Abstract
Plant growth-promoting rhizobacteria are a group of free-living bacteria that colonize plant rhizosphere and benefit plant root growth, thereby increasing host plant to cope with salinity induced stress. The aim of this study was to (1) isolate and characterize auxin-producing bacteria showing a high plant growth-promoting (PGP) potential, and (2) evaluate the PGP effects on the growth of Medicago sativa L under salinity stress (130 mM NaCl). Of thirteen isolates, Bacillus megaterium NRCB001 (NRCB001), B. subtilis subsp. subtilis NRCB002 (NRCB002) and B. subtilis NRCB003 (NRCB003) had the ability to produce auxin, which ranged from 47.53 to 154.38 μg ml−1. The three auxin-producing bacterial strains were shown multiple PGP traits, such as producing siderophore and NH3, showing ACC deaminase activity, solubilize phosphate and potassium. Furthermore, NRCB001, NRCB002, and NRCB003 could survive in LB medium containing 1750 mM NaCl. The three auxin-producing with salinity tolerance strains were selected for further analyses. In greenhouse experiments, when inoculated with NRCB001, NRCB002 and NRCB003, dry weight of alfalfa significantly (P < 0.05) increased by 24.1%, 23.1% and 38.5% respectively, compared with those of non-inoculated control seedlings under normal growth condition. When inoculated with NRCB002 and NRCB003, dry weight of alfalfa significantly (P < 0.05) increased by 96.9 and 71.6% respectively, compared with those of non-inoculated control seedlings under 130 mM NaCl condition. Our results indicated that NRCB002 and NRCB003 having PGP traits are promising candidate strains to develop biofertilizers, especially used under salinity stress conditions.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Salinity is one of most brutal environmental factors limiting the productivity of crops because most of crop plants are sensitive to salinity caused by high concentrations of salts in the soil. An area of land affected by salinity is increasing notably in intensive agricultural scenarios. For example, about 1.5 billion hectares of cultivated land is affected by salinity worldwide (Selvakumar et al. 2014). Under salinity conditions, plant growth is greatly restrained by several factors, such as nutritional imbalance, hormonal, ion toxicity, and physiological disorders (Nadeem et al. 2014).
Nowadays, soil salinization has been further brought by frequent irrigation and heavy fertilization of synthetic fertilizers. A national strategy is implemented to reduce synthetic fertilizers while increase their use efficiencies in China (Zhang et al. 2019). With the significant reduction in synthetic fertilizers, the growing demand for crop production has become a big challenge (Wang et al. 2018). In this case, it requires suitable biotechnology to increase crop yields and improve soil health through interaction between plant roots and soil microorganisms, particularly under salinity conditions.
Alfalfa (Medicago sativa L.) is an important perennial leguminous forage worldwide. It is a high-yielding and easily digested animal fodder with high nutritional value (Noori et al. 2018). However, most of commercial alfalfa cultivars showed poor ability in salt tolerance. For example, 50 mM NaCl could inhibit their normal growth (Liu et al. 2011). Several biotechnological methods have been attempted to improve the salt tolerance of alfalfa, but it is difficult to directly conduct salt tolerance breeding (Zhang and Wang 2015).
Inoculation of plant growth promoting rhizobacteria (PGPR) is a promising strategy to increase the salinity tolerance of crops and promote the growth of crops. Previous studies have reported that there are many beneficial associations between plants and PGPR, such as salinity stress alleviation, drought stress alleviation, plant growth promotion, nitrous oxide mitigation and so on (Dimkpa et al. 2009; Vaishnav and Choudhary 2019; Yaish et al. 2015; Gao et al. 2017). PGPR help plants to alleviate salinity stress via many ways, including phosphate solubilization, 1-aminocyclopropane-1-carboxylic acid (ACC)-deaminase activity and volatiles secretion, siderophores and auxin production and so on (Qin et al. 2016; Nadeem et al. 2016). Among the characteristics of PGP properties of PGPRs, auxin is a more prominent role in providing nutrients for plants under stress conditions. auxin could stimulate the division and elongation of root tissues, in order to provide the availability of nutrients under different stress conditions for plants, thereby preventing a severe decrease in stressed plant growth (Etesami and Maheshwari 2018; Etesami et al. 2016). Although a lot of PGPR has been observed to promote plant growth, there are a few reports about PGPR which could promote pasture plant growth like alfalfa under salinity conditions. (Liu et al. 2019; Noori et al. 2018; Zhang et al. 2017). Therefore, it is important to isolate for PGPR that promote the growth of alfalfa under salt stress.
The aim of this study is to (1) isolate and characterize auxin-producing bacteria showing a high plant growth-promoting (PGP) potential, and (2) evaluate the PGP effects on the growth of Medicago sativa L. under salinity conditions.
Materials and methods
Isolation of rhizobacterial isolates
The bacterial isolates were isolated from the rhizosphere of rice in Yixing, Jiangsu China (31° 12′ 23′′ N; 119° 52′ 89′′ E). Soil samples were collected on March 2018 from a long-term traditional rice alfalfa or wheat rotation land. The climate is subtropical monsoon in this area. The mean annual and accumulated temperature is 15.7 and 5418 °C, respectively. The average annual precipitation is 1177 mm, with a more than 240 frost- free days. The soil is classified as Anthrosols.
The soil adhering to the roots of rice at tillering stage was separated by gentle tapping, collected and stored in a 4 °C refrigerator. Ten grams of rice rhizosphere soil were added into a 250 ml flask containing 90 ml solution (composition in mgt l−1; K2HPO4, 100; MgSO4, 50; NaCl, 20; CaCl2·2H2O, 50; Fe-EDTA, 16.4), and shaken at 200 rpm for 30 min at 28 °C. Rhizobacterial strains were isolated using NFB medium: malic acid, 5.0 g; K2HPO4, 0.5 g; MgSO4·7H2O, 0.2 g; NaCl, 0.1 g; CaCl2·2H2O, 0.02 g; micronutrient solution (CuSO4·5H2O, 0.04 g; ZnSO4·7H2O, 0.12 g; H3BO3, 1.40 g; Na2MoO4·2H2O, 1.0 g; MnSO4·H2O, 1.175 g; up to 1000 ml with distilled water), 2 ml; bromothymol blue (5 g l−1 in 0.2 N KOH), 2 ml; Fe-EDTA (solution 16.4 g l−1), 4 ml; vitamin solution (biotin, 10 mg; pyridoxal-HCl, 20 mg; up to 100 ml with distilled water), 1 ml; KOH, 4.5 g; agar 15 g; up to 1000 ml with distilled water and adjust pH 6.5 (Baldani et al. 2014). Five serial soil dilutions were plated on NFB medium and incubated at 28 °C for 2 days. A colony that differed in appearance was purified and maintained in 25% (w/v) glycerol at − 80 °C for further characterization.
Determination of auxin production by rhizobacterial isolates
To determine the production of the auxin, the bacterial suspension (106 CFU ml−1) was used to inoculate into liquid LB medium containing 5 mM l-Tryptophan, and incubated at 28 °C for 48 h at 200 rpm. Five ml aliquots of the cultures were taken, centrifuged for 10 min at 4000 g and 1 ml of the supernatant were mixed with 2 ml of Salkowski reagent (1 ml of 0.5 M FeCl3 + 50 ml 35% HClO4). The mixture was incubated in the dark at room temperature for 30 min and the absorbance at 530 nm was measured using a spectrophotometer (UV-2700 220 V CH, Shimadzu Corp.) (Bric et al. 1991). Auxin concentrations were determined using an indoleacetic acid (IAA) (Aladdin®) standard curve from 10 to 500 μg ml−1. The auxin-producing rhizobacterial isolates biomass was evaluated by optimal density (OD600). Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate.
Molecular identification of the auxin-producing rhizobacterial isolates
The 16S rRNA genes was amplified using universal primers (Table 1). The gyrA (DNA gyrase subunit A) and gyrB (DNA gyrase subunit B) genes of auxin-producing rhizobacterial NRCB002 and NRCB003 were amplified by PCR using universal primers listed in Table 1. The gyrA and gyrB genes of auxin-producing rhizobacterial NRCB001 were amplified by PCR using custom-designed primers according to Bacillus megaterium ATCC 14581 published genome sequence (Table 1), because the universal primers did not work adequately for the species of the Bacillus megaterium being studied. The amplification reaction was performed in a final volume of 25 μl containing 2.5 μl of 10 × Taq polymerase buffer (Mg2+ plus), 2 μl dNTPs (2.5 mM each), 1 μl each primer (10 μM), 0.125 μl rTaq polymerase (5U μl−1, Takara Biotechnology (Dalian) Co., Ltd., Dalian, China), up to 25 μl with sterile distilled water and a toothpick of bacteria. PCR was performed in a S1000 Thermocycler (Bio-Rad, USA) with the following program: a 10 min initial denaturation at 95 °C, followed by 32 cycles consisting denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s, extension at 72 °C for 90 s, and final elongation at 72 °C for 7 min. The amplicons were sequenced. The obtained 16S rDNA, gyrA and gyrB gene sequences of the isolated strains were compared with the known nucleotide sequences in the GenBank database using the Blast program using default parameters except limited to sequences from type material. The amplicons were sequenced. The obtained 16S rRNA, gyrA and gyrB gene sequences were used for multilocus sequence analysis (MLSA), which can be used to resolve finer interspecific and intraspecific relationships. The nucleotide sequence alignment of concatenated genes consisted of 2538 bp (827 bp from gyrA, 825 bp from gyrB and 886 bp from 16S rRNA). Alignments for phylogenetic trees were made using MEGA software 10.0.2. The phylogenetic tree was constructed by carrying out the multiple alignments among different concatenated three gene sequences in MEGA software 10.0.2 (https://www.megasoftware.net) program by using the maximum-likelihood method. Tree topology evaluation was based on 1000 bootstrap replicates.
Morphological and biochemical characterization of auxin-producing rhizobacterial strains
The type and activity of enzymes, such as catalase and oxidase produced by the bacteria, are important characteristics for the identification of the microorganisms. auxin-producing rhizobacterial strains were cultured on LB plate for colony morphology and Gram staining (Gupta et al. 2012). Biochemical tests, including methyl red (MR), indole production, citrate utilization, catalase, oxidase and Voges-Proskaue (VP), were performed (Wang et al. 2018). Each bacterial culture experiment was conducted in triplicate.
Beneficial traits of auxin-producing rhizobacterial strains
To analyze the growth-promoting effects of the auxin-producing strains, beneficial traits were measured, including the production of ammonia and siderophores, phosphate-solubilizing, potassium-dissolving abilities, and for 1-aminocyclopropane-1-carboxyacetate (ACC) deaminase activity.
Ammonia production was analyzed using peptone water culture medium (composition in g l−1: peptone, 10; NaCl, 5; pH 7.0) (Karthik et al. 2017). Briefly, 100 μl of bacterial suspension (106 CFU ml−1) was added into 250 ml sterilized flask containing 50 ml peptone water culture medium and incubated at 28 °C and 200 rpm for 2 d. Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate. The suspension was centrifuged at 10,000 rpm for 10 min, and 1 ml of supernatant was mixed with 1 ml of Nessler’s reagent. The volume of this reaction mixture was made up to 10 ml with sterile ammonia-free distilled water. The absorbance at 450 nm was measured using a spectrophotometer (UV-2700 220 V CH, Shimadzu Corp.) with ammonium chloride as the standard.
A quantitative analysis of P-solubilizing of bacterial strains was determined with the molybdate blue color method (Mukhtar et al. 2017). Bacteria were grown in Pikovskaya broth (composition in g·l−1: Ca3(PO4)2, 5; glucose, 10; yeast extract, 0.5; MgSO4, 0.1; KCl, 0.2; MnSO4, 0.0001; FeSO4, 0.0001; (NH4)2SO4, 0.5; pH 7.0) at 28 °C and 200 rpm for 7 d (Hussein and Joo 2015). And then, cell-free culture supernatant was analyzed for solubilized phosphates. Values were calculated by using a KH2PO4 standard curve from 0.2 to 2 μg ml−1. Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate.
K-solubilizing ability of bacteria was analyzed with Aleksandrov medium (composition in g l−1: glucose, 5; MgSO4·7H2O, 0.5; FeCl3, 0.005; CaCO3, 0.1; CaPO4, 2 and KAlSi3O8, 2; pH 7.0–7.5). Briefly, 100 μl of bacterial suspension (106 CFU ml−1) was added into 250 ml sterilized flask containing 50 ml Aleksandrov medium. Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate. The flasks were incubated at 28 °C and 200 rpm for 7 d. The fermentation broth was centrifuged at 10000 rpm for 10 min, and the estimation of soluble K in the supernatant was determined by a flame photometer (Zhang and Kong 2014).
The production of siderophore was analyzed as described by Schwyn and Neilands (1987). In Brief, 100 μl of bacterial suspension (106 CFU ml−1) were added into 250 ml sterile flask containing 50 ml iron-deficient medium containing (g l−1): K2HPO4, 0.1; KH2PO4, 3.0; MgSO4·7H2O, 0.2; (NH4)2SO4, 1.0; succinic acid, 4.0 and incubated at 28 °C and 200 rpm for 48 h. And then, the cell-free culture supernatant obtained after centrifugation was analyzed for siderophore production using Universal Chrome Azurol-S (CAS) colorimetric assay. Absorbance was read at 630 nm for the loss of a blue color. Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate.
The 100 μl of bacterial suspension (106 CFU ml−1) was added into 250 ml sterilized flask containing 50 ml LB liquid medium and incubated at 28 °C and 200 rpm for 24 h. The suspension was centrifuged at 10,000 rpm for 10 min. The supernatant was extracted and washed twice in 0.86% saline water and centrifuged at 8000 rpm for 10 min at 4 °C. The collected cells were re-suspended in 0.86% saline water and adjusted to a final concentration of 108 CFU ml−1.The strain was evaluated for ACC deaminase activity by coating on a DF salt minimal medium containing ACC as the sole nitrogen source (Penrose and Glick 2003). Briefly, the strain suspension was spread on a DF plate and placed in an incubator at 28 °C for 3 to 4 days. DF medium with (NH4)2SO4 was used as a positive control, and DF medium without (NH4)2SO4 and ACC was used as a negative control. Each bacterial culture experiment was conducted in triplicate. The strain that grows well on the plate with ACC as sole nitrogen source was considered positive for ACC deaminase activity (Hmaeid et al. 2019).
Effect of NaCl on bacteria growth
To investigate the effect of salt stress on bacteria growth, 100 μl of bacterial suspensions (106 CFU ml−1) were added into 250 ml sterile flask containing 50 ml liquid NFB medium containing different concentrations of NaCl: 0% (0 mM); 2.05% (350 mM); 4.09% (700 mM); 6.14% (1050 mM); 8.18% (1400 mM); and 10.22% (1750 mM). The culture was incubated at 28 °C and 200 rpm for 24 h. The growth was evaluated by measuring the optical density at 600 nm. Autoclaved and non-inoculated broth served as control. Each bacterial culture experiment was conducted in triplicate. Following incubation, the strains that could grow on medium supplemented with ≥ 5.0% NaCl were considered salt-tolerant strains and were further characterized in vitro for PGP attributes (Hmaeid et al. 2019).
Colonization assay of auxin-producing strains
The 100 μl of bacterial suspension (106 CFU ml−1) was added into 250 ml sterilized flask containing 50 ml NFB liquid medium and incubated at 28 °C and 200 rpm for 24 h. The suspension was centrifuged at 10,000 rpm for 10 min. The supernatant was extracted and washed twice in 0.86% saline water and centrifuged at 10,000 rpm for 10 min at 4 °C. The collected cells were re-suspended in 0.86% saline water and adjusted to a final concentration of 108 CFU ml−1. The re-suspended bacterium was used for individual inoculation. Briefly, the seeds were surface- disinfected with 2% sodium hypochlorite solution for 2 min and with 75% ethanol for 4 min, and then rinsed with sterile water (Liu et al. 2019). Surface-disinfected alfalfa seeds were immersed in the bacterial solution overnight, the non-inoculated control received sterile distilled water only. The treated and control seeds were placed on non-salinized and salinized 1/5 Murashige Skoog (MS, Hope Bio-Tcehnology Co., ltd. Qingdao, China) with 0.8% agar and 1.5% sucrose. The salinized 1/5 MS medium was formed by the addition of 130 mM NaCl based on non-salinized 1/5 MS medium.
After incubation for 7 days (14 h light at 25 °C, 10 h dark at 18 °C), the roots were surface sterilized with 0.3% sodium hypochlorite solution for 20 min, and rinsed with sterile water (Jin et al. 2019). After that, the roots were weighed and ground with a mortar in sterile 0.86% saline, and the collected suspensions were diluted with sterile 0.86% saline and plated on LB agar media. After incubation at 28 °C for 12 h, the colonies were counted using plate dilution method and represented as colony forming unit log10 cfu g−1 root fresh weight. Three replicates were included for each treatment. The seedlings without PGPR treatment served as control, and the last sterile water rinsed the alfalfa root which served as negative control.
Pot experiments of alfalfa
The pot experiments were performed in a natural-light and 18–25 °C greenhouse located in Nanjing Tech University during October–November 2018. A commercial soil (pH, 7.4; total organic C, 33.6 g kg−1; alkali-hydrolysable-N, 63.7 mg kg−1; Olsen-P, 42.4 mg kg−1; exchangeable-K, 102.0 mg kg−1; soluble salt, 1.33 g kg−1) was used for pot experiments. Total organic C was determined with the potassium dichromate method (Lu 1999). Alkali-hydrolyzable N was determined with the alkali solution method (Lu 1999). Olsen-P was determined with the molybdenum-blue-colorimetric method after extraction by sodium bicarbonate (Lu 1999). Exchangeable-K was determined with the ammonium acetate extraction and flame photometric method (Lu 1999). Soluble salt content of the soil was determined with by residue drying method (Lu 1999). The soil was thoroughly mixed, passed through a 2 mm sieve, and dispensed in pots with 1 kg soil per pot (13.3 cm diameter, 17.4 cm depth).
Seeds of Medicago sativa L cv SA701, purchased from Qian lv Seed Industry (Suqian, China), were used for pot experiments. Three auxin-producing bacteria NRCB001, NRCB002, and NRCB003 were used to inoculate alfalfa. The 100 μl suspensions (106 CFU ml−1) of three auxin-producing bacteria NRCB001, NRCB002, and NRCB003 were individually inoculated into 250 ml sterile flask containing 50 ml liquid NFB medium, and incubated at 28 °C and 200 rpm for 1 day to reached the logarithmic phase (Fig S1). After incubation, the strain suspension was centrifuged at 8000 rpm for 10 min. The collected cells were re-suspended in 0.86% saline water and adjusted to a final concentration of 108 CFU ml−1. The re-suspended bacterium was used for individual inoculation. Briefly, the seeds were surface- disinfected with 2% sodium hypochlorite solution for 2 min and with 75% ethanol for 4 min, and then rinsed with sterile water (Liu et al. 2019). Surface-disinfected alfalfa seeds were immersed in the bacterial solution overnight, the non-inoculated control received sterile distilled water only (Kadmirl et al. 2018). Twenty-five treated and control seeds were sown on non-salinized and salinized soils. The salinized soil was formed by the addition of 130 mM NaCl based on non-salinized soil. Four pots of each treatment were treated daily irrigated with sterile water, and were not added any fertilizer for the 30 days after germination of seeds. The number of germination seeds was counted 7 days after sowing. And then, germination seeds were thinned to one. Plant root and shoot were carefully harvested and thoroughly washed after 30 days of growth. The number of leaves, plant height, root length and average leaf area were measured simultaneously. The root and leaves were scanned using a Microtek ScanMaker i800 plus system (WSeen, Hangzhou, China). The leaf area was calculated by an LA-S Leaf Area Analysis software and the root length, root surface, root volume was calculated by an LA-S Root System Analysis software (WSeen, Hangzhou, China), respectively. Plant dry weight was measured after oven drying at 65 °C to a constant weight.
Biochemical analysis assays of alfalfa
The leaves of alfalfa were put into the liquid nitrogen and then stored in − 80 °C refrigerator. Malondialdehyde (MDA) content, activity of superoxide dismutase (SOD) and catalase (CAT) were determined using assay kits (Comin biotechnology Co., ltd. Suzhou, China) with a Spectrophotometer (UV-2700 220 V CH, Shimadzu Corp.) according to the manufacturer’s instructions. MDA content was assayed based on the thiobarbituric acid-reactive substance assay. The measure method of SOD activity was determined based on the nitroblue tetrazolium reduced (NBT) method. The CAT activity was determined based on the decomposition of H2O2 ultraviolet (UV) absorption method. Each experiment was conducted in quadruplicate. The seedlings without PGPR treatment served as control.
Statistical analysis
The collected data were analyzed by one-way ANOVA and two-way ANOVA using SPSS version 19.0. Significant differences were calculated by Duncan’s multiple-range tests (P < 0.05) or Independent-Sample T test (P < 0.05). Two-way ANOVA were conducted to determine the significance of salt stress, strain inoculation and their interaction and determine the significance of NaCl concentrations, species of strains and their interaction in effect of NaCl on bacteria growth (Wu et al. 2016). One-way ANOVA was performed for auxin production of the isolates and plant growth-promoting characteristics of strains.
Results
Screening auxin production of the isolates
A total of 13 bacterial isolates were screened from rhizosphere of rice, named NRCB001, NRCB002, NRCB003, NRCB006, NRCB007, NRCB008, NRCB009, NRCB011, NRCB012, NRCB014, NRCB015, NRCB016 and NRCB017. All isolates produced auxin and grew well in LB medium in this study. Three isolates, NRCB001, NRCB002, and NRCB003, which secreted more than 20 μg ml−1 auxin, were selected for the following evaluations (Fig. 1).
Molecular identification of the three auxin-Producing isolates
The 16S rRNA, gyrA and gyrB genes of the three auxin-producing isolates deposited in NCBI GenBank under accession numbers: 16S rRNA (MN128363-128365), gyrA (MN662261–MN662263), gyrB (MN662264–MN662266). The 16S rRNA, gyrA and gyrB gene MLSA phylogeny is provided in Fig. 2. The results show that NRCB001 showed high similarity with Bacillus megaterium, NRCB002 showed high similarity with Bacillus subtilis subsp. subtilis and NRCB003 showed high similarity with Bacillus subtilis.
Characteristics of three strains, NRCB001, NRCB002 and NRCB003
Bacterial colonies of NRCB001, NRCB002, and NRCB003 were frequently smooth. These three strains were Gram-positive, spore-forming, positive for indole test and catalase test, and negative for methyl red test (Table 2). Thus, NRCB001 may be Bacillus megaterium, NRCB002 may be Bacillus subtilis subsp. subtilis, NRCB003 may be Bacillus subtilis.
These three strains produced a considerable quantity of ammonia (0.81 to 2.62 mg ml−1). They showed positive results for phosphate solubilization (166.3 to 235.4 μg ml−1) and potassium solubilization (5.17 to 7.46 μg ml−1). They had the ability to produce siderophore (47.2% to 59.7%), and had ACC deaminase activities (Table 3).
Effect of NaCl on bacterial growth
The growth of these three strains were tested under saline conditions. As shown in Fig. 3, they were able to grow at a concentration of 700 mM NaCl. When the NaCl concentration exceeded 700 mM, the growth of NRCB001 and NRCB002 were significantly inhibited; while the growth of the NRCB003 was notably improved. The maximum OD600 value of NRCB003 was obtained when 1750 mM NaCl was achieved in the medium. Two-way ANOVAs illustrated that the strain’s NaCl tolerance was significantly affected by salt, strain and the interaction of salt × strain.
Effects of different concentrations of NaCl on the growth of bacteria. Experiments were performed in triplicate and the values are represented as mean ± standard error. Note: *significant effect at 0.01 ≤ P ≤ 0.05; **significant effect at P < 0.01; NS no significant effect. Values with different letters (capital letters for different strains at the same NaCl concentration and lowercases for same strains at the different NaCl concentration) denote significant differences among treatment groups according to Duncan’s test (P < 0.05)
Assessment of colonization of auxin-producing strains in root of alfalfa
All of the auxin-producing bacteria NRCB001, NRCB002 and NRCB003 could colonize alfalfa root. Under 0 mM NaCl condition, the population of NRCB001, NRCB002 and NRCB003 reached 10.0 × 108 cfu g−1, 131.3 × 108 cfu g−1 and 34.3 × 108 cfu g−1, respectively. Under 130 mM NaCl condition, the population of NRCB001, NRCB002 and NRCB003 reached 7.7 × 108 cfu·g−1, 137.3 × 108 cfu g−1 and 50.3 × 108 cfu g−1, respectively (Fig. 4). In addition, roots of uninoculated plant grown with and without NaCl treatment and negative control, showed no bacterial growth on LB plates from. Two-way ANOVAs illustrated that the population of auxin-producing strains were significantly affected by strain.
Colonization of auxin-producing strains in the root of alfalfa. Experiments were performed in triplicate and the values are represented as mean ± standard error. Data are presented as the mean of 3 replicates. Note: *significant effect at 0.01 ≤ P ≤ 0.05; **significant effect at P < 0.01; NS: no significant effect. Capital letters for same strains at the different NaCl concentration (P < 0.05, Independent-Sample T Test); lowercases for different strains at the same NaCl concentration (P < 0.05, Duncan’s multiple-range tests)
Effects of strains on the growth of Medicagosativa L.
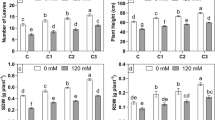
Germination rate was significantly decreased under NaCl-stressed conditions, but was not affected by bacterial inoculation (Fig. 5a). Under 0 mM NaCl condition, when inoculated with NRCB001, NRCB002 and NRCB003, dry weights of seedlings were significantly (P < 0.05) increased by 24.1%, 23.1% and 38.5%, respectively; when inoculated with NRCB001, plant height of seedlings were significantly (P < 0.05) increased by 21.8%; when inoculated with NRCB002, root length, root surface and root volume of alfalfa significantly (P < 0.05) increased by 52.2%, 49.8% and 57.8%, respectively; all of the three strains significantly (P < 0.05) increased both leaf number and area compared with those of non-inoculated control seedlings. The inoculation of NRCB002 caused significant (P < 0.05) increase in dry weight by 96.9%, plant height by 53.9%, root length by 42.2%, root volume by 55.4% and root surface by 48.0% compared with those of non-inoculated control seedlings under 130 mM NaCl condition. In addition, NRCB003 promoted the growth of seedlings and led to statistically significant (P < 0.05) increase in dry weight by 71.6%, plant height by 42.8%, root length by 50.4%, root volume by 57.4% and root surface by 54.4% compared with those of non-inoculated seedlings under 130 mM NaCl condition. All of the three strains significantly (P < 0.05) increased both leaf number and area compared with those of non-inoculated seedlings under 130 mM NaCl condition (Fig. 5). Two-way ANOVAs showed that germination rate in this study was significantly influenced by salt, plant height, leaf number and root length were significantly influenced by salt and strain, root surface was significantly affected by strain and the interaction of salt × strain, and root volume, leaf area and dry weight were significantly affected by salt, strain and the interaction of salt × strain.
Evaluation of different plant growth promoting parameters to show the effect of three strains inoculation on seedlings under non-saline and saline conditions. a Germination rate, b plant height, c root length, d leaf area of whole plant, e root surface area, f leaf area, g root volume, and h dry weight of whole plant. CK represents non-inoculated control. Error bars represent the standard error. Data are presented as the mean of 4 replicates. Note: *significant effect at 0.01 ≤ P ≤ 0.05; **significant effect at P < 0.01; NS: no significant effect. Capital letters for same strains at the different NaCl concentration (P < 0.05, Independent-Sample T Test); lowercases for different strains at the same NaCl concentration (P < 0.05, Duncan’s multiple-range tests)
Influence of auxin-producing strains on biochemical parameters of alfalfa
Under 0 mM NaCl condition, the SOD activity in alfalfa leaves inoculated with NRCB002 and NRCB003 were significantly increased by 18.9% and 25.9% (P < 0.05), respectively; the CAT activity in alfalfa leaves inoculated with NRCB001, NRCB002 and NRCB003 were significantly (P < 0.05) increased by 35.7%, 44.4% and 97.2%, respectively; the MDA content in alfalfa leaves inoculated with NRCB001, NRCB002 and NRCB003 were significantly (P < 0.05) decreased by 31.3%, 31.0% and 30.1%, respectively (Fig. 6). Under 130 mM NaCl condition, the inoculation of NRCB001,NRCB002 and NRCB003 caused significant (P < 0.05) increase in SOD activity by 11.5%, 17.4% and 21.7%, respectively; NRCB001 and NRCB002 significantly (P < 0.05) increased CAT activity by 97.7% and 95.3%, respectively; the MDA content in alfalfa leaves inoculated with NRCB001, NRCB002 and NRCB003 were significantly (P < 0.05) decreased by 20.1%, 37.6% and 41.2%, respectively. Two-way ANOVAs showed that SOD activity in this study was significantly influenced by salt and strain, CAT activity and MDA content was significantly affected by salt, strain and the interaction of salt × strain (Fig. 6).
Antioxidant levels assessed in leaves of alfalfa seedlings inoculated with auxin-producing strains under non-saline and saline conditions. a SOD activity, b CAT activity, and c MDA concentration. Error bars represent the standard error. Data are presented as the mean of 4 replicates. Note: *significant effect at 0.01 ≤ P≤0.05; **significant effect at P < 0.01; NS no significant effect. Capital letters for same strains at the different NaCl concentration (P < 0.05, Independent-Sample T Test); lowercases for different strains at the same NaCl concentration (P < 0.05, Duncan’s multiple-range tests)
Discussion
Bacteria are the most abundant microbes present in the plant rhizosphere. Bacillus species are capable of forming long-lived, stress tolerant spores and secreting metabolites that stimulate plant growth (Radhakrishnan et al. 2017). Previous research reported that genus Bacillus like B. megaterium, B. subtilis, B. azotofixans, etc. were reported as PGPR (Saxena et al. 2020). It is well known that bacteria isolated from one plant can better colonize its original plant than those isolates from other habitants. However, Bacillus species that are used for rhizosphere applications can also function as plant endophytes to increase no-original plant growth (Hashem et al. 2019). For example, B. megaterium and B. subtilis isolated from the rhizosphere of Setaria viridis and Cenchrus ciliaris respectively, have been shown to increase plant growth of alfalfa (Daur et al. 2018). Our findings are consistent with previous study, NRCB001, NRCB002 and NRCB003 are isolated from rhizosphere of rice, which can colonize the alfalfa root and significantly promote alfalfa growth (Figs. 4, 5). NaCl-induced salinity can adversely affect plant growth and development, and then lead to gradual decline in crop productivity (Saghafi et al. 2018). PGPR are a potential biological tool to alleviate abiotic constraint including salinity and to ameliorate plant productivity under abiotic constraints (Etesami and Beattie 2018). Under high-NaCl conditions, the content of reactive oxygen species (ROS) in plant cells will increase, causing oxidative damage to membrane lipids, proteins and nucleic acids (Khan et al. 2017; Li and Jiang, 2017; Yasin et al. 2018). An effective antioxidant defense system containing enzymes exists in plants, superoxide dismutase (SOD), catalase (CAT), peroxidase (POD) and ascorbate peroxidase (APX) play an important role in protecting plants from oxidative damage. Previous studies reported that APX, CAT, POD and SOD activities were significantly enhanced in various crops inoculated by PGPR under salt conditions (Khan et al. 2017; Etesami and Beattie 2018). In this study, the higher SOD and CAT activity in inoculated seedlings, thereby resisting to the effect of salt stress. Similar results were reported by Liu et al. (2019) in alfalfa plants exposed to salt stress and inoculated with Enterobacter aerogenes and Pseudomonas aeruginosa, which have maintained higher levels of activities of SOD and CAT, thereby promoting the growth of alfalfa under salt stress. Malondialdehyde is one of the products of ROS oxidative unsaturated fatty acids, which is a potential indicator of oxidative membrane injury caused by oxidative stress (Sapre et al. 2018). The malondialdehyde content of alfalfa inoculated with auxin-producing bacteria was lower than control under salt stress, which was consistent with the results of previous studies on oxidative injure of plants under salt stress. Samaddar et al. (2019) reported that the increased salt tolerance of red pepper plants may be related to Pseudomona sp significantly reducing the MDA content.
The association of plants with PGPR depends on several parameters related to the adaptation of the introduced PGPR inoculants with the root environment (root exudates and metabolites) and environmental conditions (Egamberdieva et al. 2017). Previous studies have shown that salt-tolerant PGP bacteria are physiologically adapted to abiotic stress and can increase salt tolerance and yield of crops in saline soil conditions, such as tomatoes, corn and pepper (Mayak et al. 2004; Ferreira et al. 2018; Yasin et al. 2018). Therefore, we cannot use salt-sensitive inoculants to improve the productivity of plants in saline soils. In our work, we tested the NaCl tolerance of three auxin-producing bacteria, all of which are able to grow at a concentration of 700 mM NaCl (Fig. 3). Finally, we used salt-tolerant auxin-producing bacteria to ameliorate NaCl tolerance of alfalfa.
Auxin-producing is one of the most important mechanisms for PGPR strains promoting plant growth. auxin-induced alterations in plant root architecture may lead to an increase in total root surface area. The larger root surface area can improve nutrient and water uptake, which may have a positive impact on plant growth (Etesami et al. 2015; Noori et al. 2018; Hmaeid et al. 2019). In this study, thirteen isolates were isolated from the rhizosphere of rice, and then three auxin-producing bacteria, named NRCB001, NRCB002, and NRCB003, were further identified and tested for PGP properties particularly under saline conditions. when inoculated with NRCB001, NRCB002 and NRCB003, alfalfa showed a better resistance to the presence of 130 mM NaCl. Our findings are consistent with previous studies, in which auxin production is key PGP mechanisms, and other PGP traits in inducing salt tolerance capacity also are very important for promoting various crops growth, such as pepper and maize (Maxton et al., 2017; Aslam and Ali, 2018; Samaddar et al. 2019).
Our study indicates that bacterial inoculants NRCB001, NRCB002 and NRCB003 were selected on the basis of their auxin-producing capacity and exhibiting multiple PGP features and salt tolerance capacity positively affect the growth of alfalfa under high salinity condition. Three auxin-producing strains had a series of PGP characteristics, including auxin, siderophore and NH3 production, inorganic phosphate solubilization, potassium-solubilization and ACC deaminase activity (Table 3). Plants are inhibited from growth due to low osmotic and ion injury as well as nutritional disorders under NaCl-induced salinity stress (Zhou et al. 2018). PGPR can increase the secretion of osmoprotectants in plant tissues, reduce the absorption of Na+ and Cl− by plants, and secrete exopolysaccharides in the roots to combine with sodium in the soil (Hmaeid et al. 2019). PGPR release the metal chelator siderophores in the rhizosphere of the host plant, which can affect the absorption and bioavailability of various metal ions to promote the growth and enhance the stress tolerance of plant (Dimkpa et al. 2009; Wang et al. 2014). In this study, the best siderophore producing bacteria NRCB002 significantly increased root length and dry weight in pot treatment amended with NaCl (Fig. 5c, h).
PGPR usually convert insoluble nutrients into available nutrients in the soil which can be directly absorbed by the plant. The abilities of nutrient conversion by PGPR make them promising candidates to develop bio-fertilizer (Sharma et al. 2013; Hu et al. 2006). Ammonia-production, phosphate solubilization, and potassium solubilization have been considered important traits that bacteria display to promote plant growth, as nitrogen, phosphate, and potassium are often insufficient or insoluble salts which are not available to the plant (Reyes-Castillo et al. 2019). The NRCB002 and NRCB003 have good phosphate-solubilizing, potassium- solubilizing and ammonia-producing abilities, which significantly increased most of plant growth parameters except for plant height and germination rate in this study (Fig. 5 and Table 3). The reason why bacterial strains help plants absorb minerals by solubilization and mineralization of phosphate, because it can induce more available nutrients to exist in the form of phosphate and maintain pH around neutral by proton extrusion (Hmaeid et al. 2019). ACC deaminase activity is one of the most important traits of PGPR to promote plant growth under stress conditions (Liu et al. 2019). ACC deaminase can cleave the ethylene precursor ACC into ammonia and α-ketobutyrate, which can alleviate the damage caused by stress ethylene to plants (Etesami and Maheshwari 2018). These three strains, NRCB001, NRCB002, and NRCB003, had ACC deaminase activity. In pure culture, the addition of NaCl inhibited the growth of NRCB001 severely while promoted the growth of NRCB003 notably. In the pot experiment, NRCB002 and NRCB003 show dry weight increment under salinity conditions, while NRCB001 did not alleviate salinity stress except for leaf area tested in this study. The result suggests that the activity of ACC deaminase is a critical trait for PGPR helping the plant to alleviate stress (Glick 2014; Hmaeid et al. 2019; Samaddar et al. 2019). In agreement with previous studies, our results suggest that the ammonia-production, phosphate solubilization, and potassium solubilization by these three strains play key roles in promoting plant growth besides auxin (Marques et al. 2010; Sharma et al. 2013; Zhang and Kong 2014).
This study does suggest that B. megaterium NRCB001, B. subtilis subsp. subtilis NRCB002 and B. subtilis NRCB003 had salt tolerance capacity and multiple PGP traits including auxin-producing which promote alfalfa growth under normal and salt stress conditions. However, it remains to investigate the pathogenicity of strains on alfalfa, and to evaluate the effects of strains on some physiological parameters of alfalfa, such as plant Na, K, Na/K ratio, proline, and to study legume symbiotic rhizobium.
Conclusions
Of thirteen strains isolated from rice rhizosphere, three strains, B. megaterium NRCB001, B. subtilis subsp. subtilis NRCB002 and B. subtilis NRCB003 were auxin-producing and salt-tolerant strains. Of three strains, NRCB002 and NRCB003 used for inoculation present a good PGP potential in enhancing growth and inducing salt tolerance capacity of alfalfa. In addition, NRCB003 was able to grow in the presence of NaCl concentration at 1750 mM. Thus, NRCB002 and NRCB003 may be prospective PGPRs for biofertilizers.
References
Aslam F, Ali B (2018) Halotolerant bacterial diversity associated with suaeda fruticose (L.) forssk. improved growth of maize under salinity stress. Agronomy 8(8):131
Baldani JI, Reis VM, Videira SS, Boddey LH, Baldani VLD (2014) The art of isolating nitrogen-fixing bacteria from non-leguminous plants using N-free semi-solid media: a practical guide for microbiologists. Plant Soil 384:413–431
Bric JM, Bostock RM, Silverstone SE (1991) Rapid in situ assay for indoleacetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol 57:535–538
Chun J, Bae KS (2000) Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 78:123–127
Daur I, Saad MM, Eida AA, Ahmad S, Shah ZH, Ihsan MZ, Muhammad Y, Sohrab SS, Hirt H (2018) Boosting alfalfa (Medicago sativa L.) production with rhizobacteria from various plants in saudi arabia. Front Microbiol 9:447
Dimkpa C, Weinand T, Asch F (2009) Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ 32:1682–1694
Egamberdieva D, Davranov K, Wirth S, Hashem A, Abd Allah EF (2017) Impact of soil salinity on the plant-growth-promoting and biological control abilities of root associated bacteria. Saudi J Biol Sci 24(7):1601–1608
Etesami H, Beattie GA (2018) Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front Microbiol 9:148
Etesami H, Maheshwari DK (2018) Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: action mechanisms and future prospects. Ecotox Environ Safe 156:225–246
Etesami H, Alikhani HA, Hosseini HM (2015) Indole-3-acetic acid (IAA) production trait, a useful screening to select endophytic and rhizosphere competent bacteria for rice growth promoting agents. MethodsX 2:72–78
Ferreira NC, Mazzuchelli RDCL, Pacheco AC, Araujo FFD, Antunes JEL, Araujo ASFD (2018) Bacillus subtilis improves maize tolerance to salinity. Cienc Rural 48:8
Gao N, Shen WS, Camargo E, Shiratori Y, Nishizawa T, Isobe K, He XH, Senoo K (2017) Nitrous oxide (N2O)-reducing denitrifier-inoculated organic fertilizer mitigates N2O emissions from agricultural soils. Biol Fertil Soils 53:885–898
Glick BR (2014) Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol Res 169:30–39
Gupta M, Kiran S, Gulati A, Singh B, Tewari R (2012) Isolation and identification of phosphate solubilizing bacteria able to enhance the growth and aloin-A biosynthesis of Aloe barbadensis Miller. Microbiol Res 167:358–363
Hashem A, Tabassum B, Abd Allah EF (2019) Bacillus subtilis: A plant-growth promoting rhizobacterium that also impacts biotic stress. Saudi J Biol Sci 26:1291–1297
Hmaeid N, Wali M, Metoui-Ben Mahmoud O, Pueyo JJ, Ghnaya T, Abdelly C (2019) Efficient rhizobacteria promote growth and alleviate NaCl-induced stress in the plant species Sulla carnosa. Appl Soil Ecol 133:104–113
Hongoh Y, Ohkuma M, Kudo T (2003) Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol Ecol 44:231–242
Hu XF, Chen JS, Guo JF (2006) Two phosphate- and potassium-solubilizing bacteria isolated from Tianmu Mountain, Zhejiang, China. World J Microb Biot 22:983–990
Hussein KA, Joo JH (2015) Isolation and characterization of rhizomicrobial isolates for phosphate solubilization and indole acetic acid production. J Korean Soc Appl Biol Chem 58:847–855
Jin YQ, Zhu HF, Luo S, Yang WW, Zhang L, Li SS, Jin Q, Cao Q, Sun S, Xiao M (2019) Role of maize root exudates in promotion of colonization of bacillus velezensis strain S3-1 in rhizosphere soil and root tissue. Curr Microbiol 76:855–862
Kadmirl IM, Chaouqul L, Azaroual SE, Sijilmassl B, Yaakoubl K, Wahby I (2018) Phosphate-solubilizing and IAA-producing rhizobacteria promote plant growth under saline conditions. Arab J Sci Eng 43:3403–3415
Karthik C, Elangovan N, Kumar TS, Govindharaju S, Barathi S, Oves M, Arulselvi PI (2017) Characterization of multifarious plant growth promoting traits of rhizobacterial strain AR6 under Chromium (VI) stress. Microbiol Res 204:65–71
Khan WU, Ahmad SR, Yasin NA, Ali A, Ahmad A, Akram W (2017) Application of Bacillus megaterium MCR-8 improved phytoextraction and stress alleviation of nickel in Vinca rosea. Int J Phytorem 19(9):813–824
Li HQ, Jiang XW (2017) Inoculation with plant growth-promoting bacteria (PGPB) improves salt tolerance of maize seedling. Russ J Plant Physiol 64(2):235–241
Liu ZH, Zhang HM, Li GL, Guo XL, Chen SY, Liu GB, Zhang YM (2011) Enhancement of salt tolerance in alfalfa transformed with the gene encoding for betaine aldehyde dehydrogenase. Euphytica 178:363–372
Liu JL, Tang L, Gao H, Zhang M, Guo C (2019) Enhancement of alfalfa yield and quality by plant growth-promoting rhizobacteria under saline-alkali conditions. J Sci Food Agric 99:281–289
Lu R (1999) Agricultural chemistry analysis of soil. China Agricultural Science and Technology Press, Beijing
Marques APGC, Pires C, Moreira H, Rangel AOSS, Castro Pml (2010) Assessment of the plant growth promotion abilities of six bacterial isolates using Zea mays as indicator plant. Soil Biol Biochem 42:1229–1235
Maxton A, Singh P, Masih SA (2017) ACC deaminase-producing bacteria mediated drought and salt tolerance in Capsicum annuum. J Plant Nutr 41(5):574–583
Mayak S, Tirosh T, Glick BR (2004) Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol Biochem 42(6):565–572
Mukhtar S, Shahid I, Mehnaz S, Malik KA (2017) Assessment of two carrier materials for phosphate solubilizing biofertilizers and their effect on growth of wheat (Triticum aestivum L.). Microbiol Res 205:107–117
Nadeem SM, Ahmad M, Zahir ZA, Javaid A, Ashraf M (2014) The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol Adv 32:429–448
Nadeem SM, Ahmad M, Muhammad Naveed M, Imran M, Ahmad Z, Zahir AZ, Crowley DE (2016) Relationship between in vitro characterization and comparative efficacy of plant growth-promoting rhizobacteria for improving cucumber salt tolerance. Arch Microbiol 198:379–387
Noori F, Etesami H, Zarini HN, Khoshkholgh-Sima NA, Salekdeh GH, Alishahi F (2018) Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotox Environ Safe 162:129–138
Penrose DM, Glick BR (2003) Methods for isolating and characterizing ACC deaminase-containing plant growth-promoting rhizobacteria. Physiol Plantarum 118:10–15
Qin Y, Druzhinina IS, Pan X, Yuan Z (2016) Microbially mediated plant salt tolerance and microbiome-based solutions for saline agriculture. Biotechnol Adv 34:1245–1259
Radhakrishnan R, Hashem A, Abd-Allah EF (2017) Bacillus: a biological tool for crop improvement through bio-molecular changes in adverse environments. Front Physiol 8:667
Reyes-Castillo A, Gerding M, Oyarzúa P, Zagal E, Gerding J, Fischer S (2019) Plant growth-promoting rhizobacteria able to improve NPK availability: selection, identification and effects on tomato growth. Chil J Agr Res 79(3):473–485
Saghafi D, Ghorbanpour M, Lajayer BA (2018) Efficiency of Rhizobium strains as plant growth promoting rhizobacteria on morpho-physiological properties of Brassica napus L. under salinity stress. J Soil Sci Plant Nutr 18(1):253–268
Samaddar S, Chatterjee P, Choudhury AR, Ahmed S, Sa T (2019) Interactions between Pseudomonas spp. and their role in improving the red pepper plant growth under salinity stress. Microbiol Res 219:66–73
Sapre S, Gontia-Mishra I, Tiwari S (2018) Klebsiella sp. confers enhanced tolerance to salinity and plant growth promotion in oat seedlings (Avena sativa). Microbioll Res 206:25–32
Saxena AK, Kumar M, Chakdar H, Anuroopa N, Bagyaraj DJ (2020) Bacillus species in soil as a natural resource for plant health and nutrition. J Appl Microbiol 128:1583–1594
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:46–56
Selvakumar G, Kim K, Hu S, Sa T (2014) Effect of salinity on plants and the role of arbuscular mycorrhizal fungi and plant growth-promoting bacteria in alleviation of salt stress. In: Ahmad P, Wani MR (eds) Physiological mechanisms and adaptation strategies in plants under changing environment. Springer, New York, pp 115–144
Sharma SB, Sayyed RZ, Trivedi MH, Gobi TA (2013) Phosphate solubilizing microbes: sustainable approach for managing phosphorus deficiency in agricultural soils. SpringerPlus 2:587
Vaishnav A, Choudhary DK (2019) Regulation of drought-responsive gene expression in Glycine max L. merrill is mediated through Pseudomonas simiae strain AU. J Plant Growth Regul 38:333–342
Wang W, Qiu Z, Tan H, Cao L (2014) Siderophore production by actinobacteria. Biometals 27:623–631
Wang W, Wu Z, He Y, Huang Y, Li X, Ye BC (2018) Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotox Environ Safe 164:520–529
Wu N, Li Z, Wu F, Tang M (2016) Comparative photochemistry activity and antioxidant responses in male and female Populus cathayana cuttings inoculated with arbuscular mycorrhizal fungi under salt. Sci Rep 6:37663
Yaish MW, Antony I, Glick BR (2015) Isolation and characterization of endophytic plant growth-promoting bacteria from date palm tree (Phoenix dactylifera L.) and their potential role in salinity tolerance. Antonie Van Leeuwenhoek 107:1519–1532
Yamamoto S, Harayama S (1995) PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microb 61:1104–1109
Yasin NA, Akram W, Khan WU, Ahmad SR, Ahmad A, Ali A (2018) Halotolerant plant-growth promoting rhizobacteria modulate gene expression and osmolyte production to improve salinity tolerance and growth in Capsicum annum L. Environ Sci Pollut Res 25(23):23236–23250
Zhang CS, Kong FY (2014) Isolation and identification of potassium-solubilizing bacteria from tobacco rhizospheric soil and their effect on tobacco plants. Appl Soil Ecol 82:18–25
Zhang WJ, Wang T (2015) Enhanced salt tolerance of alfalfa (Medicago sativa) by rstB gene transformation. Plant Sci 234:110–118
Zhang HH, Li X, Nan X, Sun GY, Sun ml, Cai DJ, Gu SY (2017) Alkalinity and salinity tolerance during seed germination and early seedling stages of three alfalfa (Medicago sativa L.) cultivars. Legume Res 40:853–858
Zhang G, Sun B, Zhao H, Wang X, Zheng C, Xiong K, Ouyan Z, Lu F, Yuan Y (2019) Estimation of greenhouse gas mitigation potential through optimized application of synthetic N, P and K fertilizer to major cereal crops: a case study from China. J Clean Prod 237:117650
Zhou Y, Tang N, Huang L, Zhao Y, Tang X, Wang K (2018) Effects of salt stress on plant growth, antioxidant capacity, glandular trichome density, and volatile exudates of Schizonepeta tenuifolia Briq. Int J Mol Sci 19(1):252
Acknowledgements
We appreciate anonymous reviewers very much for their positive and constructive comments and suggestions on our manuscript. This research was supported by the National Natural Science Foundation of China (31972503), the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (18KJB210007) and the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (XTE1828), China.
Author information
Authors and Affiliations
Contributions
The authors, NG and HJY designed the experiment. ZYZ, HHZ and JL performed the experiments and data analysis. ZYZ wrote the manuscript, and NG edited the manuscript critically and very carefully. HQN, XCC, DL and YC helped perform the analysis with constructive discussions. All authors have read the manuscript and approved the data of the manuscript in its present form.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflict of interest regarding the publication of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, Z., Zhang, H., Leng, J. et al. Isolation and characterization of plant growth-promoting rhizobacteria and their effects on the growth of Medicago sativa L. under salinity conditions. Antonie van Leeuwenhoek 113, 1263–1278 (2020). https://doi.org/10.1007/s10482-020-01434-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-020-01434-1