Abstract
Two strains representing a single novel yeast species were isolated from a flower of Calycoopteris floribunda Lame (SK170T) and insect frass (ST-122) collected in Thailand. On the basis of morphological, biochemical, physiological and chemotaxonomic characteristics, and the sequence analysis of the D1/D2 domain of the large subunit rRNA gene and the internal transcribed spacer region, the two strains were assigned as a single novel Candida species in the Hyphopichia clade for which the name Candida wangnamkhiaoensis sp. nov. is proposed. The type strain is SK170T=BCC 39604T=NBRC 106724T=CBS 11695T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genus Hyphopichia was first proposed to accommodate Pichia burtonii by von Arx and van der Walt (1976) but it was not accepted for the reason that its phenotypic characteristics did not distinguish it from several genera (Kurtzman 1998). Phylogenetic analysis of the D1/D2 domain of the large subunit (LSU) rRNA gene showed good separation of the genus Hyphopichia resulting in acceptance of transferring P. burtonii to this genus (Kurtzman 1998). Kurtzman (2005) proposed H. heimii as a new combination in the genus Hyphopichia and Candida pseudorhagii as a novel species. The phylogenetic placement of C. pseudorhagii and H. heimii in the Hyphopichia clade was also shown in this study (Kurtzman 2005). Groenewald and Smith (2010) demonstrated that on the basis of the nucleotide divergence in the internal transcribed spacer (ITS) region of the rRNA gene and the D1/D2 domain of the LSU rRNA gene, the species of the genus Hyphopichia can be divided into different phylogenetic groups, they introduced three phylogenetic clusters for Hyphopichia and their related anamorphic species and proposed H. pseudoburtonii as a novel species. At present the Hyphopichia clade consist of H. burtonii, H. heimii, H. pseudoburtonii, C. fennica, C. gotoi, C. homilentoma, C. khmerensis, C. pseudorhagii and C. rhagii while the placement of C. ontarioensis is uncertain (Groenewald and Smith 2010; Nagatsuka et al. 2005a; Kurtzman 2005, 2011).
Materials and methods
Yeast isolation
Strain SK170T was isolated from a flower of Calycoopteris floribunda Lame from replanted forest, Wang Nam Khiao district, Nakhon Ratchasima province, Thailand collected on 27 November 2003 by an enrichment technique using yeast extract malt extract (YM) broth (0.3% yeast extract, 0.3% malt extract, 0.5% peptone and 1% glucose) supplemented with 0.025% sodium propionate and 0.02% chloramphenicol in an Erlenmeyer flask and incubated on a rotary shaker at 25°C for 48 h (Limtong et al. 2007). Strain ST-122 was isolated from insect frass of an unknown tree in Pu Wao district, Nong Khai province collected on 3 February 2001 by an enrichment technique using YM broth supplemented with 0.2% sodium propionate and 0.01% chloramphenicol in a test tube and incubated at 25°C for 72–96 h. Purified yeast strains were suspended in YM broth supplemented with 10% glycerol and maintained at −80°C.
DNA sequencing and phylogenetic analysis
The sequences of the D1/D2 domain of the LSU rRNA gene and the ITS region were determined from PCR products amplified from genomic DNA. Methods for DNA extraction and amplification of the D1/D2 domain of the LSU rRNA gene were described previously (Limtong et al. 2007). The ITS region was amplified with primers, ITS1 and ITS4, following the method of White et al. (1990). The PCR products were checked by agarose gel electrophoresis and purified by using the QIA quick purification kit (Qiagen, Germany). The purified products were submitted to Macrogen Inc. (Korea) for sequencing with primers, NL1 and NL4, for the D1/D2 domain of the LSU rRNA gene, and with primers, ITS1 and ITS4, for the ITS region. The sequences were compared pairwise using a BLASTN search (Altschul et al. 1997) and were aligned with the sequences of related species retrieved from GenBank using the multiple alignment program CLUSTAL_X version 1.81 (Thompson et al. 1997). A phylogenetic tree was constructed from the evolutionary distance data with Kimura’s two-parameter correction (Kimura 1980), using the neighbor-joining method (Saitou and Nei 1987) by MEGA soft-ware version 4.0 (Tamura et al. 2007). Confidence levels of the clades were estimated from bootstrap analysis (1,000 replicates) (Felsenstein 1985).
Examination of taxonomic characteristics
The strains were characterized morphologically, biochemically, and physiologically according to the standard methods described by Yarrow (1998). Mycelium formation was investigated on corn meal agar in slide culture at 25°C for up to 7 days. Ascospore formation was investigated on 5% malt extract agar, Fowell’s acetate agar, Gorodkowa agar and corn meal at 15 and 25°C for up to 4 weeks. Carbon assimilation tests were conducted in liquid medium according to the method described by Yarrow (1998). Assimilation of nitrogen compounds was examined on solid media with starved inocula following the method of Nakase and Suzuki (1986). Growth at various temperatures was determined by cultivation in YM broth. Ubiquinones were extracted from cells cultivated in a 500 ml Erlenmeyer flask containing 250 ml of yeast extract peptone dextrose (YPD) broth (1% yeast extract, 2% peptone and 2% dextrose) on a rotary shaker at 28°C for 24–48 h and purified according to the method described by Yamada and Kondo (1973) and Kuraishi et al. (1985). Isoprenologues were identified by HPLC as described previously (Limtong et al. 2007).
Results and discussion
Novel species delineation and identification
Analysis of the D1/D2 domain of the LSU rRNA gene revealed that the sequences of the two strains, SK170T and ST-122, were identical. In terms of pairwise sequence similarity, the closest species to the two strains was H. pseudoburtonii but with 7.1% nucleotide divergence (30 nucleotide substitutions, 10 indels out of 564 nt). The nucleotide sequences of the ITS regions of the two strains were also analyzed, and the sequences of strains SK170T and ST-122 were deposited as AB682911 and AB682912, respectively. In this region the sequences of the two strains differed from each other by two nucleotide substitutions out of 389 nt. The ITS sequences of two strains, SK170T and ST-122, differed from the type strain of H. pseudoburtonii by 7.1% nucleotide substitutions (27 nucleotide substitutions, 48 indels out of 379 nt) and 7.7% nucleotide substitutions (29 nucleotide substitutions, 46 indels out of 379 nt), respectively.
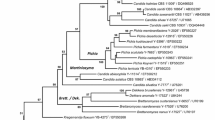
The phylogenetic tree based on the sequences of the D1/D2 domains of the LSU rRNA gene revealed that the strains SK170T and ST-122 were at the same position and formed a cluster with H. pseudoburtonii (Fig. 1). This cluster that contained the two strains and H. pseudoburtonii was separated from the H. burtonii and the H. heimii clusters of the Hyphopichia clade, a three-way distinction reported previously (Groenewald and Smith 2010; Kurtzman 2005, 2011).
Phylogenetic tree based on the sequences of the D1/D2 domain of the LSU rRNA gene, showing positions of Candida wangnamkhiaoensis sp. nov. (SK170T and ST-122) with respect to closely related species. The phylogenetic tree was constructed from evolutionary distance data corrected by two-parameter transformation of Kimura (1980), using the neighbor-joining method. The numbers at nodes indicate the percentages of bootstrap sampling, derived from 1,000 samples. The numbers in parentheses are GenBank accession numbers. Candida melibiosica was the outgroup species in the analysis
Cells of the two strains (SK170T and ST-122) were ovoid to elongate (Fig. 2a) and proliferated by multilateral budding. Pseudohyphae and true hyphae were formed (Fig. 2b). Ascospores were not produced by any of the strains either individually or when paired on 5% malt extract agar, Fowell’s acetate agar, Gorodkowa agar and corn meal agar after 4 weeks at 15 and 25°C.
On the basis of the molecular and additional taxonomic evidence presented, the existence of a single novel Candida species is assigned. The name Candida wangnamkhiaoensis sp. nov. (MB563690) is proposed.
The novel species shows similar phenotypic characteristics to its closest phylogenetic relative, H. pseudoburtonii (Groenewald and Smith 2010), except its ability to assimile of dl-lactic acid (weak). Moreover, according to CBS database the type strain of H. pseudoburtonii has no ability to grow at 30, 35, 37, 40, 42, and 45°C while the novel species grows at 30, 35, and 37°C but does not grow at 40°C.
At present there are not many yeast species in the Hyphopichia clade. The species in this clade have been reported to have been isolated from a wide variety of substrates (Kurtzman 2011). Many strains of H. burtonii have been isolated from high starch substrates e.g., corn, wheat and rice and a few strains from insects and water from fish ponds (Kurtzman 2011). Strains of H. heimii were found in decaying insect-invaded wood (Kurtzman 2011) and strains of H. pseudoburtonii were obtained from the rumen contents of animals, baker’s yeast and foods (Groenewald and Smith 2010). Some species of this clade were isolated from insects and insect frass; for example, a strain of C. rhagii was obtained from an insect while C. fennica, C. gotoi, C. homilentoma and C. pseudorhagii were isolated from insect frasses (Kurtzman 2011). One strain of the novel species in this study was isolated from a flower and another strain was obtained from insect frasses. Therefore, plants seem to be an additional habitat of yeast species in the Hyphopichia clade.
Latin diagnosis of Candida wangnamkhiaoensis Limtong, Kaewwichian, Jindamorakot Yongmanitchai et Nakase sp. nov
In medio liquido ‘cum extracto levidins et extracto malti (YM)’, post dies 3 ad 25°C cellulae ovoideae aut elongatae (2–7 × 2–19 μm), singulae, binae aut pseudohyphae fiunt, per germinationem multipolarem reproducentes. Repens pellicula formatur. In agaro ‘YM’, post dies 3 ad 25°C, cultura hebes, cremea et margo fimbriata cum mycelio. In agaro farina Zea mays post dies 7 ad 25°C, pseudohyphae et hyphae formantur. Ascosporae non formantur.
d-Glucosum, d-galactosum (infirme), sucrosum (infirme), maltosum et trehalosum (infirme) fermentantur at non lactosum nec raffinosum. d-Glucosum, d-galactosum, l-sorbosum, sucrosum, maltosum, cellobiosum, trehalosum, raffinosum (infirme), amylum solubile, d-xylosum (infirme), d-ribosum, N-acetyl-d-glucosaminum, ethanolum, glycerolum, erythritolum, ribitolum, d-mannitolum, d-glucitolum, α-methyl-d-glucosidum, salicinum, acidum d-gluconicum, acidum 2-keto-d-gluconicum, d-glucono-δ-lactonum, acidum dl-lacticum (infirme), acidum succinicum, acidum citricum, ethylaminum, l-lysinum et cadaverinum assimilantur at non lactosum, melibiosum, melezitosum, inulinum, l-arabinosum, d-arabinosum, l-rhamnosum, methanolum, galactitolum, acidum d-glucuronicum, acidum d-galacturonicum, acidum 5-keto-d-gluconicum, inositolum, kalium nitricum nec natrium nitrosum. Crescit in 50% glucosum, 60% glucosum et 10% NaCl/5% glucosum. Non crescit in 0.1% cycloheximido et 0.01% cycloheximido. Crescere potest ad temperatura 20, 25, 30, 35 et 37°C at non crescit ad temperatura 40°C. Amylum non formatur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur. Ubiquinonum majus: Q-8.
Holotypus
Stirps SK170T isolatus ex flore of C. floribunda Lame in Nakhon Ratchasima provincia, Thailandia. Cultura conservata est in Collectione Culturarum in BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailandia ut BCC 39604T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japonia conservatus ut NBRC 106724T et Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands ut CBS 11695T.
Description of Candida wangnamkhiaoensis Limtong, Kaewwichian, Jindamorakot, Yongmanitchai and Nakase sp. nov
Growth in yeast extract malt extract (YM) broth: After 3 days at 25°C, cells are ovoid to elongate (2–7 × 2–19 μm) and occur singly, in pairs or in short chain (Fig. 2a). Budding is multilateral. Creeping pellicles are present. Growth on YM agar: After 3 days at 25°C, the streak culture is dull, cream-coloured and the margin is fringed with filaments. Pseudohyphae and true hyphae are formed in slide culture on corn meal agar after 5 days at 25°C (Fig. 2b). Ascospores were not produced by any of the strains either individually or when paired on 5% malt extract agar, Fowell’s acetate agar, Gorodkowa agar and corn meal agar after 4 weeks at 15 and 25°C. Fermentation of d-glucose, d-galactose (weak), sucrose (weak), maltose and α–α trehalose are positive but negative for lactose and raffinose. d-Glucose, d-galactose, l-sorbose, sucrose, maltose, cellobiose, α–α trehalose, raffinose (weak), soluble starch, d-xylose (weak), d-ribose, N-acetyl-d-glucosamine, ethanol, glycerol, erythritol, ribitol, d-mannitol, d-glucitol, α-methyl-d-glucoside, salicin, d-gluconate, 2-keto-d-gluconate, d-glucono-δ-lactone, dl-lactic acid (weak), succinic acid, citric acid, ethylamine, l-lysine and cadaverine are assimilated, but lactose, melibiose, melezitose, inulin, l-arabinose, d-arabinose, l-rhamnose, methanol, galactitol, d-glucuronate, d-galacturonate, 5-keto-d-gluconate, inositol, potassium nitrate and sodium nitrite are non-assimilated. Growth on medium containing 50% (w/v) glucose, 60% (w/v) glucose or 10% (w/v) sodium chloride/5% (w/v) glucose are positive. Growth with 0.1% cylcloheximide and 0.01% cylcloheximide are negative. Growth at 25, 30, 35, and 37°C is positive, but at 40°C is negative. Starch like compounds are not produced. Diazonium blue B color and urease reactions are negative. The major ubiquinone is Q-8.
Holotype
SK170T is the holotype of C. wangnamkhiaoensis. The strain was isolated from a flower of C. floribunda Lame in Wang Nam Khiao district, Nakhon Ratchasima province, Thailand, collected on 27 November 2003. The living culture from type was deposited at the BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailand, as BCC 39604T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japan, as NBRC 106724T and Centraalbureau voor Schimmelcultures (CBS), Utrecht, The Netherlands as CBS 11695T.
Etymology
The species epithet wangnamkhiaoensis refers to Wang Nam Khiao district, Nakhon Ratchasima province, Thailand, where the type strain was isolated.
References
Altschul SF, Madden TL, Schäffer JZ, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Groenewald M, Smith MT (2010) Re-examination of strains formerly assigned to Hyphopichia burtonii, the phylogeny of the genus Hyphopichia, and the description of Hyphopichia pseudoburtonii sp. nov. Int J Syst Evol Microbiol 60:2675–2680
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kuraishi H, Katayama-Fujimura Y, Sugiyama J, Yokoyama T (1985) Ubiquinone systems in fungi. I. Distribution of ubiquinones in the major families of ascomycetes, basidiomycetes, and deuteromycetes, and their taxonomic implications. Trans Mycol Soc Jpn 26:383–395
Kurtzman CP (1998) Pichia E.C. Hasen emend. Kurtzman. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. The Netherlands, Elsevier, pp 273–352
Kurtzman CP (2005) New species and a new combination in the Hyphopichia and Yarrowia yeast clades. Antonie van Leewenhoek 88:121–130
Kurtzman CP (2011) Hyphopichia von Arx & van der Walt. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn, vol 2. Elsevier, Amsterdam, pp 435–438
Limtong S, Yongmanitchai W, Tun MM, Kawasaki H, Seki T (2007) Kazachstania siamensis sp. nov., an ascomycetous yeast species from forest soil in Thailand. Int J Syst Evol Microbiol 57:419–422
Nagatsuka Y, Kawasaki H, Mikata K, Seki T (2005) Candida khmerensis sp. nov., a novel cation-tolerant yeast isolated from dry salted shrimp and sewage in Cambodia. J Gen Appl Microbiol 51:235–243
Nakase T, Suzuki M (1986) Bullera megalospora, a new species of yeast forming larger ballistospores isolated from dead leaves of Oryza sativa, Miscanthus sinesis and Sasa sp. in Japan. J Gen Appl Microbiol 32:225–240
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics (MEGA) Soft-ware version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
von Arx JA, van der Walt JP (1976) The ascigerous state of Candida chodatii. Antonie van Leewenhoek 42:309–314
White TJ, Bruns T, Lee S, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Yamada Y, Kondo K (1973) Coenzyme Q system in the classification of the yeast genera Rhodotorula and Cryptococcus, and the yeast-like genera Sporobolomyces and Rhodosporidium. J Gen Appl Microbiol 19:59–77
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Acknowledgment
This study was partial supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Limtong, S., Kaewwichian, R., Jindamorakot, S. et al. Candida wangnamkhiaoensis sp. nov., an anamorphic yeast species in the Hyphopichia clade isolated in Thailand. Antonie van Leeuwenhoek 102, 23–28 (2012). https://doi.org/10.1007/s10482-012-9709-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-012-9709-z