Abstract
A novel, red-pigmented, pleomorphic and short rod-shaped haloarchaeon, designated B8T, was isolated from a salt-fermented seafood. Strain B8T was found to be able to grow at 20–45 °C, in the presence of 15–30 % (w/v) NaCl and at pH 7.0–9.0. The optimum requirements were found to be a temperature range of 35–40 °C, pH 8.0 and the presence of 25 % NaCl. The cells of strain B8T were observed to be Gram-staining negative and lysed in distilled water. Anaerobic growth did not occur in the presence of nitrate, l-arginine, dimethyl sulfoxide or trimethylamine N-oxide. The catalase and oxidase activities were found to be positive and nitrate was reduced in aerobic conditions. Tween 20, 40 and 80 were found to be hydrolyzed, whereas casein, gelatin and starch were not hydrolyzed. Indole or H2S was not formed and urease activity was not detected. A phylogenetic analysis based on the 16S rRNA gene sequences indicated that strain B8T is most closely related to members of the genus Halorubrum in the family Halobacteriaceae. Strain B8T was found to have three 16S rRNA genes, rrnA, rrnB and rrnC; similarities between the 16S rRNA gene sequences are 99.0–99.8 %. Strain B8T shared 99.0 % 16S rRNA gene sequence similarity with Halorubrum (Hrr.) lipolyticum JCM 13559T and Hrr. saccharovorum DSM 1137T, 98.8 % with Hrr. kocurii JCM 14978T, 98.3 % with Hrr. lacusprofundi DSM 5036T, 98.0 % with Hrr. arcis JCM 13916T, 97.7 % with Hrr. aidingense JCM 13560T and 97.0 % with Hrr. aquaticum JCM 14031T, as well as 93.7–96.5 % with other type strains in the genus Halorubrum. The RNA polymerase subunit B′ gene sequence similarity of strain B8T with Hrr. kocurii JCM 14978T is 97.2 % and lower with other members of the genus Halorubrum. DNA–DNA hybridization experiments showed that strain B8T shared equal or lower than 50 % relatedness with reference species in the genus Halorubrum. The genomic DNA G+C content of strain B8T was determined to be 64.6 mol%. The major isoprenoid quinone of strain B8T was identified as menaquinone-8 and the major polar lipids as phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, phosphatidylglycerol sulfate, sulfated mannosyl glucosyl diether and an unidentified phospholipid. Based on this polyphasic taxonomic study, strain B8T is considered to represent a new species in the genus Halorubrum, for which the name Hrr. halophilum sp. nov. is proposed. The type strain is B8T (=JCM 18963T = CECT 8278T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High salinity is toxic to most cells but many extremely halophilic archaea, i.e. haloarchaea, have been isolated in hypersaline environments (Grant 2004). Well known hypersaline environments for the isolation of haloarchaea are soda lakes, salt lakes and solar salterns, but some haloarchaea have been isolated from a salt-fermented seafood made from shrimp (Roh et al. 2007a, b, 2009; Roh and Bae 2009). All haloarchaea are classified within the family Halobacteriaceae in the order Halobacteriales of the phylum Euryarchaeota. This family currently contains 40 genera based on the List of Prokaryotic Names with Standing in Nomenclature (Euzéby 1997). In particular, the genus Halorubrum, which was proposed formally by McGenity and Grant (1995), currently includes 25 validly named species: Halorubrum (Hrr.) aidingense (Cui et al. 2006), Hrr. alkaliphilum (Feng et al. 2005), Hrr. aquaticum (Gutiérrez et al. 2011), Hrr. arcis (Xu et al. 2007), Hrr. californiense (Pesenti et al. 2008), Hrr. chaoviator (Mancinelli et al. 2009), Hrr. cibi (Roh and Bae 2009), Hrr. coriense (Oren and Ventosa 1996), Hrr. distributum (Oren and Ventosa 1996), Hrr. ejinorense (Castillo et al. 2007), Hrr. ezzenmoulense (Kharroub et al. 2006), Hrr. kocurii (Gutiérrez et al. 2008), Hrr. lacusprofundi (McGenity and Grant 1995), Hrr. lipolyticum (Cui et al. 2006), Hrr. litoreum (Cui et al. 2007), Hrr. luteum (Hu et al. 2008), Hrr. orientale (Castillo et al. 2006), Hrr. saccharovorum (Tomlinson and Hochstein 1976; McGenity and Grant 1995), Hrr. sodomense (McGenity and Grant 1995), Hrr. tebenquichense (Lizama et al. 2002), Hrr. terrestre (Ventosa et al. 2004), Hrr. tibetense (Fan et al. 2004), Hrr. trapanicum (McGenity and Grant 1995), Hrr. vacuolatum (Mwatha and Grant 1993; Kamekura et al. 1997) and Hrr. xinjiangense (Feng et al. 2004). Cells of the genus Halorubrum are rods or irregular-shaped, motile or non-motile, strictly aerobic and oxidase and catalase-positive. The major polar lipids are phosphatidylglycerol (PG), phosphatidylglycerol phosphate methyl ester (PGP-Me), phosphatidylglycerol sulfate (PGS) and/or sulfated mannosyl glucosyl diether (S-DGD-3) (McGenity and Grant 1995). The G+C content of the genomic DNA is in the range of 61.7–71.2 mol% (McGenity and Grant 2001; Roh and Bae 2009). The present study characterized a new haloarchaeon, strain B8T and determined the taxonomic position of this strain based on phenotypic, phylogenetic and chemotaxonomic analyses, according to the proposed minimal standards for the description of new taxa in the order Halobacteriales (Oren et al. 1997). Based on this polyphasic taxonomic study, strain B8T is considered to represent a new species in the genus Halorubrum, for which the name Hrr. halophilum sp. nov. is proposed here.
Materials and methods
Archaeal strain and culture conditions
A strain, designated B8T, was isolated from a salt-fermented seafood made from shrimp. A sample was serially diluted and spread onto a complex medium (DSM medium no. 954), which was adjusted to pH 7.0 and contained the following (l−1): 5 g casamino acids (BD), 5 g yeast extract (BD), 20 g MgCl2·6H2O, 2 g KCl, 12 g Tris, 0.2 g CaCl2·2H2O, and 200 g NaCl. A solid medium was prepared by adding 2 % (w/v) agar. A single colony was streaked repeatedly to obtain a pure culture at 37 °C. For long-term preservation, strain B8T was frozen at −80 °C in medium 954 supplemented with 5 % (v/v) dimethyl sulfoxide (DMSO). Hrr. lipolyticum JCM 13559T, Hrr. kocurii JCM 14978T, Hrr. saccharovorum DSM 1137T, Hrr. lacusprofundi DSM 5036T Hrr. aidingense JCM 13560T, Hrr. arcis JCM 13916T and Hrr. aquaticum JCM 14031T were obtained from Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ) or Japan Collection of Microorganisms (JCM) and used in the comparative taxonomic analyses.
Morphological, physiological and biochemical characterization
All of the phenotypic tests were performed using medium 954 or a halophile medium (HMD) that contained (l−1): 20 g MgCl2·6H2O, 5 g K2SO4, 0.1 g CaCl2·2H2O, 0.1 g yeast extract, 0.5 g NH4Cl, 0.05 g KH2PO4, 0.5 g casamino acids as a carbon source and 180 g NaCl (Savage et al. 2007), unless indicated otherwise. The cell morphology and size were determined using a transmission electron microscope (SUPRA 55 VP; Carl Zeiss). Gram staining was performed using the published method for haloarchaea (Dussault 1955). Cell lysis in distilled water was tested by incubating cells in distilled water for 1 week before transfer to medium 954. The optimal conditions for growth in medium 954 with various NaCl concentrations were tested using 0–30 % (w/v) NaCl at intervals of 5 %. The optimal pH range for growth was assayed from pH 5.0–11.0 at intervals of 1.0 using HMD with the following buffers: 10 mM 2-(N-morpholino) ethanesulfonic acid for pH 5.0 and 6.0, 10 mM bis–Tris propane for pH 7.0–9.0 and 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid for pH 10.0 and 11.0. Growth was also tested at different temperatures, i.e. 5–60 °C at intervals of 5 °C. The standard phenotypic tests for nitrate and nitrite reduction in aerobic conditions, indole formation, urease activity and the hydrolysis of casein and starch were performed as described by Benson (2002) using medium 954 as the basal medium. The hydrolysis of Tween 20, 40 and 80 were tested as described by González et al. (1978) and the hydrolysis of gelatin was tested according to Smibert and Krieg (1994) using medium 954 as the basal medium. Anaerobic growth was tested in filled, stoppered tubes using medium 954 in the presence of 30 mM nitrate, 5 g l-arginine, 5 g DMSO or 5 g trimethylamine N-oxide (TMAO) at 37 °C in an anaerobic chamber (Coy), where the atmosphere comprised N2/CO2/H2 (90:5:5, by vol.). H2S formation was tested according to Cui et al. (2007). Acid production from d-glucose was tested by growing strain B8T in HMD supplemented with 1 % d-glucose. Methyl red and Voges–Proskauer tests were determined using MR-VP Broth (BD) and Simmon’s citrate test was performed using Simmons citrate agar (DB), for which each medium was supplemented with 20 % (w/v) NaCl. Arginine, lysine and ornithine decarboxylases tests were carried out using Moeller Decarboxylase Broth Base (BD) containing 20 % (w/v) NaCl as the basal medium. To assess the utilization of sole carbon and energy sources, HMD was supplemented with 10 mM bis–Tris propane and 1 % of the following substrates: acetate, l-alanine, l-arginine, L-aspartate, citrate, d-fructose, fumarate, d-galactose, d-glucose, l-glutamate, glycerol, glycine, dl-lactate, lactose, l-lysine, l-malate, maltose, mannitol, d-mannose, l-ornithine, pyruvate, d-ribose, sorbitol, l-sorbose, starch, succinate, sucrose or d-xylose. For testing antibiotic sensitivity, strain B8T was inoculated on agar medium plates using antibiotic discs with the following amounts (μg per disc, unless indicated): ampicillin (20), bacitracin (0.1 IU), chloramphenicol (50), ciprofloxacin (10), erythromycin (25), neomycin (50), norfloxacin (20), novobiocin (50), penicillin G (20 IU) and rifampin (10). The strain was incubated for 2 weeks at 37 °C.
Determination of the 16S rRNA and RNA polymerase subunit B′ (rpoB′) gene sequences and phylogenetic analysis
To obtain chromosomal DNA, cells of strain B8T were harvested, extracted and purified using a DNA extraction kit (G-spin™; iNtRON Biotechnology). The 16S rRNA gene of strain B8T was amplified using a PCR pre-Mix (iNtRON Biotechnology) and the primer set Arch21F and 1492R (DeLong 1992), as described previously (Roh et al. 2008). To check the heterogeneous 16S rRNA gene sequences, PCR products were ligated and transformed using an All-in PCR cloning kit (BioFact) according to the manufacturer’s protocol. Multiple clones were picked randomly and then sequenced. PCR-mediated amplification and sequencing of the rpoB′ genes were carried out according to Minegishi et al. (2010). The almost full-length 16S rRNA and rpoB′ gene sequences were determined using SeqMan (DNASTAR). The phylogenetic neighbours and the pairwise sequence similarities were determined using EzTaxon-e (Kim et al. 2012). The 16S rRNA gene sequences of strain B8T and validly named related species were aligned using the SILVA Incremental Aligner (Pruesse et al. 2012). The rpoB′ gene sequences of the related taxa were obtained from GenBank (http://www.ncbi.nlm.nih.gov) and multiple sequence alignment was performed using the Clustal_W program. The phylogenetic trees were constructed using the neighbor-joining (NJ) (Saitou and Nei 1987), minimum-evolution (ME) (Nei et al. 1998) and maximum-likelihood (ML) (Felsenstein 1981) algorithms based on Kimura’s two-parameter model (Kimura 1980) with 1,000 randomly selected bootstrap replicates using MEGA5 (Tamura et al. 2011).
Determination of the DNA–DNA hybridization (DDH), DNA G+C content, quinones and polar lipid analysis
The DDH experiments were performed using the fluorometric method with photobiotin-labeled DNA probes and microwell plates (MaxiSorp, FluoroNunc), as described by Ezaki et al. (1989). The genomic DNA G+C content was determined as described by González and Saiz-Jimenez (2002).
Polar lipids were extracted and detected using thin layer chromatography on a silica gel 60 F254 plate (Merck), according to the method of Minnikin et al. (1984). The compositions of the polar lipid spots were determined by spraying each plate with specific detection reagents as follows: sulfuric acid–ethanol (1:2, by vol) for total lipids, molybdenum blue for phospholipids and α-naphthol-sulfuric acid for glycolipids. The quinones of strain B8T were analyzed using an HPLC system (UltiMate 3000; Dionex) coupled to a diode array detector and a single quadrupole mass spectrometer (HCT Ion-Trap MS; Bruker).
Results and discussion
Cells of strain B8T were observed to be Gram-stain negative, pleomorphic, short rod or oval shaped, 0.5–0.6 μm in width and 0.9–1.1 μm in length (Supplementary Fig. S1). The colonies were red with a smooth and rounded shape on solid medium. Strain B8T was found to grow in the presence of 15–30 % (w/v) NaCl, at 20–45 °C and at pH 7.0–9.0, with optimum growth in the presence of 25 % NaCl, at 35–40 °C and pH 8.0. The cells of strain B8T lysed in distilled water. Strain B8T was found to reduce nitrate in aerobic conditions and to be positive for acid production from d-glucose, catalase and oxidase. The novel strain was found to be able to hydrolyze Tween 20, 40 and 80, but not casein, gelatin and starch. H2S was not produced from Na2SO3. Indole was not formed and no urease activity was observed. Strain B8T could not grow in anaerobic conditions using nitrate, l-arginine, DMSO or TMAO. The Methyl red, Voges-prokauer and Simmon’s citrate tests were negative. Strain B8T did not produce arginine dihydrolase, lysine decarboxylase or ornithine decarboxylase. Strain B8T was found to be sensitive to novobiocin, bacitracin, erythromycin, rifampin, neomycin and ciprofloxacin, but resistant to penicillin G, ampicillin, chloramphenicol and norfloxacin. Detailed results of the phenotypic tests and the nutritional features of this strain are presented in the species description. Table 1 shows the different characteristics of strain B8T compared with those of closely related type strains of members of the genus Halorubrum, which indicates that strain B8T can be distinguished from closely related members of the genus Halorubrum.
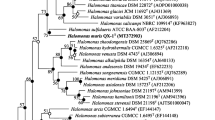
Sequence comparisons indicated that strain B8T has three rRNA genes, rrnA, rrnB and rrnC. The almost complete 16S rRNA of the three rRNA genes and the rpoB′ gene sequences of strain B8T were obtained (1,430, 1,450, 1,450 and 1,830 bp, respectively; GenBank accession numbers EF077637, KF848218, KF848217 and KF700332, respectively). Similarities between the three 16S rRNA gene sequences were 99.0–99.8 %. A comparison with related sequences showed that strain B8T (based on the 16S rRNA rrnA gene sequence) shares the highest levels of similarity with the following strains with validly published names: Hrr. lipolyticum JCM 13559T (99.0 % 16S rRNA gene sequence similarity), Hrr. saccharovorum DSM 1137T (99.0 %), Hrr. kocurii JCM 14978T (98.8 %), Hrr. lacusprofundi DSM 5036T (98.3 %), Hrr. arcis JCM 13916T (98.0 %), Hrr. aidingense JCM 13560T (97.7 %), Hrr. aquaticum JCM 14031T (97.0 %) and other type strains in the genus Halorubrum (93.7–96.5 %). Strain B8T clustered with Hrr. lipolyticum, Hrr. saccharovorum Hrr. kocurii, Hrr. lacusprofundi and Hrr. aidingense in the phylogenetic trees based on the 16S rRNA gene sequences (Fig. 1). The phylogenetic analyses based on the rpoB′ gene sequences showed that strain B8T is closely related to the following members: Hrr. kocurii JCM 14978T (97.2 % rpoB′ gene sequence similarity), Hrr. saccharovorum DSM 1137T (96.8 %), Hrr. lipolyticum JCM 13559T (96.3 %), Hrr. lacusprofundi DSM 5036T (94.9 %), Hrr. arcis JCM 13916T (94.2 %), Hrr. aidingense JCM 13560T (93.2 %) and Hrr. aquaticum JCM 14031T (92.2 %). The phylogenetic trees based on the rpoB′ gene sequences showed a similar topology compared with trees of the 16S rRNA gene (Fig. 1b).
Phylogenetic tree based on the neighbour-joining (NJ) algorithm for the 16S rRNA (a) and rpoB′ (b) gene sequences of strain B8T and closely related taxa. The numbers on the nodes indicate the bootstrap values (>70 %), which were calculated using the NJ/minimum-evolution (ME)/maximum-likelihood (ML) probabilities. The closed circles represent the nodes obtained using both the ME and ML methods, whereas the open circles indicate nodes recovered using either the ME or ML method. Methanococcus vannielii DSM 1224T and Haloquadratum walsbyi DSM 16790 were used as the outgroup for the phylogenetic trees based on the 16S rRNA and rpoB′ gene sequences, respectively. Bar, 0.02 (a) and 0.05 (b) accumulated changes per nucleotide
The DDH values of strain B8T with the type strains of Hrr. lipolyticum JCM 13559T, Hrr. saccharovorum DSM 1137T, Hrr. kocurii JCM 14978T, Hrr. lacusprofundi DSM 5036T, Hrr. arcis JCM 13916T, Hrr. aidingense JCM 13560T and Hrr. aquaticum JCM 14031T were 50 ± 9, 48 ± 11, 23 ± 2, 17 ± 3, 23 ± 4, 12 ± 2 and 14 ± 6 %, respectively. DDH values of <70 % represent distinct species according to current cut-offs employed in prokaryotic systematics (Wayne et al. 1987; Stackebrandt and Goebel 1994), so strain B8T can be considered a distinct genospecies in the genus Halorubrum. The DNA G+C content of strain B8T was determined to be 64.6 mol% (T m), which is similar to the levels found in the closely related strain Hrr. lipolyticum JCM 13559T (65.9 mol%) (Cui et al. 2006). The polar lipids detected in strain B8T comprised PG, PGP-Me, PGS, S-DGD-3 and an unidentified phospholipid (Supplementary Fig. S2). The major polar lipid profile of strain B8T resembles that of the closely related Halorubrum species Hrr. lipolyticum, Hrr. saccharovorum, Hrr. lacusprofundi and Hrr. arcis which contain PG, PGP-Me, PGS and S-DGD-3 (McGenity and Grant 1995; Cui et al. 2006; Xu et al. 2007). However, the presence of PGS and S-DGD-3 in strain B8T help distinguish the strain from Hrr. kocurii and Hrr. aidingense, respectively (Cui et al. 2006; Gutiérrez et al. 2008). The major isoprenoid quinone detected in strain B8T was menaquinone (MK)-8 and minor ones were MK-7(H4) and MK-8(H2).
In conclusion, the results of the phenotypic, phylogenetic and chemotaxonomic analyses showed that the haloarchaeal strain B8T belongs to the genus Halorubrum. Table 1 shows that strain B8T exhibits some differences compared with closely related type strains in the genus Halorubrum. Thus, based on this polyphasic taxonomic study, strain B8T is considered to represent a novel species in the genus Halorubrum, for which the name Hrr. halophilum nov. is proposed.
Description of Halorubrum halophilum sp. nov.
Halorubrum halophilum (ha.lo’phi.lum. Gr. n. hals, halos, salt; Gr. adj. philos, loving; N.L. neut. adj. halophilum salt-loving)
Cells are Gram-stain negative, pleomorphic, short rod or oval shaped and 0.5–0.6 μm in width and 0.9–1.1 μm in length. Colonies are red, smooth and round in shape. Cell lysis occurs in distilled water. Growth occurs in the presence of 15–30 % (w/v) NaCl (optimum, 25 %), at 20–45 °C (optimum, 35–40 °C) and pH 7.0–9.0 (optimum, pH 8.0). Anaerobic growth does not occur in the presence of nitrate, l-arginine, DMSO or TMAO. Cells are positive for nitrate reduction in aerobic conditions, catalase and oxidase activity, acid production from d-glucose and the hydrolysis of Tween 20, 40 and 80, but negative for nitrite reduction in aerobic conditions, indole formation, H2S production, urease activity, Methyl red, Voges–Proskauer, Simmon’s citrate and the hydrolysis of casein, gelatin or starch. Arginine dihydrolase, lysine decarboxylase and ornithine decarboxylase are not produced. Acetate, l-arginine, fumarate, d-galactose, d-glucose, l-glutamate, dl-lactate, d-mannose, l-ornithine, pyruvate and succinate are utilized as sole carbon and energy sources, whereas l-alanine, l-aspartate, citrate, d-fructose, glycerol, glycine, lactose, l-lysine, l-malate, maltose, mannitol, d-ribose, sorbitol, l-sorbose, starch, sucrose and d-xylose are not utilised. The polar lipids are phosphatidylglycerol, phosphatidylglycerol phosphate methyl ester, phosphatidylglycerol sulfate, sulfated mannosyl glucosyl diether and an unidentified phospholipid. The major isoprenoid quinone is MK-8. The DNA G+C content of the type strain is 64.6 mol%.
The type strain, B8T (=JCM 18963T = CECT 8278T), was isolated from a salt-fermented seafood made from shrimp. The GenBank/EMBL/DDBJ accession number for the 16S rRNA rrnA, rrnB and rrnC and rpoB′ gene sequences of strain B8T are EF077637, KF848218, KF848217 and KF700332, respectively.
References
Benson HJ (2002) Microbiological applications: a laboratory manual in general microbiology. McGraw-Hill, New York
Castillo AM, Gutiérrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2006) Halorubrum orientale sp. nov., a halophilic archaeon isolated from Lake Ejinor, Inner Mongolia, China. Int J Syst Evol Microbiol 56(11):2559–2563
Castillo AM, Gutiérrez MC, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2007) Halorubrum ejinorense sp. nov., isolated from Lake Ejinor, Inner Mongolia, China. Int J Syst Evol Microbiol 57(11):2538–2542
Cui HL, Tohty D, Zhou PJ, Liu SJ (2006) Halorubrum lipolyticum sp. nov. and Halorubrum aidingense sp. nov., isolated from two salt lakes in Xin-Jiang. China. Int J Syst Evol Microbiol 56:1631–1634
Cui HL, Lin ZY, Dong Y, Zhou PJ, Liu SJ (2007) Halorubrum litoreum sp. nov., an extremely halophilic archaeon from a solar saltern. Int J Syst Evol Microbiol 57(10):2204–2206
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89(12):5685–5689
Dussault HP (1955) An improved technique for staining red halophilic bacteria. J Bacteriol 70(4):484–485
Euzéby JP (1997) List of Bacterial Names with Standing in Nomenclature: a folder available on the Internet. Int J Syst Bacteriol 47(2):590–592
Ezaki T, Hashimoto H, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Fan H, Xue Y, Ma Y, Ventosa A, Grant WD (2004) Halorubrum tibetense sp. nov., a novel haloalkaliphilic archaeon from Lake Zabuye in Tibet, China. Int J Syst Evol Microbiol 54(4):1213–1216
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17(6):368–376
Feng J, Zhou PJ, Liu SJ (2004) Halorubrum xinjiangense sp. nov., a novel halophile isolated from saline lakes in China. Int J Syst Evol Microbiol 54(5):1789–1791
Feng J, Zhou P, Zhou YG, Liu SJ, Warren-Rhodes K (2005) Halorubrum alkaliphilum sp. nov., a novel haloalkaliphile isolated from a soda lake in Xinjiang, China. Int J Syst Evol Microbiol 55(1):149–152
González JM, Saiz-Jimenez C (2002) A fluorimetric method for the estimation of G+C mol% content in microorganisms by thermal denaturation temperature. Environ Microbiol 4(11):770–773
González C, Gutiérrez C, Ramirez C (1978) Halobacterium vallismortis sp. nov. An amylolytic and carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 24(6):710–715
Grant WD (2004) Life at low water activity. Philos Trans R Soc Lond B 359:1249–1267
Gutiérrez MC, Castillo AM, Pagaling E, Heaphy S, Kamekura M, Xue Y, Ma Y, Cowan DA, Jones BE, Grant WD, Ventosa A (2008) Halorubrum kocurii sp. nov., an archaeon isolated from a saline lake. Int J Syst Evol Microbiol 58(9):2031–2035
Gutiérrez MC, Castillo AM, Corral P, Kamekura M, Ventosa A (2011) Halorubrum aquaticum sp. nov., an archaeon isolated from hypersaline lakes. Int J Syst Evol Microbiol 61(5):1144–1148
Hu L, Pan H, Xue Y, Ventosa A, Cowan DA, Jones BE, Grant WD, Ma Y (2008) Halorubrum luteum sp. nov., isolated from Lake Chagannor, Inner Mongolia, China. Int J Syst Evol Microbiol 58(7):1705–1708
Kamekura M, Dyall-Smith ML, Upasani V, Ventosa A, Kates M (1997) Diversity of alkaliphilic halobacteria: proposals for transfer of Natronobacterium vacuolatum, Natronobacterium magadii, and Natronobacterium pharaonis to Halorubrum, Natrialba, and Natronomonas gen. nov., respectively, as Halorubrum vacuolatum comb. nov., Natrialba magadii comb. nov., and Natronomonas pharaonis comb. nov., respectively. Int J Syst Bacteriol 47(3):853–857
Kharroub K, Quesada T, Ferrer R, Fuentes S, Aguilera M, Boulahrouf A, Ramos-Cormenzana A, Monteoliva-Sánchez M (2006) Halorubrum ezzemoulense sp. nov., a halophilic archaeon isolated from Ezzemoul sabkha, Algeria. Int J Syst Evol Microbiol 56(7):1583–1588
Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62(3):716–721
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Lizama C, Monteoliva-Sánchez M, Suárez-García A, Rosello-Mora R, Aguilera M, Campos V, Ramos-Cormenzana A (2002) Halorubrum tebenquichense sp. nov., a novel halophilic archaeon isolated from the Atacama Saltern, Chile. Int J Syst Evol Microbiol 52(1):149–155
Mancinelli RL, Landheim R, Sánchez-Porro C, Dornmayr-Pfaffenhuemer M, Gruber C, Legat A, Ventosa A, Radax C, Ihara K, White MR, Stan-Lotter H (2009) Halorubrum chaoviator sp. nov., a haloarchaeon isolated from sea salt in Baja California, Mexico, Western Australia and Naxos, Greece. Int J Syst Evol Microbiol 59(8):1908–1913
McGenity TJ, Grant WD (1995) Transfer of Halobacterium saccharovorum, Halobacterium sodomense, Halobacterium trapanicum NRC 34021 and Halobacterium lacusprofundi to the genus Halorubrum gen. nov., as Halorubrum saccharovorum comb. nov., Halorubrum sodomense comb. nov., Halorubrum trapanicum comb. nov., and Halorubrum lacusprofundi comb. nov. Syst Appl Microbiol 18:237–243
McGenity TJ, Grant WD (2001) Genus VII. Halorubrum. In: Boone DR (ed) Bergey’s manual of systematic bacteriology, vol 1, 2nd edn. Springer, New York, pp 320–324
Minegishi H, Kamekura M, Itoh T, Echigo A, Usami R, Hashimoto T (2010) Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B’ (rpoB’) gene. Int J Syst Evol Microbiol 60(10):2398–2408
Minnikin DE, O’Donnell AG, Goodfellow M (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2(5):233–241
Mwatha WE, Grant WD (1993) Natronobacterium vacuolata sp. nov., a haloalkaliphilic archaeon isolated from Lake Magadi, Kenya. Int J Syst Bacteriol 43(3):401–404
Nei M, Kumar S, Takahashi K (1998) The optimization principle in phylogenetic analysis tends to give incorrect topologies when the number of nucleotides or amino acids used is small. Proc Natl Acad Sci USA 95(21):12390–12397
Oren A, Ventosa A (1996) A proposal for the transfer of Halorubrobacterium distributum and Halorubrobacterium coriense to the genus Halorubrum as Halorubrum distributum comb. nov. and Halorubrum coriense comb. nov., respectively. Int J Syst Bacteriol 46:1180
Oren A, Ventosa A, Grant WD (1997) Proposed minimal standards for description of new taxa in the order Halobacteriales. Int J Syst Bacteriol 47(1):233–238
Pesenti PT, Sikaroodi M, Gillevet PM, Sánchez-Porro C, Ventosa A, Litchfield CD (2008) Halorubrum californiense sp. nov., an extreme archaeal halophile isolated from a crystallizer pond at a solar salt plant in California, USA. Int J Syst Evol Microbiol 58(12):2710–2715
Pruesse E, Peplies J, Glockner FO (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28(14):1823–1829
Roh SW, Bae JW (2009) Halorubrum cibi sp. nov., an extremely halophilic archaeon from salt-fermented seafood. J Microbiol 47(2):162–166
Roh SW, Nam Y-D, Chang H-W, Kim K-H, Lee H-J, Oh H-M, Bae J-W (2007a) Natronococcus jeotgali sp. nov., a halophilic archaeon isolated from shrimp jeotgal, a traditional fermented seafood from Korea. Int J Syst Evol Microbiol 57(9):2129–2131
Roh SW, Nam YD, Chang HW, Sung Y, Kim KH, Oh HM, Bae JW (2007b) Halalkalicoccus jeotgali sp. nov., a halophilic archaeon from shrimp jeotgal, a traditional Korean fermented seafood. Int J Syst Evol Microbiol 57(10):2296–2298
Roh SW, Sung Y, Nam YD, Chang HW, Kim KH, Yoon JH, Jeon CO, Oh HM, Bae JW (2008) Arthrobacter soli sp. nov., a novel bacterium isolated from wastewater reservoir sediment. J Microbiol 46(1):40–44
Roh SW, Nam Y-D, Chang H-W, Kim K-H, Sung Y, Kim M-S, Oh H-M, Bae J-W (2009) Haloterrigena jeotgali sp. nov., an extremely halophilic archaeon from salt-fermented food. Int J Syst Evol Microbiol 59(9):2359–2363
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425
Savage KN, Krumholz LR, Oren A, Elshahed MS (2007) Haladaptatus paucihalophilus gen. nov., sp. nov., a halophilic archaeon isolated from a low-salt, sulfide-rich spring. Int J Syst Evol Microbiol 57(1):19–24
Smibert RM, Krieg NR (1994) Phenotypic characterization. In: Gerhardt P, Murray RGE, Wood WA, Kreig NR (eds) Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC, pp 607–654
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tomlinson GA, Hochstein LI (1976) Halobacterium saccharovorum sp. nov., a carbohydrate-metabolizing, extremely halophilic bacterium. Can J Microbiol 22(4):587–591
Ventosa A, Gutiérrez MC, Kamekura M, Zvyagintseva IS, Oren A (2004) Taxonomic study of Halorubrum distributum and proposal of Halorubrum terrestre sp. nov. Int J Syst Evol Microbiol 54(2):389–392
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Xu XW, Wu YH, Zhang HB, Wu M (2007) Halorubrum arcis sp. nov., an extremely halophilic archaeon isolated from a saline lake on the Qinghai–Tibet Plateau, China. Int J Syst Evol Microbiol 57(5):1069–1072
Acknowledgments
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2012R1A1A2040922) and a project fund (C33730) awarded to J. S. Choi by the Center for Analytical Research of Disaster Science of Korea Basic Science Institute. We thank Dr J. P. Euzéby (École Nationale Vétérinaire, Toulouse, France) for etymological advice.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Kyung June Yim and In-Tae Cha have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1. Transmission electron micrographs of Halorubrum halophilum B8T. Bars, 0.2 μm
Supplementary Fig. S2. Two-dimensional thin-layer chromatogram of the total polar lipids of strain B8T. The total polar lipids were detected using sulfuric acid–ethanol. Abbreviations: PG, phosphatidylglycerol; PGP-Me, phosphatidylglycerol phosphate methyl ester; PGS, phosphatidylglycerol sulfate; S-DGD-3, sulfated mannosyl glucosyl diether; PL, unidentified phospholipid
Rights and permissions
About this article
Cite this article
Yim, K.J., Cha, IT., Lee, HW. et al. Halorubrum halophilum sp. nov., an extremely halophilic archaeon isolated from a salt-fermented seafood. Antonie van Leeuwenhoek 105, 603–612 (2014). https://doi.org/10.1007/s10482-014-0115-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-014-0115-6