Abstract
Two strains HN-1T and 39 were isolated from rhizospheres of different plants grown in different regions of PR China. The two strains exhibited high nitrogenase activities and possessed almost identical 16S rRNA gene sequences. The average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values between the two strains were 99.9 and 99.8%, respectively, suggesting that they belong to one species. Phylogenetic analysis based on the 16S rRNA gene sequence showed that strains HN-1T and 39 are the members of the genus Paenibacillus and both strains exhibited 99.5% similarity to Paenibacillus stellifer DSM 14472T and the both strains represented a separate lineage from all other Paenibacillus species. However, the ANI of type strain HN-1T with P. stellifer DSM 14472T was 90.69, which was below the recommended threshold value (< 95–96% ANI). The dDDH showed 42.1% relatedness between strain HN-1T and P. stellifer DSM 14472T, which was lower than the recommended threshold value (dDDH < 70%). The strain HN-1T contain anteiso-C15:0 as major fatty acids and MK-7 as predominant isoprenoid quinone. The major polar lipids were diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, four aminophospholipids and an unidentified glycolipid. Unlike the most closely related P. stellifer DSM 14472T, strain HN-1T or 39 was positive for catalase reaction. Distinct phenotypic and genomic characterisations from previously described taxa support the classification of strains HN-1T or 39 as representatives of a novel species of the genus Paenibacillus, for which the name Paenibacillus sinensis is proposed, with type strains HN-1T (=CGMCC 1.18902, JCM 34,620), and reference strain 39 (=CGMCC 1.18879, JCM 34,616), respectively.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Paenibacillus was proposed by Ash et al. (1993) and its description was emended by Shida et al. (Shida et al. 1997). Some species of the genus Bacillus were transferred to the genus Paenibacillus (Ash et al. 1993; Heyndrickx et al. 1996; Shida et al. 1997; Lee et al. 2004; Hu et al. 2010), and further descriptions of novel members increased the number of species of the genus Paenibacillus. At the time of writing, the genus comprises 256 species and four subspecies with validly published names (www.bacterio.net/paenibacillus.html). Members of the genus Paenibacillus are rod-shaped, aerobic or facultatively anaerobic, spore-forming bacteria with anteiso-C15:0 as the major cellular fatty acid and menaquinone 7 (MK-7) as the major menaquinone, and their DNA G + C contents range from 45 to 54 mol% (Ash et al. 1993; Priest 2009).
Species of the genus Paenibacillus have been isolated from diverse environment habitats (https://lpsn.dsmz.de/genus/paenibacillus) and have diverse physiological characteristics. Presently, there are only a few species with nitrogen fixation ability in these bacteria, including Paenibacillus azotofixans (Seldin et al. 1984), Paenibacillus borealis (Elo et al. 2001), Paenibacillus brasilensis (von der Weid et al. 2002), Paenibacillus graminis (da Mota et al. 2004), Paenibacillus massiliensis (Ding et al. 2005), Paenibacillus zanthoxyli (Ma et al. 2007a), Paenibacillus sabinae (Ma et al. 2007b), Paenibacillus forsythiae (Ma and Chen 2008), Paenibacillus donghaensis (Choi et al. 2008), Paenibacillus sonchi (Hong et al. 2009), Paenibacillus riograndensis (Beneduzi et al. 2010), Paenibacillus jilunlii (Jin et al. 2011a), Paenibacillus sophorae (Jin et al. 2011b), Paenibacillus stellifer (Jin et al. 2011c), Paenibacillus triticisoli (Wang et al. 2013a), Paenibacillus polymyxa (Wang et al. 2013b), Paenibacillus beijingensis (Gao et al. 2012), Paenibacillus taohuashanense (Xie et al. 2012), Paenibacillus brassicae (Gao et al. 2013), Paenibacillus bryophyllum (Liu et al. 2018), Paenibacillus helianthi (Ambrosini et al. 2018), Paenibacillus maysiensis (Wang et al. 2018), Paenibacillus rhizophilus (Ripa et al. 2019), Paenibacillus durus (Guella et al. 2019), Paenibacillus apii (Tong et al. 2020). Most of N2-fixating Paenibacillus species isolated from plant roots have been shown to play an important role in promoting plant growth by nitrogen fixation, phosphate solubilization, production of plant phytohormones and various enzymes (Xie et al. 2016; Grady et al. 2016; Li et al. 2019). Comparative genomic analysis revealed the conservation of nif cluster comprising 9 genes (nifB nifH nifD nifK nifE nifN nifX hesA nifV) in nitrogen-fixating Paenibacillus strains (Xie et al. 2014). In addition, some N2-fixing Paenibacillus strains with additional nif and nif-like genes exhibited higher nitrogenase activities (Li et al. 2014, 2021). Such traits make the study of the regulation of the multiple nif genes of N2-fixing Paenibacillus under different environmental conditions and their adaptation to varying ecological niches interesting.
In this study, two nitrogen-fixing strains HN-1T and 39 isolated from the rhizosphere of plant were charactered by a polyphasic taxonomic approach, one more presumably novel species strain belonging to the genus Paenibacillus.
Materials and methods
Isolation of the bacterial strains and culture conditions
Strain 39 was isolated from a soil sample collected from arbor rhizosphere in Beijing of China (39°57’N, 116°17’E). 1 g soil sample was suspended in 9 mL sterile water, stirred for 30 min and heated at 80 °C for 15 min. After that, 100 µL suspension was spread on nitrogen-free medium agar plates in triplicate. After incubation at 30 °C for 3 days, single colonies were isolated by streaking plating. The nitrogen-free medium consisted 20 g sucrose, 0.1 g K2HPO4, 0.4 g KH2PO4, 0.2 g MgSO4·7H2O, 0.1 g NaCl, 0.01 g FeCl3, and 0.002 g Na2MoO4 per liter water. The strain HN-1T was previously isolated from the rhizosphere of rice on nitrogen-free medium agar plates (Liu et al. 2019). Strains were routinely cultured in LD medium (per liter contains 2.5 g NaCl, 5 g yeast, and 10 g tryptone) at 30 °C for further identification and study. The type strain of the genus Paenibacillus, P. polymyxa DSM 36T, P. sabinae DSM 17841T, P. stellifer DSM 14472T, P. zanthoxyli JH29T, P. graminis RSA19, P. triticisoli BJ-18T and P. azotofixans ATCC 35681T were obtained from our bacterial collection. Bacteria were freeze-dried or frozen using a sterile glycerol solution in cryogenic tubes to preserve the samples (30% v/v), and stored at − 80 °C and − 20 °C.
Nitrogenase activity assay
To determine nitrogenase activity, strains HN-1T and 39 and reference Paenibacillus strains, including P. polymyxa DSM 36T, P. sabinae DSM 17841T, P. stellifer DSM 14472T, P. zanthoxyli JH29T, P. graminis RSA19T, P. triticisoli BJ-18T and P. azotofixans ATCC 35681T were grown in 20 mL of LB broth medium in 50 mL flasks shaken overnight at 30 °C. The cultures were collected by centrifugation, precipitations were washed three times with sterilized water and then resuspended in nitrogen-limited medium (per liter contains 10.4 g Na2HPO4, 3.4 g KH2PO4, 26 mg CaCl2·2H2O, 30 mg MgSO4, 0.3 mg MnSO4, 36 mg Ferric citrate, 7.6 mg Na2MoO4·2H2O, 10 µg p-aminobenzoic acid, 5 µg biotin, 0.3 g glutamate and 4 g glucose). The nitrogenase activity was determined using the acetylene reduction assay and expressed as nmol C2H4 mg−1 protein h−1 as described previously (Wang et al. 2013b).
Genome sequencing and analysis
The whole genomic DNA of the strains HN-1T and 39 was extracted using the TIANamp Bacteria DNA Kit, evaluated by gel electrophoresis, and estimated using a NanoDrop 2000 (Thermo Scientific, MA, USA). The draft genome sequence was produced by using Illumina paired-end sequencing technology at the mega genomics. Assembly was conducted using SOAP de-novo v. 1.04 assembler (Li et al. 2008). Gene prediction was made using Glimmer v. 3.0 (Delcher et al. 2007). Annotation of protein-coding sequence was performed by using the Basic Local Alignment Search Tool (BLAST) against the COG, Kyoto Encyclopedia of Genes and Genomes (KEGG) databases and NCBI nr protein database.
Average nucleotide identity (ANI) was calculated in EZBioCloud [https://www.ezbiocloud.net/tools/ani] (Yoon et al. 2017a), using the algorithm published by Lee et al. (2016). Digital DNA–DNA hybridization (dDDH) values were computed at GGDC (Genome-to-Genome Distance Calculator) using GGDC 2.0 BLAST + and recommended formula 2 (Meier-Kolthoff et al. 2013).
Phylogenetic analysis of 16S rRNA gene and gyrB gene
The 16S rRNA gene sequences of 39 were acquired from a PCR product using highly specific forward primer 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and universal reverse primer 1492R (5′-GGTTACCTTGTTACGACTT-3′). The DNA sequence obtained was compared to reference 16S rRNA gene sequences available in the Genbank database using BLASTN software (Altschul et al. 1990) and the EzBioCloud server (https://www.ezbiocould.net) (Yoon et al. 2017b). The sequences of the gyrB gene were obtained from the genome of HN-1T and 39 and other type strains. Multiple sequence alignments were analysed using CLUSTAL X (Thompson et al. 1997). The phylogenetic tree calculating evolutionary distance matrices was constructed by the maximum likelihood method (Felsenstein 1981) using MEGA (version 7.0) (Kumar et al. 2016). Bootstrap analysis was conducted on 1000 replications (Felsenstein 1985).
Phenotypic characterization
Colony shape and size of strains were observed after 72 h of incubation on LD medium at 30 °C. For endospore staining, the strains grown on LD agar for 2 days at 30 °C, following 7 days at 4 °C was stained using schaeffer-fulton method (Mormak et al. 1985) and visualized by light microscopy. Cell morphology was also obtained by scanning electrical microscopy (SEM), after incubated on endospore-forming medium agar plate [yeast extract 0.07%, tryptone 0.1%, glucose 0.1%, (NH4)2SO4, 0.02%, MgSO4·7H2O, 0.02%, K2HPO4, 0.1% (w/v), pH 7.2] for 72 h. The flagellation type was determined by transmission electron microscopy (TEM) after 48 h incubation of strain HN-1 on LD medium. Cell motility was evaluated in semi-solid (0.3% agar) LD medium after incubation at 30 °C for 24 h. Physiological and biochemical characteristics were determined in comparison with P. sabinae DSM 17841T and Paenibacillus stellifer DSM 14472T. Most physiological and biochemical tests, including activities of catalase and oxidase, nitrate reduction, hydrolysis of starch, aesculin and tween 20, production of dextrin and indole, methy red reaction, Voges-Proskauer reaction, lysozyme test and production of acid from fermentation of different substrates were performed according to Zhao et al. (2014). Temperature range for growth were determined after incubation at 4, 10, 15, 25, 28, 30, 37, 40 and 45 °C on LD agar. The pH range for growth was determined in LD broth adjusted to pH 4.0–10.0 (using increments of 1.0 pH unit) by using HCl and NaOH buffers. Growth in the absence of NaCl and in the presence of 0, 0.2, 0.5, 1.0, 2.0, 3.0 and 4.0% (w/v) NaCl was investigated by using LD broth. A spectroscopic method of monitoring turbidity at OD600 was used to assess the growth at various temperature, pH values and NaCl concentration. The ability of strains to assimilate different substrates were tested using Biolog GEN III MicroPlate system (Biolog Microstation™, CA, USA) following the manufacturer’s instructions.
Chemotaxonomic characterization
Strains were incubated in LD medium at 30 °C for 2 days. The compositions of cellular fatty acid were analyzed according to the method described by Komagata and Suzuki (1987) using Sherlock Identification System (MIDI) (Sasser et al. 2005). Cellular menaquinones and respiratory quinones were extracted, purified, and analyzed by HPLC according to the method described by Collins (1980). Polar lipid was extracted by the method of Minnikin et al. (1979), and was identified by two-dimensional TLC as described by Collins et al. (1980).
Result and discussion
Bacterial isolation and acetylene-reduction assay
The two strains were isolated from rhizospheres of different plants grown in different regions of PR China. The designated type strain HN-1T was previously isolated from rhizosphere soil of rice collected from Xiangtan City, Hunan Province (Liu et al. 2019); Strain 39 was isolated from rhizosphere soil of arbor collected from Haidian District of Beijing. Since bacteria in the soil sample were cultured in nitrogen-free medium on the purpose of isolating nitrogen-fixing strain, strain 39 is possible to have nitrogen-fixing capability. Strains HN-1T isolated from the rhizosphere of rice was detected by acetylene reduction to have nitrogen-fixing capacity (Liu et al. 2019). Acetylene reduction assays were performed to verify the nitrogenase activity of HN-1T and 39. As shown in Table 1, strains HN-1T and 39 exhibited very high nitrogenase activity compared to other nitrogen-fixing Paenibacillus species, suggesting a high efficiency of the nitrogen fixation process.
Phylogenetic analysis of 16S rRNA gene and gyrB gene
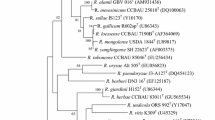
The almost-complete 16S rRNA gene sequence of strain 39 was obtained and used for initial BLAST searches of the GenBank database. Comparisons of 16S rRNA gene sequences revealed that strain 39 was shown to belong to the genus Paenibacillus and share 99.9% 16S rRNA gene sequence identity with strain HN-1T. These two strains showed highest 16S rRNA gene similarity to P. stellifer DSM 14472T (99.5%), followed by P. azotofixans ATCC 35681 T (97.1%) and P. sabinae DSM 17841T (97.0%). According to EzBiocloud database, high level of similarities included 99.5% (P. stellifer DSM 14472T), 97.1% (P. azotofixans ATCC 35681T). Others were below 97%: 96.9% (P. bryophyllum L201T), 96.7% (P. albidus Q4-3T), 96.7% (P. apii 7124T), etc. Phylogenetic trees were inferred using the maximum-likelihood (ML) methods in the software MEGA7. Phylogenetic analysis based on 16S rRNA gene sequences revealed that strains HN-1T and 39 clustered with species of the genus Paenibacillus and formed a monophyletic cluster with P. stellifer DSM 14472T, as the three strains formed a separate phylogenetic branch within the genus Paenibacillus with a high bootstrap resampling value of 100% (Fig. 1).
Maximum-likelihood phylogenetic tree based on 16S rRNA gene sequences showing the position of strains 39 and HN-1T among species of the genus Paenibacillus. Bootstrap analyses were performed with 1000 cycles. Numbers (50%) at nodes are bootstrap values. Bar 0.01 substitutions per nucleotide positions
Generally, 98.7% sequence identity on the 16S rRNA gene are considered to be within the same species (Kim et al. 2014). However, several reports have been published showing that Paenibacillus species with > 99% 16S rRNA gene sequence similarity may not belong to the same species (Kamfer et al. 2017; Kim and Cha 2018; Ghio et al. 2019; Guella et al. 2019; Velazquez et al. 2020). Thus, housekeeping genes are now routinely used to complement the 16S rRNA gene analysis for species level determination (da Mota et al. 2004; Holmes et al. 2004; Rodriguez et al. 2019). Due to the low level of discrimination based on 16S rRNA gene between closely related species, the gyrB gene (coding for the b subunit of DNA gyrase) was used as an alternative phylogenetic marker (Wang et al. 2007). The gyrB genes were retrieved from the HN-1T and 39 genomes. The gyrB gene clearly distinguishes HN-1T and 39 from other Paenibacillus species with only 93.04% gene sequence identity to P. stellifer DSM 14472T (Fig. S1). Based on the 95–96% gyrB gene sequence similarity as the interspecies gap (Lee et al. 2008; Liu et al. 2013), strains HN-1T and 39 could be assigned to novel species.
Genome sequence and similarity analysis
Genome sequencing was performed to evaluate the genomic relatedness of the strains HN-1T and 39 to its closely related recognized species in the genus Paenibacillus. Genomes of strains HN-1T and 39 were approximately 6.32 and 6.45 Mb, respectively. The DNA G + C content of the strains HN-1T and 39 were 53.36 and 52.99%, respectively. The total number of protein coding genes in HN-1T and 39 were 5631 and 5782, respectively. While, the related strain P. stellifer DSM 14472T had a complete genome of 5.66 Mb, comprising 5007 protein coding genes with a DNA G + C content of 53.5%. An overview of the genome sequences of strains HN-1T and 39 and other genome sequences from related species was given in Table 2. The high-quality draft genomes of strains HN-1T and 39 were deposited in GenBank under accession numbers JAHCMB000000000 and JAHBAZ000000000, respectively.
The average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values are widely used to define bacterial species (Konstantinidis and Tiedje 2005; Varghese et al. 2015; Chun et al. 2018; Ciufo et al. 2018). The ANI and dDDH value of genomes for strain HN-1T and strain 39 were 99.8% and 99,93%, respectively, meaning that the two strains belong to one species (Table2). But the ANI values between strain HN-1T and reference strains P. stellifer DSM 14472T, P. sabinae DSM 17841T, P. apii 7124T and P. azotofixans 35681T were 90.69, 76.96, 76.90 and 76.80%, respectively (Table 2). The dDDH values between strain HN-1T and the reference strains P. stellifer DSM 14472T, P. sabinae DSM 17841T, P. apii 7124T and P. azotofixans 35681T were 42.1, 22.0, 22.1 and 22.0%. These values are lower than the proposed and accepted species threshold value of 95–96% ANI and 70% dDDH for differentiating bacterial species (Chun et al. 2018; Richter and Rossello´-Mo´ra 2009), suggesting that the new isolate HN-1T represents a distinctive species.
Analysis of nitrogen fixation and nitrogen metabolism genes
The nitrogen fixation genes of strains HN-1T and 39 were extracted by using Prokka software from the genome sequences (Seemann 2014). The genome of strains HN-1T and 39 contain a compact nif cluster comprising ten genes nifB, nifH, nifD, nifK, nifE, nifN, nifX, orf1, hesA and nifV encoding Mo-nitrogenase, which is unique features of the Paenibacillus nitrogen fixation system. In addition to the nif cluster, the two strains have anfHDGK encoding Fe-nitrogenase and linked to additional copies of nifBENX genes, while the closely related species P. stellifer DSM 14472T contains anfHDGK preceding additional nifV gene. Beyond the nif and anf cluster, there are multiple nifHDK-like genes located at different sites in their genomes. The organization of nif, anf and nif-like genes in type strain HN-1T and the closely related species P. stellifer DSM 14472T was shown in Fig. S2. Previous studies showed that 3 nifH genes of P. sabinae DSM 17841T are functional by complementing K. oxytoca ΔnifH mutant (Hong et al. 2012). Thus, the high nitrogenase activity exhibited by these strains may be due to their additional nif genes.
Paeniacillus azotofixans ATCC 35681T can fix nitrogen even in the presence of nitrate due to the absence of nitrate reductase (Seldin et al. 1984). Whole genome sequence analysis strains HN-1T and 39 revealed that nitrate reductase gene cluster narIJHG were not detected, which suggested these two strains can also fix nitrogen in the nitrate-enriched medium. The draft genome of strains HN-1T and 39 harbor two sets of NAD(P)H-nitrite reductases (nirBD) which are involved in the reduction of nitrite to ammonium in both assimilatory and dissimilatory reduction processes. Additional searches for genes associated with nitric oxide (nirS or nirK) and nitrous oxide reduction (norBC) were performed, but these genes were not detected in their genomes. Therefore, strains HN-1T and 39 may possess dissimilatory nitrate reduction to ammonium pathway, but lack denitrification pathway.
Phenotypic characteristics
Strains HN-1T and 39 were found to be Gram-positive, facultatively anaerobic, motile and rod-shaped. Colonies grown on LD medium after 72 h of incubation at 30 °C were usually 0.8–1.2 mm in diameter, circular, moist, milky and convex (Fig. S3a). Endospores were stained with malachite green and observed under light microscope (Fig. S3b). The transmission electron micrographs of type strain HN-1T showed the presence of peritrichous flagella on cell surface (Fig. 2a). Strain HN-1T produced ellipsoidal spores in swollen sporangia in the terminal region of the cell by scanning electron microscope (Fig. 2b).
In order to determine physiological and biochemical characteristics of HN-1T and 39 in comparison with P. stellifer DSM 14472T and P. sabinae DSM 17841T, a series of tests were carried out following the proposed minimal standards for describing new taxa of facultatively anaerobic, endospore-forming bacteria (Logan et al. 2009). The strains HN-1T and 39 grew well in up to 4% NaCl (w/v), however, strain P. stellifer DSM 14472T tolerated only 3% NaCl. The pH range for growth was 5.0–9.0 and the temperature range for growth is 15–42 °C. Strains HN-1T and 39 was determined to be negative for the Voges–Proskauer reaction, and positive for the methyl red reaction. Strains HN-1T and 39 were positive for catalase reaction and can produce acid from rhamnose and sorbitol, which differentiated HN-1T and 39 from the most related P. stellifer DSM 14472T. The ability of strains to assimilate different substrates were tested using GEN III microplates by Biolog system (Biolog Microstation TM, CA, USA) (Kiran et al. 2017; Ripa et al. 2019). Strain HN-1T and P. stellifer DSM 14472T differed in the metabolization of D-Fucose, D-Maltose, 3-Methyl glucose, D-Sorbitol, Stachyose, Citric acid, α-Keto-butyric acid, Mucic acid, Methyl pyruvate, Gelatin, Inosine, D-Glucose-6-PO4, Pectin, Aztreonam, Fusidic acid, Nalidixic acid, Vancomycin, Lithium chloride, Sodium bromate, Sodium lactate 1%, Rifamycin sv and Troleandomycin as a sole carbon source. Strain HN-1T and 39 exhibited nearly identical phenotypic characteristics, indicating that they belong to one species. Table 3 shows the phenotypic properties that distinguishes the novel strains HN-1T and 39 from the other Paenibacillus species.
Chemotaxonomic characteristics
In order to determine the composition of cellular fatty acid, strains HN-1T, 39, P. stellifer DSM 14472T and P. sabinae DSM 17841T were incubated in LD medium at 30 °C for 2 days. Whole cell fatty acid analysis revealed that anteiso-C15:0, C16: 0, iso-C14: 0, iso-C16: 0 and iso-C15: 0 are present as major (> 5%) fatty acids, and anteiso-C17: 0, iso-C17: 0 and C18:1ω9c are present as minor (< 5 but > 1%) fatty acids (Table S1). Anteiso-C15:0 is the predominant fatty acid of members of the genus Paenibacillus (Ash et al. 1993), consistent with strains HN-1T and 39 being a member of this genus. However, in the closely related type strains P. stellifer DSM 14472T, the fatty acid C16:0 was found to be more abundant than anteiso-C15:0. The major menaquinone of strains HN-1T and 39 was MK-7, in conformity to genus Paenibacillus. The polar lipids of strains HN-1T and 39 detected by two-dimensional TLC are diphosphatidylglycerol (DPG), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), four aminophospholipids (APL) and unidentified glycolipid (Fig. S4).
In summary, the phylogenetic, genomic, phenotypic and chemotaxonomic data of strains HN-1T and 39 showed that they are different from all other closely related species of genus Paenibacillus. Therefore, we conclude that strain HN-1T or 39 should be recognised as a novel species of the genus Paenibacillus, for which the name Paenibacillus sinensis sp. nov. is proposed.
Description of Paenibacillus sinensis sp. nov.
Paenibacillus sinensis (sin. en’sis. L.gen. n. sinensis of China, where the type strain HN-1T was isolated).
Cells are Gram-positive, facultative anaerobic, rod-shaped (0.4–0.5 µm × 2.0–3.2 µm) and motile by means of peritrichous flagella. In slightly swollen sporangia, an ellipsoidal spore is formed and located in terminal position of cells. Colonies on LD medium are circular, convex, cream white, with diameter 1.0–2.0 mm. Nitrogen fixation positive and multiple nifH genes are present. The growth temperature is 15–42 °C, optimal at 30 °C. The growth pH range is 5.0–9.0, optimal at pH 7.0. NaCl concentration of 0–4% (w/v) is tolerable for growth, optimal at 0–0.2%. Positive tests for catalase, methyl red test, starch and aesculin hydrolysis, but negative for oxidase, Voges–Proskauer reaction, nitrate reduction. The various substrates are assimilated examined using Biolog GEN III microplates: dextrin, D-maltose, D-trehalose, D-cellobiose, D-gentiobiose, sucrose, D-turanose, stachyose, D-raffinose, α-D-lactose, D-melibiose, β-Methyl-D-glucoside, D-salicin, α-D-glucose, D-mannose, D-fructose, D-galactose, 3-methyl glucose, 1% sodium lactate, D-serine, D-sorbitol and pectin were utilized. Strains are resistant to inhibitory chemicals: aztreonam, nalidixic acid, vancomycin, lithium chloride, potassium tellurite, sodium bromate, sodium butyrate, sodium lactate 1% and sensitive to troleandomycin, lincomycin, guanidine HCl, niaproof 4, tetrazolium blue. The major menaquinone is MK-7. The predominant fatty acid is anteiso-C15:0. The major polar lipids are DPG, PE, and PG. The DNA G + C contents for strains HN-1T and 39 are 53.36 and 52.99 mol%, respectively.
The type strain, HN-1T (= CGMCC 1.18902, JCM 34,620), was isolated from the rhizosphere soil of rice in Hunan P. R. China. The GenBank (EMBL) accession number for the 16S rRNA gene sequence of strain HN-1T is MF967304 and the GenBank accession number for the draft genome sequence is JAHCMB000000000.
Availability of data and material
The GenBank accession numbers for 16S rRNA gene sequences of strains HN-1T and 39 are MF967304 and MZ153121, respectively. The draft genome sequences of strains HN-1T and 39 have been deposited at NCBI under the accession no. JAHCMB000000000 and JAHBAZ000000000.
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ambrosini A, Sant’Anna FH, Heinzmann J, Fernandes GD, Bach E et al (2018) Paenibacillus helianthi sp. Nov., a nitrogen fixing species isolated from the rhizosphere of Helianthus annuus L. Antonie Van Leeuwenhoek 111(12):2463–2471
Ash C, Priest FG, Collins MD (1993) Molecular identification of rRNA group 3 bacilli (ash, farrow, wallbanks and collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64:253–260
Beneduzi A, Costa PB, Parma M, Melo IS, Bodanese-Zanettini MH et al (2010) Paenibacillus riograndensis sp nov., a nitrogen-fixing species isolated from the rhizosphere of Triticum aestivum. Int J Syst Evol Microbiol 60:128–133
Choi JH, Im WT, Yoo JS, Lee SM, Moon DS et al (2008) Paenibacillus donghaensis sp. nov., a xylan-degrading and nitrogen-fixing bacterium isolated from East Sea sediment. J Microbiol Biotechnol 18(2):189–193
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S et al (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Ciufo S, Kannan S, Sharma S, Badretdin A, Clark K, Turner S, Brover S, Schoch CL, Kimchi A, DiCuccio M (2018) Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int J Syst Evol Microbiol 68:2386–2392
Collins MD, Goodfellow M, Minnikin DE (1980) Fatty acid, isoprenoid quinone and polar lipid composition in the classification of Curtobacterium and related taxa. J Gen Microbiol 118:29–37
da Mota FF, Gomes EA, Paiva E, Rosado AS, Seldin L (2004) Use of rpoB gene analysis for identification of nitrogen-fixing Paenibacillus species as an alternative to the 16S rRNA gene. Lett Appl Microbiol 39:34–40
Delcher AL, Bratke KA, Powers EC, Salzberg SL (2007) Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679
Ding Y, Wang J, Liu Y, Chen S (2005) Isolation and identification of nitrogen-fixing bacilli from plant rhizospheres in Beijing region. J Appl Microbiol 99(5):1271–1281
Elo S, Suominen I, Kampfer P, Juhanoja J, Salkinoja-Salonen M et al (2001) Paenibacillus borealis sp, nov., a nitrogen-fixing species isolated from spruce forest humus in Finland. Int J Syst Evol Microbiol 51:535–545
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence -limits on phylogenies-an approach using the bootstrap. Evolution 39:783–791
Gao M, Xie LQ, Wang YX, Chen J, Xu J et al (2012) Paenibacillus beijingensis sp. Nov., a novel nitrogen-fixing species isolated from jujube garden soil. Antonie Van Leeuwenhoek 102(4):689–694
Gao M, Yang H, Zhao J, Liu J, Sun YH et al (2013) Paenibacillus brassicae sp nov., isolated from cabbage rhizosphere in Beijing China. Antonie Van Leeuwenhoek 103(3):647–653
Ghio S, Sauka DH, Ferrari AE, Piccini RE, Ontanon OM, Campos D (2019) Paenibacillus xylanivorans sp. nov., a xylan-degrading bacterium isolated from decaying forest soil. Int J Syst Evol Microbiol 69:3818–3823
Grady EN, MacDonald J, Liu L, Richman A, Yuan ZC (2016) Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact 15:203
Guella F, Porto RZ, Sant’Anna FH, Ambrosini A, Passaglia LMP (2019) Genomic metrics analyses indicate that Paenibacillus azotofixans is not a later synonym of Paenibacillus durus. Int J Syst Evol Microbiol 69(9):2870–2876
Heyndrickx M, Vandemeulebroecke K, Scheldeman P, Kersters K, DeVos P, Logan NA, Aziz AM, Ali N, Berkeley RCW et al (1996) A polyphasic reassessment of the genus Paenibacillus, reclassification of Bacillus lautus (Nakamura 1984) as Paenibacillus lautus comb nov and of Bacillus peoriae (Montefusco et al 1993) as Paenibacillus peoriae comb nov, and emended descriptions of P. lautus and of P. peoriae. Int J Syst Bacteriol 46:988–1003
Holmes DE, Nevin KP, Lovley DR (2004) Comparison of 16S rRNA, nifD, recA, gyrB, rpoB and fusA genes within the family Geobacteraceae fam. nov. Int J Syst Evol Microbiol 54:1591–1599
Hong YY, Ma YC, Zhou YG, Gao F, Liu HC et al (2009) Paenibacillus sonchi sp nov., a nitrogen-fixing species isolated from the rhizosphere of Sonchus oleraceus. Int J Syst Evol Microbiol 59:2656–2661
Hong Y, Ma YC, Wu LX, Maki M, Qin WS, Chen SF (2012) Characterization and analysis of nifH genes from Paenibacillus sabinae T27. Microbiol Res 16:596–601
Hu XF, Li SX, Wu JG, Wang JF, Fang QL, Chen JS (2010) Transfer of Bacillus mucilaginosus and Bacillus edaphicus to the genus Paenibacillus as Paenibacillus mucilaginosus comb. nov and Paenibacillus edaphicus comb. nov. Int J Syst Evol Microbiol 60:8–14
Jin HJ, Zhou YG, Liu HC, Chen SF (2011a) Paenibacillus jilunlii sp nov., a nitrogen-fixing species isolated from the rhizosphere of Begonia semperflorens. Int J Syst Evol Microbiol 61:1350–1355
Jin HJ, Lv J, Chen SF (2011b) Paenibacillus sophorae sp. nov., a nitrogen-fixing species isolated from the rhizosphere of Sophora japonica. Int J Syst Evol Microbiol 61:767–771
Jin HJ, Tu R, Xu F, Chen SF (2011c) Identification of nitrogen-fixing Paenibacillus from different plant rhizospheres and a novel nifH gene detected in the P. stellifer. Microbiology 80(1):117–124
Kamfer P, Busse HJ, McInroy JA, Hu CH, Kloepper JW, Glaeser SP (2017) Paenibacillus rhizoplanae sp nov., isolated from the rhizosphere of Zea mays. Int J Syst Evol Microbiol 67:1058–1063
Kim M, Oh HS, Park SC, Chun J (2014) Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol 64:346–351
Kim YS, Cha CJ (2018) Paenibacillus translucens sp nov., isolated from tidal flat sediment. Int J Syst Evol Microbiol 68:936–941
Kiran S, Swarnkar MK, Mayilraj S, Tewari R, Gulati A (2017) Paenibacillus ihbetae sp nov., a cold-adapted antimicrobial producing bacterium isolated from high altitude Suraj Tal Lake in the Indian trans-Himalayas. Syst Appl Microbiol 40:430–439
Konstantinidis KT, Tiedje JM (2005) Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA 102:2567–2572
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lee F, Tien CJ, Tai CJ, Wang LT, Liu YC, Chern LL (2008) Paenibacillus taichungensis sp nov., from soil in Taiwan. Int J Syst Evol Microbiol 58:2640–2645
Lee I, Kim YO, Park SC, Chun J (2016) OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66:1100–1103
Lee JS, Pyun YR, Bae KS (2004) Transfer of Bacillus ehimensis and Bacillus chitinolyticus to the genus Paenibacillus with emended descriptions of Paenibacillus ehimensis comb. nov and Paenibacillus chitinolyticus comb. nov. Int J Syst Evol Microbiol 54:929–933
Li Q, Zhang HW, Zhang LQ, Chen SF (2021) Functional analysis of multiple nifB genes of Paenibacillus strains in synthesis of Mo-. Fe- and V-Nitrogenases Microb Cell Fact 20(1):139–139
Li RQ, Li YR, Kristiansen K, Wang J (2008) SOAP: short oligonucleotide alignment program. Bioinformatics 24:713–714
Liu XM, Li Q, Li YB, Guan GH, Chen SF (2019) Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth. PeerJ 7:e744
Liu LH, Yuan T, Yang F, Liu ZW, Yang MY et al (2018) Paenibacillus bryophyllum sp. Nov., a nitrogen-fixing species isolated from Bryophyllum pinnatum. Antonie Van Leeuwenhoek 111(12):2267–2273
Li XX, Deng ZP, Liu Z, Yan YL, Wang TS, Xie JB, Lin M, Cheng Q, Chen SF (2014) The genome of Paenibacillus sabinae T27 provides insight into evolution, organization and functional elucidation of nif and nif-like genes. BMC Genomics 15:723
Li YB, Li YL, Zhang HW, Wang MY, Chen SF (2019) Diazotrophic Paenibacillus beijingensis BJ-18 provides nitrogen for plant and promotes plant growth, nitrogen uptake and metabolism. Front Microbiol 10:1119
Liu Y, Lai QL, Dong CM, Sun FQ, Wang LP, Li GY, Shao ZZ (2013) Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis. PLoS ONE 8:e80097
Logan NA, Berge O, Bishop AH, Busse HJ, De Vos P, Fritze D, Heyndrickx M, Kampfer P, Rabinovitch L, Salkinoja-Salonen MS et al (2009) Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int J Syst Evol Microbiol 59:2114–2121
Ma YC, Zhang J, Chen SF (2007a) Paenibacillus zanthoxyli sp nov, a novel nitrogen-fixing species isolated from the rhizosphere of Zanthoxylum simulans. Int J Syst Evol Microbiol 57:873–877
Ma YC, Xia ZQ, Liu XM, Chen SF (2007b) Paenibacillus sabinae sp nov., a nitrogen-fixing species isolated from the rhizosphere soils of shrubs. Int J Syst Evol Microbiol 57:6–11
Ma YC, Chen SF (2008) Paenibacillus forsythiae sp nov., a nitrogen-fixing species isolated from rhizosphere soil of Forsythia mira. Int J Syst Evol Microbiol 58:319–323
Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf 14:60
Minnikin DE, Collins MD, Goodfellow M (1979) Fatty acid and polar lipid composition in the classification of Cellulomonas, Oerskovia and related taxa. J Appl Bacteriol 47:87–95
Mormak DA, Casida LE (1985) Study of Bacillus subtilis endospores in soil by use of a modified endospore stain. Appl Environ Microbiol 49:1356–1360
Priest FG (2009) Genus I, Paenibacillus. In: De Vos P, Garrity G, Jones D, Krieg NR, Ludwig W, Rainey FA, Schleifer KH, Whitman WB (eds) The firmicutes, Bergey’s manual of systematic bacteriology, vol 2, 2nd edn. Springer, New York, pp 269–296
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131
Ripa FA, Tong S, Cao WD, Wang ET, Wang TY, Liu HC, Gao JL, Sun JG (2019) Paenibacillus rhizophilus sp. nov., a nitrogen-fixing bacterium isolated from the rhizosphere of wheat (Triticum aestivum L.). Int J Syst Evol Microbiol 69:3689–3695
Rodriguez M, Reina JC, Bejar V, Llamas I (2019) Paenibacillus lutrae sp. Nov., a chitinolytic species Isolated from a river otter in castril natural park, Granada Spain. Microorganisms 7:637
Sasser M, Kunitsky C, Jackoway G, Ezzell JW, Teska JD, Harper B, Parker S, Barden D, Blair H, Breezee J et al (2005) Identification of Bacillus anthracis from culture using gas chromatographic analysis of fatty acid methyl esters. J AOAC Int 88:178–181
Seemann T (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069
Seldin L, Vanelsas JD, Penido EGC (1984) Bacillus azotofixans sp. nov., a nitrogen-fixing species from Brazilian soils and grass Roots. Int J Sys Bacteriol 34(4):451–456
Shida O, Takagi H, Kadowaki K, Nakamura LK, Komagata K (1997) Transfer of Bacillus alginolyticus, Bacillus chondroitinus, Bacillus curdlanolyticus, Bacillus glucanolyticus, Bacillus kobensis, and Bacillus thiaminolyticus to the genus Paenibacillus and emended description of the genus Paenibacillus. Int J Syst Bacteriol 47:289–298
Suominen I, Sproer C, Kampfer P, Rainey FA, Lounatmaa K et al (2003) Paenibacillus stellifer sp. nov., a cyclodextrin-producing species isolated from paperboard. Int J Syst Evol Microbiol 53:1369–1374
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tong S, Wang LW, Sun YC, Khan MS, Gao JL et al (2020) Paenibacillus apii sp. nov., a novel nifH gene-harbouring species isolated from the rhizospheres of vegetable plants grown in different regions of northern China. Int J Syst Evol Microbiol 70:5531–5538
Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K et al (2015) Microbial species delineation using whole genome sequences. Nucleic Acids Res 43:6761–6771
Velazquez LF, Rajbanshi S, Guan SH, Hinchee M, Welsh A (2020) Paenibacillus ottowii sp. nov., isolated from a fermentation system processing bovine manure. Int J Syst Evol Microbiol 70(3):1463–1469
von der Weid I, Duarte GF, van Elsas JD, Seldin L (2002) Paenibacillus brasilensis sp nov., a novel nitrogen-fixing species isolated from the maize rhizosphere in Brazil. Int J Syst Evol Microbiol 52:2147–2153
Wang LT, Lee FL, Tai CJ, Kasai H (2007) Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA-DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol 57:1846–1850
Wang LY, Li J, Li QX, Chen SF (2013a) Paenibacillus beijingensis sp. nov., a nitrogen-fixing species isolated from wheat rhizosphere soil. Antonie Van Leeuwenhoek 104(5):675–683
Wang L, Zhang LH, Liu ZZ, Zhao DH, Liu XM, Zhang B, Xie JB, Hong YY, Li PF, Chen SF et al (2013b) A minimal nitrogen fixation gene cluster from Paenibacillus sp. WLY78 enables expression of active nitrogenase in Escherichia coli. PLoS Genet 9:1003865
Wang TS, Xie JY, Wang LY, Chen SF (2018) Paenibacillus maysiensis sp. nov., a nitrogen-fixing species isolated from the rhizosphere soil of maize. Curr Microbiol 75(10):1267–1273
Xie JB, Zhang LH, Zhou YG, Liu HC, Chen SF (2012) Paenibacillus taohuashanense sp. nov.,, a nitrogen-fixing species isolated from rhizosphere soil of the root of Caragana kansuensis Pojark. Antonie Van Leeuwenhoek 102(4):735–741
Xie JB, Du ZL, Bai LQ, Tian CF, Zhang YZ, Xie JY, Wang TS, Liu XM, Chen X, Cheng Q et al (2014) Comparative genomic analysis of N2-fixing and non-N2-fixing Paenibacillus spp.: organization, evolution and expression of the nitrogen fixation genes. PLoS Genet 10:e1004231
Xie JB, Shi HW, Du ZL, Wang TS, Liu XM et al (2016) Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci Rep 6:21329
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017a) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017b) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhao B, Lin H, He SJ (2014) Microbiology experiment, 2nd edn. Science Press, Beijing
Zhuang J, Xin D, Zhang YQ, Guo J, Zhang J (2017) Paenibacillus albidus sp. nov., isolated from grassland soil. Int J Syst Evol Microbiol 67:4685–4691
Funding
This work was supported by the National Key Research and Development Program of China (No. 2019YFA0904700) and the National Natural Science Foundation of China (No. 32000048).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10482_2021_1677_MOESM1_ESM.tif
Fig. S1 Maximum-likelihood phylogenetic tree based on gyrB gene sequences showing the position of strains HN-1T and 39 among species of the genus Paenibacillus. Bootstrap analyses were performed with 1000 cycles. Only bootstrap values >50 % are shown at the branch points. Bar 0.01 substitutions per nucleotide positions (TIF 13883 KB)
10482_2021_1677_MOESM3_ESM.tif
Fig. S3 Colony and strain morphology, (a) Colony morphology of strain HN-1T and light micrograph of endospores which stained with specific endospores staining method. Endospores-staining was performed by the modified schaeffer-fulton method according to Mormak et al. (1985) (TIF 13947 KB)
10482_2021_1677_MOESM4_ESM.tif
Fig. S4 Two-dimensional TLC plate of polar lipids extracted from strains HN-1T (a) and 39 (b). The plate was sprayed with 10% (v/v) molybdophosphoric aicd to show all polar lipids present. PE, phosphatidylethanolamine; PG, phosphatidylglycerol; DPG, diphosphatidylglycerol; PL, unidentified phosphoglycolipid; APL, aminophospholipid; GL, unidentified glycolipid (TIF 12719 KB)
10482_2021_1677_MOESM5_ESM.docx
Table S1 Fatty acid content (%) of strain HN-1T and 39 and some other type strains of the Paenibacillus genus (DOCX 15 KB)
Rights and permissions
About this article
Cite this article
Li, Q., Li, Y., Liu, X. et al. Paenibacillus sinensis sp. nov., a nitrogen-fixing species isolated from plant rhizospheres. Antonie van Leeuwenhoek 115, 7–18 (2022). https://doi.org/10.1007/s10482-021-01677-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01677-6