plotted in the inset plot showing % Cell Viability of MTT versus photonic crystal biosensor (The MTT assay data was obtained from previously published work [26])
The label-free cell assay presented in this work represents a new method for direct visual observation of cell behavior without the use of labels or stains. Because the cells are maintained within their culture environment, they remain viable for repeated measurements that may extend over the course of several days. While the MCF-7 human breast cancer cells studied in this work adhere readily to the TiO2 surface of the photonic crystal, it is also possible to attach suspension cells to the sensor through an immobilized protein that will form a high affinity bond with proteins expressed on the outer surface of the cell.
Conclusion
The methods described are broadly applicable to a wide range of cell types, both diseased and healthy, to study the effects of exposure to potential pharmaceutical treatments or toxins. Although the cell-attachment images are useful for direct visualization of cell behavior, the image data is easily translated to quantitative measures such as a simple cell count. The ability to gather repeated images of the same cells enables not only two-step before/after comparison, but also multi-stepped “movies” of cell behavior including proliferation and chemotaxis. Although the method does not have sufficient optical resolution to gather detailed imageswithin a single cell, the method can easily detect a single cell, and is useful for studying large populations of cells. The plastic-based photonic crystal biosensor design allows the sensor to be inexpensively fabricated over a large enough surface area to be incorporated into standard cell assay laboratory formats such as 96-well microplates. The method represents a new approach for visualization and quantitative measurement of a broad range of cell types to the effects of the cells’ environment, including chemical exposure.
References
Dive C et al (1992) Analysis and discrimination of necrosis and apoptosis (programmed cell-death) by multiparameter flow-cytometry. Biochimica Et Biophysica Acta 1133:275–285
Schwartzman RA, Cidlowski JA (1993) Apoptosis—the biochemistry and molecular-biology of programmed cell-death. Endocr Rev 14:133–151
Huang P, Plunkett WA (1992) Quantitative assay for fragmented Dna in apoptotic cells. Anal Biochem 207:163–167
Bortner CD, Oldenburg NBE, Cidlowski JA (1995) The role of Dna fragmentation in apoptosis. Trends Cell Biol 5:21–26
Verhoven B, Schlegel RA, Williamson P (1995) Mechanisms of phosphatidylserine exposure, a phagocyte recognition signal, on apoptotic T-lymphocytes. J Exp Med 182:1597–1601
Petit PX, Susin SA, Zamzami N, Mignotte B, Kroemer G (1996) Mitochondria and programmed cell death: Back to the future. FEBS Lett 396:7–13
Zou H, Henzel WJ, Liu XS, Lutschg A, Wang XD (1997) Apaf-1, a human protein homologous to C-elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 90:405–413
Ellis HM, Horvitz HR (1986) Genetic-control of programmed cell-death in the nematode C-elegans. Cell 44:817–829
Takagi S, McFadden ML, Humphreys RE, Woda BA, Sairenji T (1993) Detection of 5-bromo-2-deoxyuridine (brdurd) incorporation with monoclonal anti-brdurd antibody after deoxyribonuclease treatment. Cytometry 14:640–648
Zourob M et al (2005) Bacteria detection using disposable optical leaky waveguide sensors. Biosens Bioelectron 21:293–302
Cooper MA (2003) Label-free screening of bio-molecular interactions. Nature 377:834–842
Cunningham BT, Li P, Lin B, Pepper J (2002) Colorimetric resonant reflection as a direct biochemical assay technique. Sensors and Actuators B 81:316–328
Haes AJ, Duyne RPV (2002) A nanoscale optical biosensor: sensitivity and selectivity of an approach based on the localized surface plasmon resonance spectroscopy of triangular silver nanoparticles. J Am Chem Soc 124:10596–10604
Joannopoulos JD, Meade RD, Winn JN (1995) Photonic crystals. Princeton University Press, Princeton, NJ
Munk BA (2000) Frequency selective surfaces. John Wiley & Sons, New York
Lin B, Li P, Cunningham BT (2005) A label-free biosensor-based cell attachment assay for characterization of cell surface molecules. Sensors and Actuators B (Accepted April 2005)
Cunningham BT et al (2004) Label-free assays on the BIND system. J Biomol Screen 9:481–490
Coleman RE et al (2006) A randomised phase II study of two different schedules of pegylated liposomal doxorubicin in metastatic breast cancer (EORTC-10993). Eur J Cancer 42:882–887
Joyner DE, Bastar JD, Randall RL (2006) Doxorubicin induces cell senescence preferentially over apoptosis in the FU-SY-1 synovial sarcoma cell line. J Orthop Res 24:1163–1169
Syng-ai C, Kumari AL, Khar A (2004) Effect of curcumin on normal and tumor cells: Role of glutathione and bcl-2. Mol Can Ther 3:1101–1108
Suwei Wang EAK, Kotamraju Srigiridhar, Joseph Joy, Kalivendi Shasi, Kalyanaraman B (2004) Doxorubicin induces apoptosis in normal and tumor cells via distincly different mechanisms intermediacy of H202-AND p53-dependent pathways. J Biol Chem 279:25535–25543
Sun JR, Cheng KC, Pan TY, Si XM (2002) A new acyclic sesquiterpene oligoglycoside from pericarps of Sapindus mukurossi. Chin Chem Lett 13:555–556
Yoshiharu Fujii SSP, Mohammad Masud Parvez, Yoshio Ohmae, Osamu Iida (2003) Screening of 239 medicinal plant species for allelopathic activity using the sandwich method. Weed Biol Manag 3:233–241
Dhar JD, BV, Setty BS, Kamboj VP (1989) Morphological changes in human spermatozoa as examined under scanning electron microscope after in vitro exposure to saponins isolated from Sapindus mukorossi. Contraception 39:563–568
Yata N, SN, Yamajo R, Murakami T, Higashi Y, Kimata H, Nakayama K, Kuzuki T, Tanaka O (1985) Enhanced rectal absorption of beta-lactam antibiotics in rat by monodesmosides isolated from pericarps of Sapindus mukurossi (Enmei-hi). J Pharmacobiodyn 8:1041–1047
Chi KN et al (2000) Effects of Bcl-2 modulation with G3139 antisense oligonucleotide on human breast cancer cells are independent of inherent Bcl-2 protein expression. Breast Can Res Treat 63:199–212
Acknowledgments
This material is based upon work supported by the National Science Foundation under Grant no. 0427657. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation. The authors would like to thank Professor R. Chowdhury from University of Dhaka, Bangladesh for providing the plant extracts. The authors gratefully acknowledge SRU Biosystems for the providing the photonic crystal biosensor microplates. The authors also extend their gratitude to the support staff of the Micro and Nanotechnology Laboratory at the University of Illinois at Urbana-Champaign.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chan, L.L., Gosangari, S.L., Watkin, K.L. et al. A label-free photonic crystal biosensor imaging method for detection of cancer cell cytotoxicity and proliferation. Apoptosis 12, 1061–1068 (2007). https://doi.org/10.1007/s10495-006-0031-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10495-006-0031-y