Abstract
A white bacterial strain, designated EGI 650022T, was isolated from the roots of Salsola affinis C. A. Mey, collected from Urumqi City, Xinjiang, north-western China. The strain was found to be aerobic, Gram-stain positive, oxidase-positive and catalase-positive. Cells were non-motile and irregular rods. Growth occurred at NaCl concentrations between 0 and 7 % (w/v), temperatures between 5 and 45 °C, and pH 6.0–9.0. Phylogenetic analysis based on 16S rRNA gene sequence indicated that strain EGI 650022T belongs to a clade with the genera Okibacterium and Plantibacter in the family Microbacteriaceae. The novel strain EGI 650022T showed highest levels of 16S rRNA gene sequence similarity with members of the genera Okibacterium and Plantibacter (97.2–98.0 %). The cell-wall peptidoglycan contained glutamate, homoserine, glycine, alanine and lysine. The predominant menaquinones (MKs) were MK-11, MK-12 and MK-12 (H4). The polar lipid pattern comprised phosphatidylglycerol, diphosphatidylglycerol, two unknown glycolipids and two unknown phospholipids. The major fatty acids were anteiso-C15:0 and anteiso-C17:0. The DNA G+C content was 66.0 mol%. The DNA–DNA relatedness values of strain EGI 650022T with Okibacterium fritillariae DSM 12584T, Plantibacter flavus DSM 14012T and Plantibacter auratus DSM 19586T were 39.7, 19.7 and 22.0 %. Based on phylogenetic, phenotypic, chemotaxonomic and DNA–DNA hybridization data, strain EGI 650022T is considered to represent a novel species of the genus Okibacterium, for which the name Okibacterium endophyticum sp. nov. is proposed; the type strain is EGI 650022T (=JCM 30086T = KCTC 29492T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
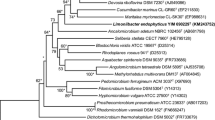
The genus Okibacterium was first proposed by Evtushenko et al. (2002) and comprises only one species, Okibacterium fritillariae, which was isolated from seeds of Fritillaria ruthenica Wikstr and Clematis recta L. Okibacterium strains contain lysine, alanine, glycine, glutamate and homoserine in their cell wall peptidoglycan, which enable them to be differentiated from members of the genus Plantibacter (Behrendt et al. 2002). The genus Plantibacter currently contains two recognized species and can be clustered with Okibacterium in the phylogenetic tree based on 16S rRNA gene sequence analysis (Lin and Yokota 2006; as shown in Fig. 1). In this study, we describe a bacterial strain, designated EGI 650022T, which was isolated from surface-sterilized root of Salsola affinis C. A. Mey, collected from Urumqi City, Xinjiang. Comparative 16S rRNA gene sequence analysis indicated that strain EGI 650022T is closely related to O. fritillariae JCM 12284T, Plantibacter flavus DSM 14012T and Plantibacter auratus DSM 19586T (highest levels of 16S rRNA gene sequence similarity of 98.0, 97.5 and 97.2 %, respectively). The aim of this work was to determine the exact taxonomic position of strain EGI 650022T by using a polyphasic approach.
Materials and methods
Isolation and maintenance of organism
The halophyte plant S. affinis C. A. Mey was collected from the suburban district of Urumqi, Xinjiang Province and used as a source for isolation of bacterial strains. Strain EGI 650022T was isolated from the roots of healthy halophyte plant S. affinis C. A. Mey. The healthy plant samples were washed in running tap water to remove adhered epiphytes and surface-sterilized according to the five-step sterilization procedure (Qin et al. 2008). The surface-sterilized roots were aseptically crumbled into smaller fragments using a commercial Joyoung blender (Qin et al. 2009), spread onto glycerol-asparagine agar (ISP 5; Shirling and Gottlieb 1966) amended with 3 % NaCl, and incubated at 30 °C for 4–6 weeks. The purified strain was maintained on yeast extract-malt extract agar (ISP 2; Shirling and Gottlieb 1966) slants at 4 °C for short-term preservation and as 25 % (v/v) glycerol suspension at −80 °C for long-term preservation. Strain EGI 650022T has been deposited in the Korean Collection for Type Cultures (KCTC; South Korea) and the Japan Collection of Microorganisms (JCM; Japan) under the accession numbers KCTC 29492T and JCM 30086T, respectively.
Phenophytic characterization
Gram-stain was carried out by using the standard Gram stain procedure. The morphological, physiological and biochemical characteristics of strain EGI 650022T were investigated using cultures grown on ISP 2 medium, at 30 °C for 5 days. Cell morphology was observed using light microscopy (BH-2; Olympus) and scanning electron microscopy (QUANTA200; FEI). Colours of colonies were determined by using colour chips from the ISCC-NBS colour charts standard (Kelly 1964). Cell motility was tested by monitoring the degree of turbidity on motility test medium as described by MacFaddin (1980). Growth at different temperatures (0, 5, 10, 15, 20, 25, 30, 37, 40, 45, 50, 55 and 60 °C), different NaCl concentrations (0–10 %, w/v) (at intervals of 1 %) was tested on ISP 2 medium at 30 °C for 14 days. The pH range (4.0–12.0 at intervals of 1.0 pH units) for growth was tested in ISP 2 liquid medium with the pH adjusted as described by Xu et al. (2005). Catalase and oxidase activities were determined in 3 % (v/v) H2O2 and 1 % (w/v) tetramethyl-p-phenylenediamine, respectively. Decomposition of test substances was performed by using the media and methods of Gonzalez et al. (1978). H2S production was tested as described by Lee et al. (2001). Other biochemical tests including methyl red and indole tests were detected according to Goodfellow (1986). Nutritional features were determined using Biolog GN III MicroPlate according to manufacturer’s instructions and the reactions were observed after incubating the plates at 30 °C for 12–48 h. Enzyme activities were examined using the API ZYM kit (bioMérieux) following the manufacturer’s instructions. Other physiological and biochemical tests were performed using API 20NE and API 50CH.
O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T, which were used as reference strains for phenotypic characterization, fatty acid and DNA–DNA hybridization, were obtained from the JCM (Japan) and the Deutsche Sammlung von Mikroorganismen and Zellkulturen (DSMZ, Germany).
Chemotaxonomy
Cell biomass of strain EGI 650022T for DNA extraction and for the analysis of menaquinones (MKs), cell wall peptidoglycan and polar lipids were obtained from cultures grown on ISP 2 medium at 30 °C for 5 days. MKs were extracted and analysed as described previously (Collins et al. 1977; Kroppenstedt 1982), using reversed-phase HPLC with an Agilent ZORBAX Eclipse XDB-C18 (150 × 4.6 mm) column. A purified cell-wall preparation was obtained and hydrolysed as described by Kim and Lee (2011). Amino acids in cell-wall hydrolysates were analysed by using precolumn derivatization with o-phthalaldehyde (OPA) by using HPLC as described by Tang et al. (2009). Polar lipids were extracted and separated by two-dimensional TLC following the method of Minnikin et al. (1984), and identified by spraying the plates with 10 % ethanolic molybdophosphoric acid, molybdenum blue, ninhydrin and α-naphthol reagents (Minnikin et al. 1984; Komagata and Suzuki 1987) and with Dragendorff’s reagent (Sigma). For cellular fatty acid analysis, cell biomass of strain EGI 650022T, O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T was harvested from TSA plates [tryptic soy agar (soybean-casein digest agar medium); BD] at 30 °C for 5 days. Cellular fatty acids analysis was performed as described by Sasser (1990) according to the standard protocol of the MIDI/Hewlett Packard microbial identification system (Sherlock Version 6.1; MIDI database TSBA6). For identification of the mycolic acids present in the cell wall, one-dimensional TLC was carried out following the standard procedure by Minnikin et al. (1975).
Molecular analysis
Extraction of chromosomal DNA and the amplification of the 16S rRNA gene by PCR were performed as described by Li et al. (2007). The DNA G+C content of strain EGI 650022T was determined by the method of Mesbah et al. (1989). The 16S rRNA gene was sequenced at Sangon Biotech (Shanghai) Co., Ltd. (http://www.sangon.com), and was identified using the Eztaxon-e server database (Kim et al. 2012). Alignment of sequences was carried out with CLUSTAL X 1.83 software (Thompson et al. 1997). Phylogenetic tree was constructed according to the neighbor-joining (Saitou and Nei 1987) methods by using MEGA version 5.0 (Tamura et al. 2011). Evolutionary distances were computed using the Kimura two-parameter method (Kimura 1983). Bootstrap analysis was used to evaluate reliability of tree topology of the neighbor-joining data by performing 1,000 replications (Felsenstein 1985). DNA–DNA hybridization tests were carried out by the fluorometric micro-well method (Ezaki et al. 1989; Christensen et al. 2000). Hybridization was performed with five replications for each example, and the two extreme values (highest and lowest) for each sample were excluded. DNA–DNA relatedness values are expressed by calculating the means of the remaining three values. The hybridization temperature was 46 °C.
Results and discussion
Phenotypic characteristics
Strain EGI 650022T was found to be Gram-staining positive, aerobic, non-spore-forming, non-motile and irregular rod-shaped (Supplementary Fig. S2). Colonies on ISP 2 medium were observed to be smooth, circular, convex, translucent and white-coloured. Growth of strain EGI 650022T was observed at 5–45 °C, pH 6.0–9.0 and 0–7 % NaCl (w/v), with optimal growth at 25–30 °C, 2–4 % NaCl (w/v) and pH 7.0–8.0. In contrast to strains O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T, EGI 6500322T tolerated wider temperature range (5–45 °C) and higher salt concentration (up to 7 % NaCl), which distinguished the novel strains from its closest related reference strains. The strain was oxidase-positive, which is the same as the type strain of O. fritillariae JCM 12284T and significantly different to the type strains of P. flavus DSM 14012T and P. auratus DSM 19586T. The main characteristics that differentiate strain EGI 650022T from species of the genera Okibacterium and Plantibacter are listed in Table 1.
Chemotaxonomic characteristics
The predominant MKs detected in strain EGI 650022T were determined to be MK-12 (51.6 %), MK-11 (22.0 %), MK-12(H4) (16.0 %) and MK-10 (10.4 %). The amino acids in the peptidoglycan layer of strain EGI 650022T comprised glutamic acid, glycine, alanine, homoserine and lysine. The peptidoglycan type of EGI 650022T was similar to those of the type strain of O. fritillariae JCM 12284T, but differs significantly compared with other Plantibacter species; l-2,4-diaminobutyric acid was not detected. Mycolic acids were absent. The polar lipids detected in strain EGI 650022T were diphosphatidylglycerol, phosphatidylglycerol, two unidentified glycolipids and two unidentified phospholipids (Supplementary Fig. S1). The major fatty acid profile mainly consisted of (>10 % of the total fatty acids) anteiso-C15:0 (37.5 %), anteiso-C17:0 (36.7 %) and Sum in Feature 4 (iso-C17:1 and/or anteiso-C17:1; 10.4 %). Qualitative and quantitative differences were noted between the cellular fatty acid composition and proportion of strain EGI 650022T and its closest phylogenetic neighbours (Supplementary Table S1). In comparison with strains P. flavus DSM 14012T and P. auratus DSM 19586T, strains EGI 650022T and O. fritillariae JCM 12284T contained larger amounts of anteiso-C17:1, and also showed smaller amounts of anteiso-C15:0. However, strain EGI 650022T could also be distinguished from O. fritillariae JCM 12284T by the presence of anteiso-C17:1 A.
Phylogenetic analysis
The DNA G+C content of strain EGI 650022T was determined to be 66.0 mol% which is similar to the range of the genus Okibacterium (66.6–67.2 %; Evtushenko et al. 2002), but a value lower than those reported for Plantibacter species (68.0–70.0 %; Behrendt et al. 2002; Lin and Yokota 2006). The almost complete 16S rRNA gene sequence of strain EGI 650022T determined in this study (Gen Bank accession number KM114213) comprised 1,529 nucleotides. The results from the EzTaxon server (http://www.eztaxon.org/) indicated that the closest phylogenetic neighbours were O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T, which showed 98.0, 97.5 and 97.2 % of 16S rRNA gene sequence similarities to strain EGI 650022T, respectively. Analysis based on the neighbor-joining method showed that strain EGI 650022T grouped with the type species of the genus Okibacterium, and formed a cluster with O. fritillariae DSM 12584T, P. flavus DSM 14012T and P. auratus DSM 19586T (Fig. 1). Furthermore, strains O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T were selected as the representative strains to perform DNA–DNA hybridization studies. The experiments showed that DNA–DNA relatedness values with O. fritillariae JCM 12284T, P. flavus DSM 14012T and P. auratus DSM 19586T were 39.7, 19.7 and 22.0 %, respectively (Supplementary Table S2), which is significantly less than 70 % cut-off point according to the criterion recommended for the delineation of bacterial species by Stackebrandt and Goebel (1994).
Conclusion
The results obtained from the chemotaxonomic and phylogenetic analysis clearly indicate that strain EGI 650022T is a member of the genus Okibacterium (Table 1; Fig. 1). Strain EGI 650022T could be distinguished from the type strain of O. fritillariae by differences in phenotypic and chemotaxonomic features, including colony color, decomposition of starch and Tween 60, H2S production, nitrate reduction, utilization and product acid of some substrates, activity of some enzymes and susceptibility to some chemical substrates, MKs composition (Table 1). These differences, in combination with the phylogenetic analysis and DNA–DNA hybridization data of strain EGI 650022T, suggest that the novel strain EGI 650022T is differentiated from the type strain of the O. fritillariae. Therefore, strain EGI 650022T is considered to represent a new species of the genus Okibacterium, for which the name Okibacterium endophyticum sp. nov. is proposed.
Description of O. endophyticum sp. nov.
Okibacterium endophyticum (en.do.phy’ti.cum. Gr. pref. endo, within; Gr. n. phyton, plant; L. fem. suff. –icum, adjectival suffix used with the sense of belonging to; N.L. neut. adj. endophyticum, within plant, endophytic, pertaining to the original isolation from plant tissues).
Cells are Gram-stain positive, catalase-positive, oxidase-positive, aerobic and non-motile, irregular rods (length 0.78–1.25 µm, width 0.27–0.29 µm). Colonies grown on ISP 2 agar are white-colored, convex and round. Growth occurs at 5–45 °C on ISP 2 medium, with optimum growth occurring at 25–30 °C. Cells grow in the presence of 0–7 % (w/v) NaCl (optimum 2–4 %) and at pH 6.0–9.0 (optimum pH 7.0–8.0). Hydrolyses Tween 20, Tween 40 and Tween 60, but not starch, casein and Tween 80. H2S production and methyl red test are negative. In the API 20NE test system, positive for aesculin hydrolysis, nitrate reduction, assimilation of arabinose, glucose, malic acid, maltose, mannitol and mannose, but negative for arginine dihydrolase, β-galactosidase, gelatin hydrolysis, glucose fermentation, indole production, urease and assimilation of N-acetyl-glucosamine, adipic acid, capric acid, citrate, phenylacetic acid and potassium gluconate. In the API ZYM test system, positive for acid phosphatase, cystine arylamidase, esterase (C4), esterase lipase (C8), α-galactosidase, α-glucosidase, β-glucosidase, leucine arylamidase, naphthol-AS-BI-phosphohydrolase and valine arylamidase, but negative for N-acetyl-β-glucosaminidase, alkaline phosphatase, chymotrypsin, β-fucosidase, β-glucuronidase, lipase (C14), α-mannosidase and trypsin. In the API 50CH test system, acid is produced only from aesculin, l-arabinose, cellobiose, fructose, d-glucose, maltose, d-mannose, melibiose, l-rhamnose, d-ribose, salicin, d-sorbitol, l-sorbose, sucrose, trehalose and d-xylose. According to the Microplates (Biolog GN III) assay, positive for acetic acid, N-acetyl-d-glucosamine, l-alanine, l-arginine, l-aspartic acid, aztreonam, d-cellobiose, formic acid, d-fructose, d-fructose-6-phosphate, d-fucose, l-fucose, d-galactose, l-galactonic acid lactone, d-galacturonic acid, gelatin, d-gluconic acid, d-glucose-6-phosphate, l-glutamic acid, glycerol, glycyl-l-proline, α-hydroxy-butyric acid, β-hydroxy-d,l-butyric acid, p-hydroxy-phenylacetic acid, inosine, α-keto-butyric acid, α-keto-glutaric acid, l-lactic acid, d-lactic acid methyl ester, lithium chloride, d-melibiose, methyl pyruvate, β-methyl-d-glucoside, nalidixic acid, pectin, potassium tellurite, propionic acid, d-raffinose, l-rhamnose, rifamycin sv, d-salicin, l-serine, sodium butyrate, 1 % sodium lactate, d-sorbitol, stachyose, sucrose,d-trehalose, troleandomycin and Tween 40; but negative for acetoacetic acid, N-acetyl-d-galactosamine, N-acetyl-β-d-mannosamine, N-acetyl neuraminic acid, γ-amino-butyric acid, d-arabitol, d-aspartic acid, bromo-succinic acid, dextrin, fusidic acid, gentiobiose, glucuronamide, d-glucuronic acid, guanidine HCl, l-histidine, myo-inositol, α-d-lactose, lincomycin, 3-methyl glucose, minocycline, mucic acid, niaproof 4, l-pyroglutamic acid, quinic acid, d-saccharic acid, d-serine, sodium bromate, tetrazolium blue, tetrazolium violet, d-turanose and vancomycin. The major cellular fatty acids are anteiso-C15:0 and anteiso-C17:0. The predominant MKs are MK-12, MK-11 and MK-12 (H4). The cell wall peptidoglycan contains the amino acids glutamic acid, glycine, alanine, homoserine and lysine. The polar lipid profile comprises diphosphatidylglycerol, phosphatidylglycerol, two unidentified glycolipids and two unidentified phospholipids. The G+C content of the genomic DNA of the type strain is 66.0 mol%.
The type strain, EGI 650022T (=KCTC 29492T = JCM 30086T), was isolated from the roots of a healthy halophyte sample of S. affinis C. A. Mey, collected from suburban district of Urumqi, Xinjiang province, north-west China. The 16S rRNA gene sequence of strain EGI 650022T has been deposited in GenBank under the accession number KM114213.
References
Behrendt U, Ulrich A, Schumann P, Naumann D, Suzuki K-l (2002) Diversity of grass-associated Microbacteriaceae isolated from the phyllosphere and litter layer after mulching the sward; polyphasic characterization of Subtercola pratensis sp. nov., Curtobacterium herbarum sp. nov. and Plantibacter flavus gen. nov., sp. nov. Int J Syst Evol Microbiol 52:1441–1454
Christensen H, Angen O, Mutters R, Olsen JE, Bisgaard M (2000) DNA–DNA hybridization determined in micro-wells using covalent attachment of DNA. Int J Syst Evol Microbiol 50:1095–1102
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and Corynebacteria. J Gen Microbiol 100:221–230
Evtushenko LL, Dorofeeva LV, Krausova VL, Gavrish EY, Yashina SG, Takeuchi M (2002) Okibacterium fritillariae gen. nov., sp. nov., a novel genus of the family Microbacteriaceae. Int J Syst Evol Microbiol 52:987–993
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid–deoxyribonucleic acid hybridization in micro-dilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gonzalez C, Gutierrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov., an amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24:710–715
Goodfellow M (1986) Genus Rhodococcus Zopf 1891, 28 AL. In: Sneath PHA, Mair NS, Sharpe NE, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 2. Williams and Wilkins, Baltimore, pp 1472–1481
Kelly KL (1964) Color-name charts illustrated with centroid colors. Inter-Society Color Council-National Bureau of Standards, Chicago (Published in US)
Kim SJ, Lee SS (2011) Amnibacterium kyonggiense gen. nov., sp. nov., a new member of the family Microbacteriaceae. Int J Syst Evol Microbiol 61:155–159
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Komagata K, Suzuki K-I (1987) Lipid and cell-wall analysis in bacterial systematics. In: Colwell RR, Grigorova R (eds) Methods in microbiology. Academic Press, Orlando
Kroppenstedt RM (1982) Separation of bacterial menaquinones by HPLC using reverse phase (RP18) and a silver loaded ion exchanger as stationary phases. J Liq Chromatogr 5:2359–2387
Lee KC, Kim KK, Eom MK, Kim MJ, Lee JS (2001) Fontibacillus panacisegetis sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol 61:369–374
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China) and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Lin YC, Yokota A (2006) Plantibacter auratus sp. nov., in the family Microbacteriaceae. Int J Syst Evol Microbiol 56:2337–2339
MacFaddin JF (1980) Biochemical tests for identification of medical bacteria, 2nd edn. Williams and Wilkins, Baltimore
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, Alshamaony L, Goodfellow M (1975) Differentiation of Mycobacterium, Nocardia, and related taxa by thin-layer chromatographic analysis of whole-organism methanolysates. J Gen Microbiol 88:200–204
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal K, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Qin S, Wang HB, Chen HH, Zhang YQ, Jiang CL, Xu LH, Li WJ (2008) Glycomyces endophyticus sp. nov., an endophytic actinomycete isolated from the root of Carex baccans Nees. Int J Syst Evol Microbiol 58:2525–2528
Qin S, Li J, Chen HH, Zhao GZ, Zhu WY, Jiang CL, Xu LH, Li WJ (2009) Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl Environ Microbiol 75:6176–6186
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic tree. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. Microbial ID, Inc., Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Tang SK, Wang Y, Chen Y, Lou K, Cao LL, Xu LH, Li WJ (2009) Zhihengliuella alba sp. nov., and emended description of the genus Zhihengliuella. Int J Syst Evol Microbiol 59:2025–2033
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Xu P, Li WJ, Tang SK, Zhang YQ, Chen GZ, Chen HH, Xu LH, Jiang CL (2005) Naxibacter alkalitolerans gen. nov., sp. nov., a novel member of the family ‘Oxalobacteraceae’ isolated from China. Int J Syst Evol Microbiol 55:1149–1153
Acknowledgments
The authors are grateful to Prof. Dr. Aharon Oren (The Hebrew University of Jerusalem, Israel) for his kind help with the Latin etymology for the new species, Prof. Hans-Peter Klenk (DSMZ, Germany) and Prof. Takuji Kudo (JCM, Japan) for their kindly providing reference type strains. This research was supported by the National Natural Science Foundation of China (Nos. 81102806, 31200008 and 31400009). The Hundred Talents Program of Chinese Academy of Sciences, the High-level Talents Program of Xinjiang Autonomous Region and the West Light Foundation of Chinese Academy of Sciences. WNH and W-J L extend their appreciation to the Deanship of Scientific Research at King Saud University for funding the work through the research group project no. RGP-205. W-J Li was also supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, HF., Zhang, YG., Li, L. et al. Okibacterium endophyticum sp. nov., a novel endophytic actinobacterium isolated from roots of Salsola affinis C. A. Mey. Antonie van Leeuwenhoek 107, 835–843 (2015). https://doi.org/10.1007/s10482-014-0376-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0376-0