Abstract
A novel endophytic actinomycete strain, designated KM-1-2T, was isolated from seeds of Ginkgo biloba at Yangling, China. A polyphasic approach was used to study the taxonomy of strain KM-1-2T and it was found to show a range of phylogenetic and chemotaxonomic properties consistent with those of members of the genus Streptomyces. The diamino acid of the cell wall peptidoglycan was identified as LL-diaminopimelic acid. No diagnostic sugars were detected in whole cell hydrolysates. The predominant menaquinones were identified as MK-9(H6) and MK-9(H8). The diagnostic phospholipids were found to be phosphatidylethanolamine and phosphatidylcholine. The DNA G + C content of the novel strain was determined to be 72.9 mol%. The predominant cellular fatty acids (> 10.0 %) were identified as iso-C14 : 0, iso-C16 : 0, C16 : 0 and C17 : 0 cyclo. Phylogenetic analysis based on the 16S rRNA gene sequence revealed that the strain is closely related to Streptomyces carpaticus JCM 6915T (99.3%), Streptomyces harbinensis DSM 42076T (98.9%) and Streptomyces cheonanensis JCM 14549T (98.5%). DNA-DNA hybridizations with these three close relatives gave similarity values of 39.1 ± 1.9, 35.8 ± 2.3, and 47.4 ± 2.7%, respectively, which indicated that strain KM-1-2T represents a novel species of the genus Streptomyces. This is consistent with the morphological, physiological and chemotaxonomic data. Cumulatively, these data suggest that strain KM-1-2T represents a novel Streptomyces species, for which the name Streptomyces ginkgonis sp. nov. is proposed, with the type strain KM-1-2T (= CCTCC AA2016004T = KCTC 39801T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Streptomyces, the largest genus in the domain Bacteria (Hain et al. 1997; http://www.bacterio.net/streptomyces.html), was established by Waksman and Henrici (1943). The members of this genus are widely distributed in different environments, not only in soil but also in plant rhizospheres (Xiao et al. 2009), the stems of plants (Liu et al. 2014) and marine sediment (Haritha et al. 2012). Streptomyces species are an important source of medicines, especially antibiotics (Loria et al. 1997; Hwang et al. 2014). Members of the genus Streptomyces carry an extremely versatile group of biosynthetic machineries capable of producing complex molecules of medical, agricultural and economical significance.
Endophytes are microorganisms that colonise within plant cells or tissues at some or at all stages of their life cycle. They are integral parts of the plant micro-ecosystem. Their metabolic activities and products can promote host plant adaptation to various external (biological, non-biological) environmental stresses, maintaining ecological balance (Stone et al. 2000). Endophytes exploit an unusual habitat (i.e. internal living tissues of plants), and many possess the potential to produce bioactive compounds similar to their hosts. This makes endophytic actinomycetes a potential resource for isolating new active substances, which can be widely used in medicine, agriculture, and industrial production (Cao et al. 2004). Some endophytic Streptomyces have been found previously (Liu et al. 2014; Kirkpatrick and Dugatkin 2015; Passari et al. 2016).
During our investigations of the diversity of endophytic actinomycetes, we isolated a novel actinomycete, KM-1-2T, which showed the morphological characteristics typical of members of the genus Streptomyces. In this report, we described the isolation and taxonomic characterisation of strain KM-1-2T, based on a polyphasic approach.
Materials and methods
Strain KM-1-2T was isolated from the aril of a seed of Ginkgo biloba. Healthy seed samples of G. biloba, collected from the schoolyard of Northwest A&F University in Yangling, Shaanxi Province P. R. China, were used as sources for the isolation of endophytic actinomycetes. Seeds were surface sterilised with serial washes of 75% ethanol for 1 min, 10% sodium hypochlorite for 5 min and several rinses with distilled water. Strain KM-1-2T was isolated using Gause’s Synthetic agar medium supplemented with streptomycin sulphate (10 μg/mL) and actidione (50 μg/mL) and incubated at 28 °C for 21 days. The final distilled water rinse was smeared on the same medium as negative control and no culturable microorganisms were recovered. Strain KM-1-2T was routinely cultured in the same medium and the stock culture were maintained as a glycerol suspension (20%, w/v) at − 80 °C. The reference strains Streptomyces carpaticus JCM 6915T, Streptomyces harbinensis DSM 42076T and Streptomyces cheonanensis JCM 14549T were obtained from Japan Collection of Microorganisms and cultured under comparable conditions for parallel testing.
Morphological, cultural, physiological and biochemical characteristics of strain KM-1-2T were determined by following the standard protocol of the International Streptomyces Project (ISP). Colour production was determined by comparison with chips from the colour charts of the Inter-Society Colour Council-National Bureau of Standards (Kelly 1964). After 14 days incubation on Gause’s Synthetic agar, morphological properties of the isolate were characterised using both light microscopy (BH2; Olympus) and scanning electron microscopy (Stereoscan JSM-6360LV). The temperature, concentrations of NaCl and pH tolerance for growth were determined on Bennett’s agar plates incubated for up to 14 days. For the carbon utilisation tests, 1% of each substrate was added to the Pridham and Gottlieb carbon utilisation medium (Pridham and Gottlieb 1948). Inoculated Petri dishes were incubated at 28 °C. Plates were checked for growth after 2 weeks. Additional physiological and biochemical properties were determined using standard media and methods (Shirling and Gottlieb 1966).
The isomer of diaminopimelic acid of the cell wall and the whole cell sugar composition were analysed using TLC according to the procedures described by Lechevalier and Lechevalier (1980). Menaquinones were extracted and determined by HPLC using the methods of Collins (1985). The analysis of phospholipids was carried out according to the method of Lechevalier et al. (1981). Fatty acids were analysed by GC–MS using the method of Kämpfer and Kroppenstedt (1996).
The extraction of genomic DNA, PCR amplification of the 16S rRNA gene and sequencing of the purified PCR products were carried out as described by Rainey et al. (1996). A preliminary sequence comparison with those from GenBank indicated that strain KM-1-2T is closely related to members of the genus Streptomyces. The sequence was then aligned manually with reference sequences of the genus Streptomyces. The 16S rRNA gene sequence of the strain was aligned by Clustal_X version 1.8 (Thompson et al. 1997) with the 16S rRNA gene sequences of reference species of the genus Streptomyces obtained from the GenBank/EMBL/DDBJ database. Phylogenetic trees were constructed using neighbor-joining (Saitou and Nei 1987), maximum-likelihood and maximum-parsimony (Fitch 1971) methods in the TREECON software package version 1.3b (Van de Peer and de Wachter 1994) and the MEGA software version 7.0 (Kumar et al. 2016). The genetic distance matrices were estimated by the Kimura two-parameter model (Kimura 1980). The topology of the trees was evaluated by bootstrap analysis based on 1000 replicates (Felsenstein 1985).
DNA G+C content was determined by reversed-phase HPLC (Mesbah et al. 1989). DNA–DNA relatedness was determined as described by Rong et al. (2009), using photobiotin-labelled DNA probes and micro-dilution wells.
Results and discussion
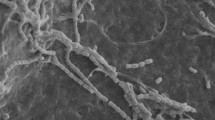
Strain KM-1-2T was observed to form straight or wavy, long chains of non-motile spores with rough surfaces (Fig. 1). The strain was found to grow well on ISP 2, 4, 5, Gause’s Synthetic agar and Glucose- asparagine agar media, as well as Czapek’s agar. The aerial mycelium and substrate mycelium were observed to develop without fragmentation. The aerial mycelium is transparent white to yellowish on all media tested and the substrate mycelium is light yellow to deep yellow. Strain KM-1-2T was found to grow between pH 6.0 and 9.0, with optimum growth at pH 7–8. The optimum growth temperature was found to be 28–30 °C. The strain was observed to grow in the presence of 0–6 % NaCl (w/v). Detailed physiological and biochemical properties are shown in Table 1 and the species description.
Strain KM-1-2T was found to contain LL-diaminopimelic acid in cell wall hydrolysates, whilst no diagnostic sugars were detected in the whole cell hydrolysate, which is a type I cell wall as defined by Lechevalier et al. (1970). The phospholipid profile was found to contain phosphatidylethanolamine and phosphatidylcholine i.e. the phospholipid type II pattern of Lechevalier et al. (1981). The predominant menaquinones were identified as MK-9 (H6) and MK-9 (H8). The predominant cellular fatty acids (> 10.0 %) were identified as iso-C14 : 0 (10.6%), iso-C16 : 0 (37.2%), C16 : 0 (14.7%) and C17 : 0 cyclo (12.1%). The morphological and chemotaxonomic characteristics described above clearly supports placement of strain KM-1-2T within the genus Streptomyces. Strain KM-1-2T is physiologically different from other closely related Streptomyces species, as can be seen from the differential physiological characteristics presented in Table 1.
The phylogenetic relationship between strain KM-1-2T and the other recognised species of Streptomyces can be seen in the neighbour-joining dendrogram shown in Fig. 2. The 16S rRNA gene sequence of strain KM-1-2T was found to be similar to that of S. carpaticus JCM 6915T (99.3%), S. harbinensis DSM 42076T (98.9%) and S. cheonanensis JCM 14549T (98.5%). Strain KM-1-2T was observed to form a coherent cluster with S. harbinensis NEAU-Da3T (JQ750974), S. cheonanensis VC-A46T (AY822606) and S. carpaticus NBRC 15390T (AB184641). These data suggest the strain belongs to Clade 128 as defined by Labeda et al. (2012). However, the levels of DNA–DNA relatedness of strain KM-1-2T with these three close relatives, S. carpaticus JCM 6915T, S. harbinensis DSM 42076T and S. cheonanensis JCM 14549T were 39.1 ± 1.9, 35.8 ± 2.3, and 47.4 ± 2.7%, respectively, values which are much lower than the 70% accepted threshold value for prokaryotic genomic species delineation (Stackebrandt et al. 1997). The genomic DNA G +C content of KM-1-2T was determined to be 72.9 mol %.
Phylogenetic relationships based on neighbour-joining analysis (Saitou and Nei 1987) of 16S rRNA gene sequences of strain KM-1-2T and related species of the genus Streptomyces. Amycolatopsis albidoflavus IMSNU 22139T was used as an outgroup. Numbers at branch nodes indicate bootstrap percentages derived from 1000 replications (only values > 50 % are shown). Bar, 0.005 substitutions per nucleotide position
Based on its morphologic, physiologic, chemotaxonomic and phylogenetic characteristics, strain Km-1-2T was identified as a member of the genus Streptomyces. Accordingly, we propose a novel species, Streptomyces ginkgonis sp. nov., with the type strain KM-1-2T (= CCTCC AA2016004T = KCTC 39801T). The Digital Protologue database (Rosselló-Móra et al. 2017) TaxoNumber for strain Km-1-2T is TA00320.
Description of Streptomyces ginkgonis sp. nov.
Streptomyces ginkgonis (gink.go’nis. N.L. gen. n. ginkgonis of Ginkgo).
Aerobic, Gram-positive bacterium. Grows well on ISP 2, 4, 5, Gause’s Synthetic agar, glucose asparagine agar and Czapek’s agar, poorly on ISP 3 and shows no growth on potato dextrose agar. Growth occurs at pH 6–9 and 15–35 °C, respectively, but not at or above 37 °C. Tolerates concentrations of NaCl up to 6%. Optimal temperature and pH for growth are at 28–30 °C and pH 7–8, respectively. Utilises sodium citrate, d-fructose, d-glucose, inositol, maltose, d-mannitol, d-mannose, l-arabinose, sucrose and raffinose as sole carbon sources. Utilises l-alanine, l-arginine, glycine, l-asparagine and l-hydroxyprolin as sole nitrogen sources. Hydrolysis of starch and cellulose are positive. Negative in tests for hydrolysis of gelatin, production of H2S and reduction of nitrate. The cell wall contains LL-diaminopimelic acid and no diagnostic sugars are present in whole cell hydrolysates. The phospholipid profile contains phosphatidylethanolamine and phosphatidylcholine (type II). The DNA G+C content of the type strain is 72.9 mol %.
The type strain is KM-1-2T (= CCTCC AA2016004T = KCTC 39801T), which was isolated from seeds of Ginkgo biloba at Yangling, China. The GenBank accession number for the 16S rRNA gene sequence of strain KM-1-2T is KU758899. The species description is based on a single strain and hence serves as the type strain description.
References
Cao LX, Qiu ZQ, You JL, Tan HM, Zhou S (2004) Isolation and characterization of endophytic Streptomyces antagonists of Fusarium wilt pathogen from surface sterilized banana roots. FEMS Microbiol Lett 247:147–152
Collins MD (1985) Isoprenoid quinone analysis in classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics. Academic Press, London, pp 267–287
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Hain T, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E, Rainey FA (1997) Discrimination of Streptomyces albidoflavus strains based on the size and number of 16S-23S ribosomal DNA intergenic spacers. Int J Syst Bacteriol 47:202–206
Haritha R, Sivakumar K, Swathi A et al (2012) Characterization of marine Streptomyces carpaticus and optimization of conditions for production of extracellular protease. Appl Environ Microbiol 78:4826–4834
Hwang KS, Kim HU, Charusanti P, Palsson BØ, Sang YL (2014) Systems biology and biotechnology of Streptomyces, species for the production of secondary metabolites. Biotechnol Adv 32:255–268
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kelly KL (1964) Inter-Society Color Council-National Bureau of Standards color name charts illustrated with centroid colors. US Government Printing Office, Washington
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirkpatrick M, Dugatkin LA (2015) Antimicrobial and antioxidant activities of a new benzamide from endophytic Streptomyces sp. YIM 67086. Nat Prod Res 29:331–335
Kumar S, Stecher G, Tamura K (2016) Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M et al (2012) Phylogenetic study of the species within the family streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Lechevalier HA, Lechevalier MP (1980) The chemotaxonomy of actinomycetes. In: Dietz A, Thayer DW (eds) Actinomycete taxonomy. (Special Publication 6). Society of Industrial Biology, Arlington, pp 277–284
Lechevalier HA, Lechevalier MP, Gerber NN (1970) Chemical composition as a criterion in the classification of actinomycetes. Int J Syst Evol Microbiol 20(4):435–443
Lechevalier MP, Stern AE, Lechevalier HA (1981) Phospholipids in the taxonomy of actinomycetes. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg. Abt. 1(Suppl. 11):111–116
Liu M, Abdel-Mageed WM, Ren B et al (2014) Endophytic Streptomyces sp. Y3111 from traditional Chinese medicine produced antitubercular pluramycins. Appl Microbiol Biotechnol 98:1077–1085
Loria R, Bukhalid RA, Fry BA, King RR (1997) Plant pathogenecity in the genus Streptomyces. Plant Dis 81:836–846
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Passari AK, Mishra VK, Gupta VK et al (2016) Distribution and identification of endophytic Streptomyces species from Schima wallichii as potential biocontrol agents against fungal plant pathogens. Pol Soc Microbiol 65:319–329
Pridham TG, Gottlieb D (1948) The utilization of carbon compounds by some actinomycetales as an aid for species determination. J Bacteriol 56:107
Rainey FA, Ward-Rainey N, Kroppenstedt RM, Stackebrandt E (1996) The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol 46:1088–1092
Rong X, Guo Y, Huang Y (2009) Proposal to reclassify the Streptomyces albidoflavus clade on the basis of multilocus sequence analysis and DNA-DNA hybridization, and taxonomic elucidation of Streptomyces griseus subsp. solvifaciens. Syst Appl Microbiol 32:314–322
Rosselló-Móra R, Trujillo ME, Sutcliffe IC (2017) Introducing a digital protologue: a timely move towards a database-driven systematics of archaea and bacteria. Syst Appl Microbiol 110:455–456
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stackebrandt E, Rainey FA, Wardrainey NL (1997) Proposal for a new hierarchic classification system, Actinobacteria classis nov. Int J Syst Bacteriol 47:479–491
Stone JK, Bacon CW, White JF (2000) An overview of endophytic microbes: endophytism defined. Microb Endophytes 3:29–33
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Van de Peer PY, de Wachter R (1994) TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput Appl Biosci 10:569–570
Waksman SA, Henrici AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46:337–341
Xiao J, Wang Y, Luo Y, Xie SJ, Ruan JS, Xu J (2009) Streptomyces avicenniae sp. nov., a novel actinomycete isolated from the rhizosphere of the mangrove plant Avicennia mariana. Int J Syst Evol Microbiol 59:2624–2628
Acknowledgements
This work was supported by Foundation Research Project of Shaanxi Province (No. 2017JZ006); National Natural Science Foundation of China (No. 31101476); Science and Technology Program of Yangling Demonstration Zone (No. 2014NY-41).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors do not claim any conflict of interest.
Rights and permissions
About this article
Cite this article
Yan, X., Li, Y., Wang, N. et al. Streptomyces ginkgonis sp. nov., an endophyte from Ginkgo biloba . Antonie van Leeuwenhoek 111, 891–896 (2018). https://doi.org/10.1007/s10482-017-0987-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-017-0987-3