Abstract
Varroa destructor is one of the scourges of global beekeeping. It was detected for the first time in Benin in 2011 on the honeybee Apis mellifera adansonii. The aim of this study was to identify the strain of Varroa sp. found and study its genetic diversity. In total 183 Varroa mites were sampled in 21 municipalities in Benin. The COI intergenic region of each mite mtDNA was amplified by PCR. The SacI restriction enzyme was used to determine the strains of Varroa sp. Only the Korean (K) haplotype, identical to the most prevalent strain in Africa, was detected. Analysis of the genetic diversity of Varroa mites with eight microsatellite loci (Simple Sequence Repeats) indicated a very low diversity of genotypes. Thus, V. destructor populations from Benin appear to make up a single group. Their clonal wealth ranges from 0.00 to 0.47. This study is an important step forward in the monitoring of the infestation of V. destructor.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the main problems facing the beekeeping industry is the infestation of bee colonies by the parasitic mite Varroa destructor (Anderson and Trueman 2000). Varroa destructor lives as a haematophagous ectoparasite on bees (Wendling 2014). It is an ecto-parasite on Apis mellifera honeybees (Colin et al. 1997). In addition to the high rates of V. destructor mite infestation in A. mellifera colonies, V. destructor is also a vector of various infectious diseases, such as the acute paralysis virus, Kashmir virus and deforming wing virus (Bakonyi et al. 2002; Chen et al. 2004; Tentcheva et al. 2006). In Africa, beekeeping activity is mainly based on swarms of trapped bees. The presence of V. destructor in sub-Saharan Africa was mentioned for the first time in South Africa in 1997. The mite was detected on the two local subspecies A. m. capensis and A. m. scutellata. It was also detected in 2009 in several countries of East Africa (Kenya, Tanzania, and Uganda) and recently in part of West Africa (Viniwanou 2009; Fazier et al. 2010). Varroa destructor was found on the endemic bee A. m. adansonii in Benin where most of the honey production is the result of bee hunting (Paraiso et al. 2011, 2012a).
Africa is the only continent with a large natural population of bees (Paraiso et al. 2012b). The presence of V. destructor then raises specific concerns. The spread of this parasite is not only a threat to health and the monitoring of colonies but also to wild bee populations that pollinate 40–70% of native plants (Dietmann et al. 2009). Indeed, major losses of bee colonies due to the actual parasite were reported in Benin. But in actual fact, V. destructor is a parasite which beekeepers from Benin know little about and as a result, it is often found late when it arrives in a region. It was identified in the colonies from Benin by Paraiso et al. (2011). Its infestation rate varies from 5 to 7% in Benin hives and this parasite was found in all the hives sampled during the present study. Further, the Varroa found in Benin originates from natural spread and movement of bees from neighboring countries. Its distribution in the hives is done especially through the exchange of bee colonies between close beekeepers in quest of colonies producing more honey. The control of Varroa in Benin is a major cause for concern because of the importance of bees in crop pollination. Many tropical crops, like cashew, rely on pollination by bees (Reddi 1987; Allsopp 2006). The colony losses could have disastrous economic impacts on small beekeepers from Benin who often associate beekeeping with other farming activities (De Holanda-Neto et al. 2002).
Several mitochondrial haplotypes of V. destructor were described in previous studies, but only two of them are able to reproduce on A. mellifera (Solignac et al. 2003). These are the Korean (K) and Japanese (J) polymorphic haplotypes (De Guzman et al. 1997, 1999). These mites were found parasitizing their original host A. cerana. The K type infests A. mellifera almost everywhere in the world, but the J type was observed on A. mellifera in Japan, the Americas (including Brazil), Thailand and Island (De Guzman et al. 1999; Garrido et al. 2003; Solignac et al. 2003, Brettell and Martin 2017). No data concerning the genetic diversity of this parasite exist in Benin.
The aim of this study was to identify the strain of V. destructor present in Benin and analyze its genetic diversity.
Materials and methods
Sample collection
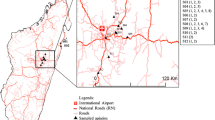
Honeybee (A. m. adansonii) samples were collected from 20 municipalities located in Northwest, Northeast and Central Benin (Fig. 1). Between 100 and 300 bees per hive were collected in containers with 96% ethanol and transported to the laboratory for further analysis. All receptacles with the bees were stirred for about 2 min and the mites were collected separately in 96% ethanol and stored at 4 °C for subsequent DNA analysis.
DNA extraction
To extract DNA each mite was ground and added to 300 µl of Chelex (10%) heated at 60 °C (Walsh et al. 1991). Ten μl of proteinase K (0.25 mg/ml) was added to the mixture, and the whole was placed at 50 °C for 1 h and then at 95 °C for 30 min in a heating appliance. The eluates containing the DNA were centrifuged for 10 min at 12,000 rpm and stored at −20 °C until use.
Amplification of mtDNA and enzyme restriction with SacI
The mitochondrial (mt) DNA of Varroa was PCR-amplified using the primers COXF (5′-TACAAAGAGGGAAGAAGCAGCC-3′) and COXR (5′GCCCCTATTCTTAATACATAGTGAAAAATG-3′). PCR was performed based on the method proposed by Anderson and Fuchs (1998).
The COI intergenic region of Varroa mtDNA was amplified according to the following program: initial denaturation (92 °C for 4 min) followed by 35 cycles of denaturation (92 °C for 1 min), annealing (52 °C for 90 s), initial elongation (72 °C for 90 s) and final elongation (72 °C for 3 min), and finally steadiness at 15 °C. The product was electrophoresed on 2% agarose gel stained with ethidium bromide. The rest of the PCR product was digested for 4 h at 37 °C with the SacI restriction enzyme to identify the strain of Varroa present in Benin (De Guzman et al. 1997). The digested products were examined on 2% agarose gel stained with ethidium bromide and visualized by ultraviolet light.
Analysis of nuclear DNA
To study the genetic diversity within the mites investigated, the analysis of their nuclear genome was carried out through the amplification of eight microsatellite loci VD163, VD001, VD114, VD016, VD112, VD119, VD126 and VD152 (Solignac et al. 2005). PCR was performed using the method of Dynes et al. (2016). In order to identify the genotypes, the PCR products were then prepared for capillary electrophoresis by mixing 1 µl of the PCR product with 9 µl of formamide (GENSCAN). The mixtures were genotyped using an automated DNA Sequencer ‘Applied Biosystems (ABI 3130XL)’. The identification of alleles at each locus was performed by comparison with the size standard (Rox) using the GeneMapper software (AB) 4.0 V.
Statistical analysis
The data were compiled in an Excel 2010 spreadsheet and Varroa strains were identified. The statistical analysis of the population genetics was performed using the GeneAlex package v.6.5 and the Power Markers package v.3.2. Individuals with missing genotype data for more than 40% of all loci were excluded from statistical analysis. The diallelic data elicited from the 08 SSR markers were used to estimate genetic diversity within the parasites by means of the GeneAlex software package. The polymorphic information content (PIC) was calculated and the genetic diversity was estimated with the Power Marker and GeneAlex packages. The structure of the genetic diversity tree was constructed using the PowerMarker software package based on Nei’s distance method and UPGMA. The dissimilarity matrix that generated SSR diallelic data was subjected to principal component analysis (PCoA) of coordinates with the GeneAlex package v.6.2.
Results
Variability of fragment lengths of the COI region and polymorphism restriction
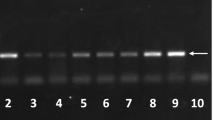
The fragments obtained on agarose gel reveal that all the Varroa samples tested have 563 bp as size (Fig. 2). None of the DNA samples was digested by the SacI enzyme. This suggests that the Varroa samples are not polymorphic and thus belong to the same K haplotype.
Genetic diversity parameters
Data on the genetic diversity parameters indicate that 7 out of the 8 markers used were amplified. It also appears that in 99% of cases, the same alleles were obtained for all samples analyzed at all loci. The majority of the primers used have a rate of Polymorphic Information Content (PIC) which is very low or nil. The allele frequency obtained ranges from 0.60 to 1 with an average of 0.94.
Grouping of Varroa colonies based on their similarities
The analysis indicates that all the Varroa mites analyzed are identical at a genetic distance of 1 with 0 as genetic distance between the individuals analyzed. However, only two samples collected in Central Benin can be considered as different from all other mites. In general, this analysis shows that all the Varroa samples are clones and do not vary from one municipality to another.
Discussion
The analysis of mitochondrial and microsatellite DNA helped to identify the subspecies of Varroa present in Benin. The PCR with mtDNA showed that there is no size variability, indicating that all the Varroa analyses belong to a single clone and corroborate those of Solignac et al. (2005). Indeed, those researchers indicated that the sub-species of Varroa (Japanese and Korean) have a clonal distribution.
The enzyme restriction did not show digested samples among the analyzed Varroa mites. According to Solignac et al. (2005), this showed that the analyzed Varroa samples belong to the Korean (K) haplotype. This haplotype is the most common sub-species in the world and is also present in Africa (Fazier et al. 2010). Indeed, the actual haplotype infests races of European bees and of pure African and Africanized races. The K haplotype is able to reproduce on all the subspecies of A. mellifera and on the other Apis species from Europe, Middle East, Asia, America and Africa (Solignac et al. 2003; Maggi et al. 2012).
The microsatellite DNA analysis of Varroa showed that, in 99% of cases, the same alleles were obtained for all samples, which indicates that 99% of the analyzed mites are genetically identical. Solignac et al. (2005) suggested that the eight markers used in this study are among the most polymorphic ones for analysis of genetic diversity of Varroa. The markers VD001, VD114, VD163, VD119, VD126 appeared monomorphic for the Varroa analyzed and therefore do not discriminate them, whereas VD 016 and VD112 are both polymorphic for the samples analyzed. The allelic frequency obtained ranged from 0.60 to 1 with an average of 0.94. These results confirm those of Solignac et al. (2005), i.e., a very low diversity within each sub-species of Varroa (Japanese and Korean). Similar results—low genetic variability—were observed with Varroa of A. m. iberiensis in Spain (Muñoz et al. 2008). In general, this analysis shows that all Varroa samples are clones and do not vary from one municipality to another.
References
Allsopp M (2006) Analysis of Varroa destructor infestation of southern African honeybee populations. M.Sc. thesis (Zoology), University of Pretoria, 2006. http://www.repository.up.ac.za/dspace/bitstream/handle/2263/27094/dissertation.pdf?sequence=1
Anderson D, Fuchs S (1998) Two genetically distinct populations of Varroa jacobsoni with contrasting reproductive abilities on Apis mellifera. J Apic Res 37(2):69–78. doi:10.1080/00218839.1998.11100957
Anderson DL, Trueman JWH (2000) Varroa jacobsoni (Acari: Varroidae) is more than one species. Exp Appl Acarol 24:165–189. doi:10.1023/A:1006456720416
Bakonyi T, Farkas R, Szendroi A, Dobos-Kovacs M, Rusvai M (2002) Detection of acute bee paralysis virus by RT-PCR in honey bee and Varroa destructor field samples: rapid screening of representative Hungarian apiaries. Apidologie 33:63–74. doi:10.1051/apido:2001004
Brettell LE, Martin SJ (2017) Oldest Varroa tolerant honey bee population provides insight into the origins of the global decline of honey bees. Sci Rep 7:45953. doi:10.1038/srep45953
Chen Y, Pettis JS, Evans JD, Kramer M, Feldlaufer MF (2004) Transmission of Kashmir bee virus by the ectoparasitic mite Varroa destructor. Apidologie 35:441–448. doi:10.1051/apido:2004031
Colin ME, Vandame R, Jourdan P, Dipasquale S (1997) Fluvalinate resistance of Varroa jacobsoni Oudemans (Acari: Varroidae) in Mediterranean apiaries of France. Apidologie 28:375–384. doi:10.1051/apido:19970605
De Guzman LI, Rinderer TE, Stelzer JA (1997) DNA evidence of the origin of Varroa jacobsoni Oudemans in the Americas. Biochem Genet 35:327–335. doi:10.1023/A:1021821821728
De Guzman LI, Rinderer TE, Stelzer JA (1999) Occurrence of two genotypes of Varroa jacobsoni Oud in North America. Apidologie 30:31–36. doi:10.1051/apido:19990104
De Holanda-Neto JP, Freitas BM, Bueno DM, De Araujo ZB (2002) Low seed/nut productivity in Cashew (Anacardium occidentale): effects of self-incompatibility and honey bee (Apis mellifera) foraging behaviour. J Hortic Sci Biotechnol 77:226–231. doi:10.1080/14620316.2002.11511484
Dietmann V, Pirk CWW, Crewe R (2009) Is there a need for conservation of honey bees in Africa? Apidologie 40:285–295. doi:10.1051/apido/2009013
Dynes TL, De Roode JC, Lyons JI, Berry JA, Delaplane KS, Brosi BJ (2016) Fine scale population genetic structure of Varroa destructor, an ectoparasitic mite of the honey bee (Apis mellifera). Apidologie 48:93–101. doi:10.1007/s13592-016-0453-7
Fazier M, Muli E, Conklin T, Schmehl D, Torto B, Frazier J, Tumlinson J, Evans DJ, Raina S (2010) A scientific note on Varroa destructor found in East Africa; threat or opportunity? Apidologie 41:463–465. doi:10.1051/apido/2009073
Garrido C, Rosenkranz P, Paxton RJ, Gonçalves LS (2003) Temporal changes in Varroa destructor fertility and haplotype in Brazil. Apidologie 34:535–541. doi:10.1051/apido:2003041
Maggi M, Medici S, Quintana S, Ruffinengo S, Marcangeli J, Gimenez Martinez P, Fuselli S, Equaras M (2012) Genetic structure of Varroa destructor populations infesting Apis mellifera colonies in Argentina. Exp Appl Acarol 56:309–318. doi:10.1007/s10493-012-9526-0
Muñoz I, Garrido-Bailón E, Martín-Hernández R, Meana A, Higes M, De la Rúa P (2008) Genetic profile of Varroa destructor infesting Apis mellifera iberiensis colonies. J Apicul Res Bee World 47:310–313. doi:10.3896/IBRA.1.47.4.13
Paraiso A, Olodo GP, Tokoudagba S, Sanni A (2011) Déterminants et contraintes de la production de miel dans le Nord-Ouest du Bénin: Cas des communes de Natitingou et de Tanguiéta. J Rech Sci Univ Lomé 14:69–84
Paraiso A, Sossou A, Iz-Haquou D, Yegbemey NH, Sanni A (2012a) Perceptions and adaptations of beekeepers and honey hunters to climate change: the case of the communes of Natitingou and Tanguieta in Northwest of Benin. Afr Crop Sci J 20:523–532
Paraiso A, Tokoudabga SF, Olodo GP, Yegbemey RN, Sanni A (2012b) Beekeeping and honey production in Norwest Benin: a case study of the communes of Natitingou and Tanguiéta. Int J Sci Adv Techonol 12:46–53
Reddi EUB (1987) Under-pollination: a major constraint of cashew nut production. Proc Indian Natn Sci Acad B53:249–252
Solignac M, Vautrin D, Pizzo A, Navajas M, Le Conte Y, Cornuet JM (2003) Characterization of microsatellite markers for the apicultural pest Varroa destructor (Acari: Varroidae) and its relatives. Mol Ecol Notes 3:556–559. doi:10.1046/j.1471-8286.2003.00510.x
Solignac M, Cornuet J-M, Vautrin D, Le Conte Y, Anderson D, Evans J, Cros-Arteil S, Navajas M (2005) The invasive Korea and Japan types of Varroa destructor, ectoparasitic mites of the Western honeybee (Apis mellifera), are two partly isolated clones. Proc Biol Sci 272:411–419. doi:10.1098/rspb.2004.2853
Tentcheva D, Gauthier L, Bagny L, Flevet J (2006) Comparative analysis of deformed wing virus (DWV) RNA in Apis mellifera and Varroa destructor. Apidologie 36:41–50. doi:10.1051/apido:2005057
Viniwanou N (2009) Caractéristiques morphométriques de l’abeille (Apis mellifera adansonii) et identification de quelques parasites et maladies dans les ruchers au Nord-Est du Bénin. Engineering Degree Dissertation, Faculty of Agronomy, University of Abomey-Calavi, Benin
Walsh PS, Metzqer DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Bio Techniques 10:506–513
Wendling S (2014) Reproductive features of Varroa destructor, agent of honeybee varroosis. Prospects for control. Bull Acad Vét France 167(4):309–3015
Acknowledgments
We thank the West African Agricultural Production Project (WAAPP) for the scholarship which was granted to us for our doctoral studies. We are also grateful to The International Foundation for Science (IFS) for its support by the grant B/5812-1. We thank all beekeepers who allowed us to collect samples of bees from their hives. Equally, we thank the Centre National de Recherche Scientifique (CNRS) of Gif-Sur-Yvette in France for the various analyses.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no Conflict of interests exists.
Rights and permissions
About this article
Cite this article
Kelomey, A.E., Paraiso, A., Sina, H. et al. Genetic characterization of the honeybee ectoparasitic mite Varroa destructor from Benin (West Africa) using mitochondrial and microsatellite markers. Exp Appl Acarol 72, 61–67 (2017). https://doi.org/10.1007/s10493-017-0141-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-017-0141-y