Abstract
Hundung Limestone habitat, Manipur, India is an unexplored site for microbial diversity studies. Using polyphasic taxonomy, a Streptomyces strain, MBRL 172T, has been characterized. The strain was found to show highest 16S rRNA gene sequence similarity with Streptomyces coeruleofuscus NBRC 12757T (99.2 %). The DNA relatedness between MBRL 172T and S. coeruleofuscus NBRC 12757T, and between MBRL 172T and Streptomyces nogalater NBRC 13445T, were 36.8 ± 4.4 and 52.5 ± 2.7 %, respectively. Strain MBRL 172T was found to contain ll-diaminopimelic acid as the diagnostic diamino acid and glucose, mannose and xylose as the major sugars in whole cell hydrolysates. The polar lipids in the cell membrane were identified as diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, phosphatidylinositol and phosphatidylinositolmannoside. The predominant menaquinones detected were MK-9(H6) and MK-9(H8). The cellular fatty acids identified were mainly saturated fatty acids: anteiso-C15:0, iso-C16:0 and iso-C15:0. Based on differences in the biochemical and molecular characteristics from its closest relatives, the strain can be proposed to represent a novel taxon in the genus Streptomyces, for which the name Streptomyces canchipurensis is proposed, with the type strain MBRL 172T (=JCM 17575T = KCTC 29105T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Limestone habitats usually have high deposits of CaCO3 salts and therefore may be considered as unique habitats. Although microbes have been reported to play a major role in the formation of caves by dissolution of limestone and other calcareous rocks, and through extracellular precipitation of calcium carbonate (Danielli and Edington 1983; Engel et al. 2001; Riding 2000), relatively few studies have been performed on microbial diversity, including actinobacterial diversity, in limestone habitats (Groth et al. 1999a; Jurado et al. 2009; Kim et al. 1998; Nakaew et al. 2009; Niyomvong et al. 2012). To date, three new genera, Beutenbergia, Hoyosella and Knoellia, have been reported from limestone habitats and related limestone ecosystems, such as cave biofilms (Groth et al. 1999b, 2002; Jurado et al. 2009).
The genus Streptomyces is the currently most diverse known group of bacteria with more than 650 species reported (http://www.bacterio.net/streptomycesa.html; Parte 2014). Streptomyces species have been found to have diverse roles in nature, from being antibiotic producers to being pathogens (Keiser et al. 2000). The genus is characterized by high G+C content in DNA, formation of extensively branched substrate and aerial mycelia, presence of LL-diaminopimelic acid (LL-DAP) and absence of characteristic sugars in the cell wall (cell wall type I) (Kämpfer 2012). The present study reports the polyphasic characterization of a novel strain isolated from a limestone environment, MBRL 172T, which is proposed as representative of a novel species of the genus Streptomyces.
Materials and methods
Strain and culture conditions
Strain MBRL 172T was isolated from a soil sample collected from a limestone quarry site at Hundung, Manipur, India (25.05°N, 94.33°E). Isolation was performed on Gauze’s Medium No. 1 using a procedure described earlier (Nimaichand et al. 2012). Strain MRBL 172T was preserved as lyophilized spore suspensions in skim milk at room temperature and as glycerol suspensions (20 %, v/v) at −80 °C.
The related type strains Streptomyces coeruleofuscus NBRC 12757T and Streptomyces nogalater NBRC 13445T were obtained from Biological Resource Center (NBRC), National Institute of Technology and Evaluation, Japan and cultured under comparable conditions as reference strains.
Phenotypic characterization
To observe its morphological characteristics, strain MBRL 172T was cultivated aerobically in Gauze’s Medium No. 1 (28 °C) for 2 weeks. Morphology of spores and mycelia was observed by light microscopy (Olympus BH2) and scanning electron microscopy (SEM) (Quanta 200, FEI) (Williams and Davies 1967). Growth on various International Streptomyces Project media (ISP, Shirling and Gottlieb 1966), Tryptic Soy Agar (TSA, Difco), Starch Casein Nitrate Agar (SCNA), Gauze’s Medium No. 1, Czapek’s Dox agar and Nutrient agar (NA) were observed. The colony colour was determined using the ISCC-NBS colour chart (Kelly 1964).
Utilization of sole carbon and nitrogen sources was determined as described by Shirling and Gottlieb (1966). Tests for decomposition of casein and acid production from carbohydrates were performed following the methods of Gordon et al. (1974). Hydrolysis of gelatin and starch was determined as described by Collins et al. (2004) and that of Tweens 20, 40, 60 and 80 according to Sierra (1957). Growth at different temperatures (5, 15, 20, 28, 37, 42, 50 and 60 °C), pH (4, 5, 6, 7, 8, 9 and 10) and NaCl concentrations (0, 2, 5, 7 and 10 % w/v) was determined on TSA as described by Goodfellow (1986). Catalase activity was observed by assessing bubble production in 3 % (v/v) H2O2 and oxidase activity was determined by using a 1 % (w/v) solution of tetramethyl-p-phenylenediamine (Kovacs 1956). Other biochemical tests including Voges-Proskauer, methyl red and indole tests were performed according to Goodfellow (1986).
Chemotaxonomy
The amino acid content of the cell wall was determined according to Staneck and Roberts (1974) and the sugars in the whole cell hydrolysates were analyzed as described by Tang et al. (2009). For other chemotaxonomic analyses, cell biomass from a two week old culture on Tryptic Soy Broth (TSB, Difco) was harvested by centrifugation, washed with distilled water and lyophilized. Polar lipids were extracted and analyzed by two-dimensional TLC as described by (Minnikin et al. 1984). The extraction of menaquinones was performed according to Collins et al. (1977) and analyzed by HPLC (Tamaoka et al. 1983). Cellular fatty acids were extracted, methylated and analyzed by using Sherlock Microbial Identification System (MIDI) according to the method of Sasser (1990) and the manufacturer’s instructions. Fatty acid methyl esters were analysed by GC (Agilent Technologies 7890A GC System) and identified using the Microbial Identification Software Package (Sherlock Version 6.1; MIDI database: TSBA6).
Molecular analysis
Genomic DNA extraction and PCR amplification of the 16S rRNA gene was performed as described by Li et al. (2007) with a slight modification. The strain was given a brief ultrasonic wave shock (53 kHz, 30 s) prior to lysozyme treatment. The almost complete 16S rRNA gene sequence (1,534 bp) of the strain was identified using the EzTaxon-e server database (Kim et al. 2012) and aligned with the 16S rRNA gene sequences of other Streptomyces species using CLUSTAL X version 2.1 (Larkin et al. 2007). Phylogenetic analyses were performed using the software package MEGA version 6.0 (Tamura et al. 2013). Phylogenetic distances were calculated with the Kimura two-parameter model (Kimura 1983) and tree topologies were inferred using the maximum-likelihood (Felsenstein 1981), maximum-parsimony (Fitch 1971) and neighbour-joining (Saitou and Nei 1987) methods. To determine the support of each clade, bootstrap analysis was performed with 1,000 resamplings (Felsenstein 1985).
The G + C content of the genomic DNA prepared by the method of Marmur (1961) was determined as described by Mesbah et al. (1989). DNA–DNA relatedness was studied by a fluorimetric method (Ezaki et al. 1988, 1989). One of the two DNA molecules for hybridization was labeled while the other was immobilized and vice versa. Six replications were done for each sample and the two extreme values (highest and lowest) for each sample were excluded. The relatedness values are expressed by calculating the means of the remaining four values and the DNA–DNA hybridization was taken from the two means of relatedness values.
Results and discussion
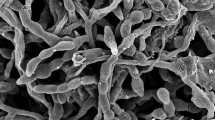
Strain MBRL 172T was observed to form extensive substrate and white coloured aerial mycelia on Gauze’s Medium No. 1. The strain was observed to form spiralis spore chains with warty spores (approximately 50 spores per chain, each spore measuring ~600 × 900 nm in dimensions, Fig. 1). The strain was found to grow well in all the media tested and formed brown and blue diffusible pigments on ISP 7 and Gauze’s Medium No. 1, respectively (Table 1). The differentiating properties of strain MBRL 172T relative to the related type strains Streptomyces coeruleofuscus NBRC 12757T and Streptomyces nogalater NBRC 13445T are listed in Table 2 and the detailed phenotypic characteristics are mentioned in the species description.
Strain MBRL 172T was found to have LL-DAP as the diagnostic cell wall diamino acid and xylose (61 %), glucose (23 %) and mannose (13 %) were detected as the major sugars in whole cell hydrolysates, along with small amounts of ribose (4 %). The major polar lipids detected were diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositolmannoside, along with other unknown phospholipids and lipids (see Supplementary Fig. S1). MK-9(H8) (47.5 %) and MK-9(H6) (41.2 %) were identified as the predominant menaquinones, along with small amounts of MK-9(H4) (9.8 %) and MK-9(H2) (1.5 %). The fatty acid methyl ester profile (>1 %) was found to contain anteiso-C15:0 (19.7 %), iso-C16:0 (19.5 %), iso-C15:0 (14.9 %), anteiso-C17:0 (8.5 %), Summed Feature 9 (as defined by MIDI) containing iso-C17:1ω9c and/or 10-methyl C16:0 (6.5 %), iso-C16:1- H (5.9 %), anteiso-C17:1ω9c (5.8 %), C16:0 (4.8 %), iso-C17:0 (4.2 %), Summed Feature 3 containing C16:1ω7c and/or C16:1ω6c (3.6 %) and iso-C14:0 (3.1 %). The major fatty acids in S. coeruleofuscus NBRC 12757T are unsaturated fatty acids iso-C15:0 (26.0 %), iso-C16:0 (24.8 %) and anteiso-C15:0 (15.7 %), and in the case of S. nogalater NBRC 13445T are iso-C15:0 (34.4 %), anteiso-C15:0 (32.3 %) and anteiso-C17:0 (14.9 %). The fatty acids C16:0 (7.6 %) and iso-C14:0 (5.3 %) in S. coeruleofuscus NBRC 12757T were not detected or detected in lesser quantities (<5 %) in the other two strains, indicating the variability in the fatty acids profile (Table 2).
The G + C content of the genomic DNA of strain MBRL 172T was found to be 69.4 %. Based on the 16S rRNA gene sequence similarities and the neighbour-joining tree (Fig. 2), the strain forms a clade with S. coeruleofuscus NBRC 12757T (99.2 %; 12/1462 nucleotide differences), Streptomyces chromofuscus NBRC 12851T (99.0 %; 15/1460), Streptomyces cinereospinus NBRC 15397T (98.8 %; 17/1462), S. nogalater JCM 4799T (98.6 %; 21/1469) and Streptomyces pluripotens MUSC 135T (98.5 %; 22/1483). This indicated that the strain belongs to clade 114 as defined by Labeda et al. (2012). The maximum-likelihood tree and maximum-parsimony tree are shown in Supplementary Fig. S2 and Fig. S3, respectively. As Streptomyces strains sharing more than 99.5 % 16S rRNA gene sequence similarities with their closest homologues have been reported among novel species (Bouchek-Mechiche et al. 2000; Dastager et al. 2007; Goodfellow et al. 2007; Nimaichand et al. 2012, 2013), strains S. coeruleofuscus NBRC 12757T and S. nogalater NBRC 13445T were selected as the representative strains to perform DNA–DNA hybridization studies. The experiments showed that DNA–DNA relatedness values with S. coeruleofuscus NBRC 12757T and S. nogalater NBRC 13445T (Table 3) are well below the 70 % cut off point for species identification (Wayne et al. 1987).
Neighbour-joining tree, based on 16S rRNA gene sequences, showing the relationships between strain MBRL 172T and other type strains of the genus Streptomyces. Hash and asterisks indicate the branches that were also recovered in the maximum-likelihood analysis, and both maximum-likelihood and maximum parsimony trees, respectively. Numbers at nodes are levels of bootstrap support (%) for branch points (1,000 resamplings). Bar 0.002 substitutions per nucleotide position
The chemotaxonomic data and the phylogenetic analysis clearly indicates the affiliation of strain MBRL 172T to the genus Streptomyces. The genotypic and phenotypic features suggest that strain MBRL 172T could be clearly distinguished from its closest phylogenetic relatives. Besides low DNA–DNA relatedness values, the strain is also distinguished from S. coeruleofuscus NBRC 12757T and S. nogalater NBRC 13445T by several phenotypic properties as listed in Table 2. Therefore, strain MBRL 172T, isolated from Hundung, is considered to represent a new species of the genus Streptomyces, for which the name Streptomyces canchipurensis sp. nov. is proposed.
Description of Streptomyces canchipurensis sp. nov
Streptomyces canchipurensis (can.chi.pur.en´sis. N.L. masc. adj. canchipurensis of or belonging to Canchipur, Manipur University, India, where the MBRL research group which isolated the type strain is located).
Gram-stain positive, aerobic, with Spiralis spore chains containing up to 50 spores per chain. Each spore on maturity measures ~600 × 900 nm. Growth occurs at 20–37 °C and pH 6–9, with optimum growth at 28 °C and pH 7. Growth occurs in presence of up to 2 % NaCl. Able to utilise dulcitol, galactose, inositol, inulin, lactose, maltose, mannose, melibiose, raffinose, sorbitol and sucrose as sole carbon sources; and l-alanine, l-arginine, l-asparagine, l-glutamine, l-hydroxyproline, l-leucine, l-lysine, dl-methionine, l-tryptophan, l-tyrosine and l-valine as sole nitrogen sources. Does not utilise arabinose, cellobiose, salicin and l-cysteine as either sole carbon or nitrogen sources. Acid is produced from lactose, maltose and sucrose but not from mannose. Hydrolyzes starch and Tweens 20, 40, 60 and 80 but not casein, gelatin and urea. Positive in catalase, oxidase and citrate utilization tests but negative in methyl red, Voges-Proskauer and indole production tests. Contains LL-diaminopimelic acid, xylose, glucose and mannose with small amount of ribose in the whole cell hydrolysates. MK-9(H6) and MK-9(H8) are the predominant menaquinones, while the polar lipids consist of diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol and phosphatidylinositolmannoside along with other unknown phospholipids and lipids. The fatty acid profile (>1 %) contains anteiso-C15:0, iso-C16:0, iso-C15:0, anteiso-C17:0, Summed Feature 9 containing iso-C17:1ω9c and/or 10-methyl C16:0; iso-C16:1- H, anteiso-C17:1ω9c, C16:0, iso-C17:0, Summed Feature 3 containing C16:1ω7c and/or C16:1ω6c and iso-C14:0. The G + C content of the genomic DNA of the type strain is 69.4 %.
The type strain MBRL 172T (=JCM 17575T = KCTC 29105T) was isolated from a limestone quarry site at Hundung, Manipur, India. The 16S rRNA gene sequence of strain MBRL 172T has been deposited in GenBank under the accession number JN560154. The strain belongs to clade 114 of the genus Streptomyces as defined by Labeda et al. (2012).
References
Bouchek-Mechiche K, Gardan L, Normand P, Jouan B (2000) DNA relatedness among strains of Streptomyces pathogenic to potato in France: description of three new species, S. europaeiscabiei sp. nov. and S. stelliscabiei sp. nov. associated with common scab, and S. reticuliscabiei sp. nov. associated with netted scab. Int J Syst Evol Microbiol 50:91–99
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Collins CH, Lyne PM, Grange JM, Falkinham JO (2004) Microbiological methods, 8th edn. Arnold, London
Danielli HMC, Edington MA (1983) Bacterial calcification in limestone caves. Geomicrobiol J 3:1–16
Dastager SG, Li WJ, Agasar D, Sulochana MB, Tang ST, Tian XP, Zhi XY (2007) Streptomyces gulbargensis sp. nov., isolated from soil in Karnataka, India. Antonie van Leeuwenhoek 91:99–104
Engel AS, Porter ML, Kinkle BK, Kane TC (2001) Ecological assessment and geological significance of microbial communities from Cesspool Cave, Virginia. Geomicrobiol J 18:259–274
Ezaki T, Hashimoto Y, Takeuchi N, Yamamoto H, Liu SL, Miura H, Matsui K, Yabuuchi E (1988) Simple genetic method to identify viridans group streptococci by colorimetric dot hybridization and fluorometric hybridization in microdilution wells. J Clin Microbiol 26:1708–1713
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fitch WM (1971) Towards defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Goodfellow M (1986) Genus Rhodococcus Zopf 1891, 28AL. In: Sneath PHA, Mair NS, Sharpe NE, Holt JG (eds) Bergey’s manual of systematic bacteriology, vol 2. Williams & Wilkins, Baltimore, pp 1472–1481
Goodfellow M, Kumar Y, Labeda DP, Sembiring L (2007) The Streptomyces violaceusniger clade: a home for streptomycetes with rugose ornamented spores. Antonie Van Leeuwenhoek 92:173–199
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Groth I, Vettermann R, Schuetze B, Schumann P, Saiz-Jimenez D (1999a) Actinomycetes in karstic caves of northern Spain (Altamira and Tito Bustillo). J Microbiol Methods 36:115–122
Groth I, Schumann P, Schuetze B, Augsten K, Kramer I, Stackebrandt E (1999b) Beutenbergia cavernae gen. nov., sp. nov., an l-lysine containing actinomycete isolated from a cave. Int J Syst Bacteriol 49:1733–1740
Groth I, Schumann P, Schuetze B, Augsten K, Stackebrandt E (2002) Knoellia sinensis gen. nov., sp. nov., and Knoellia subterranean sp. nov., two novel actinobacteria isolated from a cave. Int J Syst Evol Microbiol 52:77–84
Jurado V, Kroppenstedt RM, Saiz-Jimenez C, Klenk HP, Mouniee D, Laiz L, Couble A, Potter G, Boiron P, Rodriquez-Nava V (2009) Hoyosella altamirensis gen. nov., sp. nov., a new member of the order Actinomycetales isolated from a cave biofilm. Int J Syst Evol Microbiol 59:3105–3110
Kämpfer P (2012) Genus I. Streptomyces Waksman and Henrici 1943, 339 emend. Witt and Stackebrandt 1990, 370 emend. Wellington, Stackebrandt, Sanders, Wolstrup and Jorgensen 1992, 159. In: Goodfellow M, Kämpfer P, Busse H-J et al (eds) Bergey’s manual of systematic bacteriology, Part B, vol 5, 2nd edn. Springer, New York, pp 1455–1767
Keiser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (2000) In: Practical Streptomyces genetics. The John Innes Foundation, Norwich
Kelly KL (1964) Inter-society color council—national bureau of standards color-name charts illustrated with centroid colors. US Government Printing Office, Washington
Kim BS, Lee JY, Hwang BK (1998) Diversity of actinomycetes antagonistic to plant pathogenic fungi in cave and sea-mud soils of Korea. J Microbiol 36:86–92
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Kimura M (1983) The neutral theory of molecular evolution. Cambridge University Press, Cambridge
Kovacs N (1956) Identification of Pseudomonas pyocyanea by the oxidase reaction. Nature 178:703–770
Labeda DP, Goodfellow M, Brown R, Ward AC, Lanoot B, Vanncanneyt M, Swings J, Kim SB, Liu Z, Chun J, Tamura T, Oguchi A, Kikuchi T, Kikuchi H, Nishii T, Tsuji K, Yamaguchi Y, Tase A, Takahashi M, Sakane T, Suzuki KI, Hatano K (2012) Phylogenetic study of the species within the family Streptomycetaceae. Antonie Van Leeuwenhoek 101:73–104
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Marmur J (1961) A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol 3:208–218
Mesbah M, Premachandran U, Whitman WB (1989) Precise measurement of the G + C content of deoxyribonucleic acid by high-performance liquid chromatography. Int J Syst Bacteriol 39:159–167
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaal A, Parlett JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Nakaew N, Pathom-aree W, Lumyong S (2009) Generic diversity of rare actinomycetes from Thai cave soils and their possible use as new bioactive compounds. Actinomycetol 23:21–26
Nimaichand S, Zhu WY, Yang LL, Ming H, Nie GX, Tang SK, Ningthoujam DS, Li WJ (2012) Streptomyces manipurensis sp. nov., a novel actinobacterium isolated from a limestone deposit site in Manipur, India. Antonie van Leeuwenhoek 102:133–139
Nimaichand S, Tamrihao K, Yang LL, Zhu WY, Zhang YG, Li L, Tang SK, Ningthoujam DS, Li WJ (2013) Streptomyces hundungensis sp. nov., a novel actinomycete with antifungal activity and plant growth promoting traits. J Antibiot 66:205–209
Niyomvong N, Pathom-aree W, Thamchaipenet A, Duangmal K (2012) Actinomycetes from tropical limestone caves. Chiang Mai J Sci 39:373–388
Parte AC (2014) LPSN—list of prokaryotic names with standing in nomenclature. Nucleic Acids Res 42:D613–D616
Riding R (2000) Microbial carbonates: the geological record of calcified bacterial-algal mats and biofilms. Sedimentology 47:179–214
Saitou N, Nei M (1987) The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl 20:16
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Sierra GA (1957) A simple method for the detection of lipolytic activity of micro-organisms and some observations on the influence of the contact between cells and fatty substrates. Antonie Van Leeuwenhoek 23:15–22
Staneck JL, Roberts GD (1974) Simplified approached to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol 54:31–36
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA 6: molecular evolutionary genetics analysis version 6.0. Mol Bio Evol 30:2725–2729
Tang SK, Wang Y, Chen Y, Lou K, Cao LL, Xu LH, Li WJ (2009) Zhihengliuella alba sp. nov., and emended description of the genus Zhihengliuella. Int J Syst Evol Microbiol 59:2025–2031
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, KandlerO Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) International committee on systematic bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematic. Int J Syst Bacteriol 37:463–464
Williams ST, Davies FL (1967) Use of a scanning electron microscope for the examination of actinomycetes. J Gen Microbiol 48:171–177
Acknowledgments
The authors are grateful to Dr. Tomohiko Tamura (NBRC) for providing the reference type strains and to Dollyca Ningombam, Khumukcham Saratchandra Singh and Langam Ibochouba Singh (Manipur University) for their technical assistance. This research was supported by Yunnan Provincial Natural Science Foundation (2013FA004), the Deanship of Scientific Research at King Saud University for funding this work through the research group no RGP-205 and a grant NRF-2006-08790 funded by Ministry of Science, ICT and Future Planning of Korean government. W-J Li was also supported by Guangdong Province Higher Vocational Colleges & Schools Pearl River Scholar Funded Scheme (2014). SN gratefully acknowledges the University Grants Commission (UGC), Government of India, for the award of the Rajiv Gandhi National Fellowship which partly enabled this work. DSN and SN also thankfully acknowledge the financial help and facilities extended by Department of Biotechnology (DBT), Government of India, through the State Biotech Hub (SBTHub) Program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, WJ., Nimaichand, S., Jiang, Z. et al. Streptomyces canchipurensis sp. nov., isolated from a limestone habitat. Antonie van Leeuwenhoek 106, 1119–1126 (2014). https://doi.org/10.1007/s10482-014-0281-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0281-6