Abstract
Species of the family Phytoseiidae are known as predatory mites, some of them being used in crops to control mite pests, all around the world. Neoseiulus (=Cydnodromus) californicus is among the most commonly used Phytoseiidae species in biological control programs, especially in vineyards, orchards and vegetable fields. This species is distributed world-wide but has never been reported from Australia. On the other hand, specimens morphologically close to N. californicus have been assigned to a species called Neoseiulus wearnei, only reported from Australia. Investigations based on morphological and molecular comparisons were carried out to investigate whether these two taxa are conspecific. Morphological analyses showed no significant difference between specimens identified as N. wearnei and N. californicus. Similarly, genetic distances between these taxa were null, showing that all these specimens belong to the same species. Although it is not yet possible to conclude that all the specimens identified as N. wearnei are N. californicus, we can conclude that N. californicus is present in Australia. The information about the biology of N. californicus can thus now be applied to the Australian population of this species for biological control purposes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mite family Phytoseiidae Berlese contains several predatory species known to be efficient in controlling mite pests and small phytophagous insects, all over the world (Kostiainen and Hoy 1996; McMurtry and Croft 1997). One of the most commonly used species in biological control programs is Neoseiulus californicus (McGregor), recently re-included in the genus Cydnodromus (Tsolakis et al. 2012). This species was described from a lemon tree at Riverside, CA, USA (McGregor 1954) but it is now reported from all over the world (de Moraes et al. 2004). It has been commercialized and is currently used to control several mite pests in orchards, vineyards and vegetable fields (i.e. Castagnoli et al. 1999; Escudero and Ferragut 2005; Gotoh et al. 2006; Palevsky et al. 2008). It also occurs naturally in uncultivated areas (Ragusa et al. 1995; Tixier et al. 2000; Ragusa and Vargas 2002).
To ensure its success in biological control programs, the correct identification of this species is required. Yet, this is not so easy as (1) several species have been reported to be morphologically close to N. californicus (Athias-Henriot 1977; Chant and McMurtry 2003; de Moraes et al. 2004; Tixier et al. 2008); (2) attempts to obtain the type specimens of N. californicus by several authors were unsuccessful (Beard pers. com.; Ragusa 2003; Tixier et al. 2008); and (3) the description of N. californicus was based on male specimens (McGregor 1954), with the female specimens being initially identified as Neoseiulus mungeri (McGregor) on the same host plant and locality in Riverside on lemon tree in the same publication and synonymized by Chant (1959) and then by Schuster and Pritchard (1963).
Recent studies have thus been carried out to provide morphological and molecular diagnostics for this species (Tixier et al. 2008; Okassa et al. 2011). Tixier et al. (2008) provided a morphological re-description of this species based on 300 specimens collected in nine countries. Okassa et al. (2011) provided both molecular (three genes: 12S rRNA, CytB mtDNA, ITS) and morphological characterization of intraspecific variations of this species, based on 333 specimens collected in 11 countries. These works thus provide a sufficiently large data set with which to compare specimens that are morphologically close to this species. Neoseiulus wearnei (Schicha 1987), a species described from Australia and found nowhere else (de Moraes et al. 2004), is such a species. The skeleton weed, Chondrilla juncea L. (Asteraceae), on which the first specimens of this species were found, was imported from France (Beard 2001). It is therefore possible that this species was introduced into Australia when this plant was imported. Beard (2001) also wondered about the origin of this species in Australia since it is apparently the only native Australian species of Neoseiulus known so far with three teeth on the movable digit of the chelicerae, just like the specimens of N. californicus reported by Athias-Henriot (1977) and Tixier et al. (2008).
In light of the morphological similarities between N. wearnei and N. californicus, the present study aims to investigate the possible synonymy between these two species using molecular and morphological approaches.
Materials and methods
The populations and species studied
The specimens of N. wearnei used in this study were taken from a colony of mites maintained by Biological Services in Loxton (South Australia, Australia). The culture was established several years ago from specimens collected in Renmark, South Australia, Australia. They were identified as N. wearnei by Beard who had re-described this species from the type material (Beard 2001).
The specimens of N. californicus compared with those of N. wearnei were the same analysed by Tixier et al. (2008) and Okassa et al. (2011), which comprise 300 and 54 adult females collected from 11 countries and 6 host plants (15 populations).
For morphological analyses, we tried to obtain the holotype of N. wearnei deposited in Agricultural Scientific Collections Unit (Acarology) at the Orange Agricultural Institute, Forest Road, Orange, NSW, Australia. However, according to the curator, Dr. Danuta Knihinicki, the holotype and paratypes for this species were lost about 10 years ago. For morphological comparisons, we used the measurements given in the original description (Schicha 1987) and the indications given on the morphology of the holotype material in Beard (2001).
The voucher specimens
For such small organisms (<500 μm in length and 300 μm in width), it is quite difficult to retain voucher specimens after DNA extraction. We used the method proposed by Tixier et al. (2010b) to retrieve the carcass of Phytoseiidae females for which DNA had been extracted using a Qiagen kit. However, although this method allows identification, morphological measurements were carried out on intact specimens of N. wearnei, because setae on dorsal shield and legs could be broken or deformed during the centrifugations and heating phases of DNA extraction.
Morphological analyses
Terminologies used in this paper for chaetotaxy follow those proposed by Lindquist and Evans (1965) as adapted by Rowell et al. (1978) for the Phytoseiidae, and those for poroidotaxy and adenotaxy follow Athias-Henriot (1975).
Fifteen adult females identified as N. wearnei from Australia were mounted on slides in Hoyer’s medium. According to Tixier (2012), this number would be sufficient to estimate seta mean lengths with an error of 15 %. Measurements were then performed with a phase contrast microscope at 400× magnification (Leica DMLB, Leica Microsystèmes SAS, Reuil-Malmaison, France). The lengths of the 19 dorsal idiosomal and interscutal setae were assessed: j1, j3, j4, j5, j6, J2, J5, s4, S2, S4, S5, z2, z4, z5, Z1, Z4, Z5, r3 and R1. Other characters, such as the lengths of the seta JV5, the macroseta on the basitarsus IV, the dimensions (length and width) of the dorsal shield and the three ventral shields were also taken into account. A total of 33 characters were considered. Student comparison tests (StatSoft 2008) were performed to determine differences in measurements between specimens identified as N. wearnei and N. californicus. A multifactorial analysis was carried out to determine whether the combination of the morphological characters would differentiate these specimens (StatSoft 2008).
Molecular analyses
DNA sequences were obtained for ten specimens identified as N. wearnei. The DNA marker used was the fragment 12S rRNA as it has been found to be useful for testing synonymies between Phytoseiidae species (Jeyaprakash and Hoy 2002; Okassa et al. 2009, 2010, 2011; Tixier et al. 2010a, 2011, 2012). The sequences were compared to those already published for N. californicus by Okassa et al. (2011) and deposited in Genbank for 124 specimens [for accession numbers and origin of the specimens sequenced see Okassa et al. (2011)]. We also assessed the interspecific distances using the 12S rDNA sequences of two species morphologically close to N. californicus: Neoseiulus idaeus Denmark and Muma collected in Argentina on Sida sp. (Malvaceae) and Neoseiulus fallacis (Garman) (sequence retrieved in Genbank, accession number AY099364, Jeyaprakash and Hoy 2002). For the phylogenetic analyses, a species belonging to another genus within the same subfamily as the genus Neoseiulus was used as an out-group: Kampimodromus aberrans (Oudemans). The DNA sequence was obtained in our lab and deposited in the Genbank database (accession number: HQ404832).
DNA extraction
Total DNA was individually extracted using the DNeasy Tissue DNA extraction Kit (Qiagen, Hilden) according to the DNA extraction protocol Purification of Total DNA from Animal Blood or Cells (Spin-Column Protocol) adapted for extracting total DNA from mites (Kanouh et al. 2010a, b; Tixier et al. 2010a).
DNA amplification
Primers for the amplification of the 12S rRNA fragment were as follows: 5′–3′ TACTATGTTACGACTTAT and 3′–5′ AAACTAGGATTAGATACCC (Jeyaprakash and Hoy 2002). The PCR reactions were performed in a 25 μl volume, containing 4 μl of mite DNA, 2.5 μl (1 mM) of buffer 10X, 1 μl (1.5 mM) of MgCl2, 0.5 μl (0.05 mM for each) DNTPs, 0.175 μl (0.7 μM) for each primer, 0.125 μl (0.625 U) of Taq Qiagen and 16.525 μl of water. Thermal cycling conditions were as follows: 95 °C for 1 min, followed by 35 cycles of 94 °C for 30 s, 40 °C for 30 s, 72 °C for 1 min, and 72 °C for 5 min. Electrophoresis was carried out on a 1.5 % agarose gel in 0.5 X TBE buffer during 20 min at 135 V.
DNA sequencing
PCR products were sequenced using CEQ Dye Terminator Cycle Sequencing with Quick Start Kit (CEQ[TM] 2000XL DNA Analysis System, Beckman, Beckman Coulter, USA). All DNA fragments were sequenced along both strands. Sequences were analysed and aligned with Geneious v3.5.4 (Drummond et al. 2007). The accession numbers of the sequences in the Genbank database are as follows: KC810036, KC810037, KC810038, KC810039, KC810040, KC810041, KC810042, KC810043, KC810044 and KC810045.
Data analyses
A parsimony analysis was carried out, using PAUP*, v.4.0b.10 (Swofford 2002). A heuristic search procedure repeated 100 times was applied, with randomized taxa additions and branch-swapping algorithm (TBR). Node support was determined using 1,000 bootstrap replicates.
Results and discussion
Morphological comparison of specimens identified as Neoseiulus wearnei and N. californicus
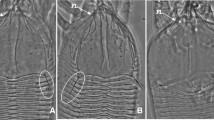
Table 1 reports the mean values for the 33 characters measured for N. californicus and N. wearnei and the results of the t Student test. Significant differences between N. wearnei and N. californicus were observed for 12 characters among the 33 considered. The highest differences were observed for the dimensions of the dorsal, genital and ventrianal shields (width and lengths), with mean values being higher for N. wearnei than for N. californicus. However, even if significant, these differences were too small to support reliable diagnosis. According to the works carried out by Tixier (2012, 2013), these differences could be considered intraspecific variations. On the two axes of the multifactorial analyses explaining 43 % of the variation (Fig. 1), the 15 specimens of N. wearnei are included in the cloud containing the specimens of N. californicus, showing thus that specimens identified as N. wearnei (including the holotype) are not morphologically differentiated from the specimens of N. californicus. Furthermore the observation of chelicerae dentition and spermatheca shape did not reveal any difference between these two taxa. The pictures of dorsal, ventral shields, chelicera dentition and spermatheca of females identified as N. wearnei and N. californicus (from California) are shown on the Fig. 2.
Slides-mounted females of specimens identified as Neoseiulus californicus (California population) (a dorsal shield, c spermatheca, e ventral shields, i chelicerae dentition) and N. wearnei (b dorsal shield, d spermatheca, f ventral shields, g dentition of the fixed digit of the chelicerae, h dentition of the mobile digit of the chelicerae)
Molecular comparison of specimens identified as Neoseiulus wearnei and N. californicus
Fragments of 432 bp were aligned for the 12S rRNA. DNA analyses showed quite similar and constant rates of nucleotide substitutions for all the populations and species studied. A BLAST search in the Genbank database showed that the sequences aligned with other 12S rRNA sequences of Phytoseiidae.
No genetic difference (0 %) between the specimens identified as N. wearnei and N. californicus (Table 2) was observed. The specimens of N. wearnei are included within the clade of the phylogenetic tree that contains specimens of N. californicus. Furthermore, intra-specific distances among specimens of N. wearnei and N. californicus were also null. By contrast, interspecific genetic distances with N. idaeus and N. fallacis were of 12 and 10 % for these respective species. When describing N. wearnei, Schicha (1987) mentioned that this species is morphologically close to N. fallacis, certainly because this latter was the only other species of Neoseiulus present in Australiai. Indeed, it is clear from our observations that N. fallacis and N. wearnei are not similar. Our molecular comparisons also show significant differences between these two latter species (Fig. 3).
Strict consensus trees obtained after a parsimony analysis (100 trees obtained) carried out on the 12S rRNA fragment of Neoseiulus californicus, Neoseiulus wearnei (marked with stars), Neoseiulus fallacis, Neoseiulus idaeus and Kampimodromus aberrans as an out-group. Values below branches indicate bootstrap support
These results thus suggest that specimens herein identified as N. wearnei belong to the species N. californicus. Indeed, in previous studies on N. californicus, the intraspecific genetic distances assessed with the 12S rRNA were usually below 2 %, while interspecific distances exceeded 10 % (Tixier et al. 2010a, 2012; Kanouh et al. 2010a, b; Okassa et al. 2009, 2010, 2011). Such a synonymy is also supported by what we know about the geographic origin of the specimens originally described as N. wearnei. They were found in conjunction with plant material imported from France where N. californicus is common, especially in South of France. The conclusion that N. wearnei in Australia is conspecific with specimens identified around the world as N. californicus clearly questions the correct identification of the Australian material. Because the type material of N. wearnei is now considered lost, unequivocal verification that the material subsequently identified as N. wearnei, including the specimens used in the present study, is conspecific with the type series is not possible, though it is strongly suggested. What can be said for certain though is that the commercially available species known world-wide under the name of N. californicus is present in Australia and commercially available as a biocontrol agent. As a consequence, the results of biological studies on this species carried out elsewhere could be useful for developing biological control programs in Australia.
To conclude this work and the previous ones on N. californicus (Tixier et al. 2008; Okassa et al. 2011), as the original material (holotype) of N. californicus is impossible to observe, and as some of the specimens considered in Tixier et al. (2008) and herein analysed have been originated from California, the locality where the original specimens have been collected, we here propose that those specimens could be considered as the neotypes of this species. A complete description is provided in Tixier et al. (2008) and material is deposited in the acaralogical collection of Montpellier SupAgro/CBGP (Centre de Biologie pour la Gestion des Populations) in France. We also propose N. wearnei to be considered synonym of N. californicus.
References
Athias-Henriot C (1975) Nouvelles notes sur les Amblyseiini. II. Le relevé organotaxique de la face dorsale adulte (Gamasides, Protoadéniques, Phytoseiidae). Acarologia 27:20–29
Athias-Henriot C (1977) Nouvelles notes sur les Amblyseiini. III. Sur le genre Cydnodromus: Redéfinition, composition (Parasitiformes, Phytoseiidae). Entomophaga 22:61–73. doi:10.1007/BF02372991
Beard JJ (2001) A review of Australian Neoseiulus Hughes and Typhlodromips de Leon (Acari: Phytoseiidae: Amblyseiinae). Invert Syst 15:73–158
Castagnoli M, Simoni S, Biliotti N (1999) Mass-rearing of Amblyseius californicus (Acari: Phytoseidae) on two alternative food sources. In: Bruin J, Geest LPS, Sabelis MW (eds) Ecology and evolution of the Acari. Kluwer Academic Publishers, Dordrecht, Boston, London, pp 425–431
Chant DA (1959) Phytoseiid mites (Acarina: Phytoseiidae). Part I. Bionomics of seven species in south-eastern England. Part II. A taxonomic review of the family Phytoseiidae, with descriptions of thirty-eight new species. Can Entomol Suppl 12:166
Chant DA, McMurtry JA (2003) A review of the subfamilies Amblyseiinae (Acari: Phytoseiidae): Part II. Neoseiulini new tribe. Int J Acarol 29:3–46
de Moraes GJ, McMurtry JA, Denmark HA, Campos CB (2004) A revised catalog of the mite family Phytoseiidae. Zootaxa 434:1–494
Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A (2007) Geneious v3.5. Available from http://www.geneious.com/. Accessed Nov 2010
Escudero LA, Ferragut F (2005) Life-history of predatory mites Neoseiulus californicus and Phytoseiulus persimilis (Acari: Phytoseiidae) on four spider mite species as prey, with special reference to Tetranychus evansi (Acari: Tetranychidae). Biol Control 32:378–384
Gotoh T, Tsuchiya A, Kitashima Y (2006) Influence of prey on developmental performance, reproduction and prey consumption of Neoseiulus californicus (Acari: Phytoseiidae). Exp Appl Acarol 40:189–204
Jeyaprakash A, Hoy MA (2002) Mitochondrial 12S rRNA sequences used to design a molecular ladder assay to identify six commercially available phytoseiids (Acari: Phytoseiidae). Biol Control 25:136–142
Kanouh M, Tixier MS, Okassa M, Kreiter S (2010a) Phylogenetic and biogeographic analysis of the genus Phytoseiulus (Acari: Phytoseiidae). Zool Scr 39:450–461. doi:10.1111/j.1463-6409.2010.00439.x
Kanouh M, Tixier MS, Guichou S, Cheval B, Kreiter S (2010b) Two synonymy cases within the genus Neoseiulella (Acari: Phytoseiidae): is the molecular evidence so evident? Biol J Linn Soc 101:323–344
Kostiainen T, Hoy MA (1996) The Phytoseiidae as biological control agents of pest mites. A bibliography (1960–1994). University of Florida, IFAS Publication, Florida Agricultural Experiment Station, Monograph 17
Lindquist E, Evans GW (1965) Taxonomic concepts in the Ascidae, with a modified setal nomenclature for the idiosoma of the Gamasina Acarina: Mesostigmata. Memorium Entomol Soc Can 47:64
McGregor EA (1954) Two new mites in the genus Typhlodromus (Acarina: Phytoseiidae). South Calif Acad Sci Bull 53:89–92
McMurtry JA, Croft BA (1997) Life-styles of phytoseiid mites and their roles in biological control. Annu Rev Entomol 42:291–321
Okassa M, Tixier M-S, Cheval B, Kreiter S (2009) Molecular and morphological evidence for a new species status within the genus Euseius (Acari: Phytoseiidae): consequences for taxonomy. Can J Zool 87:689–698
Okassa M, Tixier M-S, Kreiter S (2010) The use of two mitochondrial DNA fragments (12S rDNA and Cytb mtDNA) for assessing the diagnosis of Phytoseiulus persimilis Athias-Henriot and Phytoseiulus macropilis (Banks) (Acari: Phytoseiidae). Exp Appl Acarol 52:291–303
Okassa M, Kreiter S, Guichou S, Tixier M-S (2011) Molecular and morphological boundaries of the predatory mite Neoseiulus californicus (McGregor) (Acari: Phytoseiidae). Biol J Linn Soc 104:393–406
Palevsky E, Walzer A, Gal S, Schausberger P (2008) Evaluation of dry-adapted strains of the predatory mite Neoseiulus californicus for spider mite control on cucumber, strawberry and pepper. Exp Appl Acarol 45:15–27
Ragusa S (2003) Description of a new genus and of two new species of phytoseiid mites (Parasitiformes, Phytoseiidae) collected in Chile. Acarologia 43:337–344
Ragusa S, Vargas R (2002) On some phytoseiid mites (Parasitiformes, Phytoseiidae) from Chile. Phytophaga 12:129–139
Ragusa S, Papaionnou-Souliotis P, Tsolakis H, Tsagarakou N (1995) Acari fitoseidi (parasitiformes, Phytoseiidae) della Grecia associati a piante forestali a diverse altitudini. Boll Zool Agric Bachicol 27:85–91
Rowell HJ, Chant DA, Hansell RIC (1978) The determination of setal homologies and setal patterns on the dorsal shield in the family Phytoseiidae (Acarina: Mesostigmata). Can Entomol 110:859–876
Schicha E (1987) Phytoseiidae of Australia and neighboring areas. Indira Publishing House, West Bloomfield
Schuster RO, Pritchard AE (1963) Phytoseiid mites of California. Hilgardia 34:191–285
StatSoft France (2008) STATISTICA version 7.1. www.statsoft.fr
Swofford DL (2002) PAUP—Phylogenetic analysis using parsimony (*and other methods), Ver. 4. 0. Beta. [computer software]. Sinauer Associates, Sunderland, MA
Tixier M-S (2012) Approaches to assess intraspecific variations of morphological continuous characters: the case study of the family Phytoseiidae (Acari: Mesostigmata). Cladistics 28:489–502. doi:10.1111/j.1096-0031.2012.00394.x
Tixier M-S (2013) Statistical approaches to assess interspecific differences for morphological continuous characters: the case study of the family Phytoseiidae (Acari: Mesostigmata). Zool Scr 42(3):327–334. doi:10.1111/zsc.12004
Tixier M-S, Kreiter S, Auger P (2000) Colonization of vineyards by phytoseiid mites: their dispersal patterns in the plot and their fate. Exp Appl Acarol 24(3):191–211
Tixier M-S, Guichou S, Kreiter S (2008) Morphological variation of the species Neoseiulus californicus (McGregor) (Acari: Phytoseiidae): importance for diagnostic reliability and synonymies. Invertebr Syst 22:453–469
Tixier M-S, Ferrero M, Okassa M, Guichou S, Kreiter S (2010a) On the specific identity of specimens of Phytoseiulus longipes Evans (Mesostigmata: Phytoseiidae) showing different feeding behaviours: morphological and molecular analyses. Bull Entomol Res 17:1–11
Tixier M-S, Okassa M, Liguori M, Poinso A, Salerno B, Kreiter S (2010b) Voucher specimens for DNA sequences of Phytoseiid mites (Acari: Mesostigmata). Acarologia 50:487–494
Tixier MS, Tsolakis H, Ragusa S, Poinso A, Ferrero M, Okassa M, Kreiter S (2011) An integrative taxonomical approach demonstrates the synonymy between Cydnodromus idaeus and C. picanus (Acari: Phytoseiidae). Invertebr Syst 25:273–281
Tixier M-S, Okassa M, Kreiter S (2012) An integrative morphological and molecular diagnostics for Typhlodromus pyri (Acari: Phytoseiidae). Zool Scr 41:68–78
Tsolakis H, Tixier M-S, Kreiter S, Ragusa S (2012) The genus concept within the family Phytoseiidae (Acari: Parasitiformes). Historical review and phylogenetic analyses of the genus Neoseiulus Hughes. Zool J Linn Soc 165:253–273
Acknowledgments
Data used in this paper were (partly) produced through the technical facilities of the “Montpellier Environnement Biodiversité” Research Federation. We thank James Altmann from Biological Services for providing the populations of N. wearnei from Australia.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tixier, MS., Otto, J., Kreiter, S. et al. Is Neoseiulus wearnei the Neoseiulus californicus of Australia?. Exp Appl Acarol 62, 267–277 (2014). https://doi.org/10.1007/s10493-013-9740-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10493-013-9740-4