Abstract
Five strains (LN12, LN14T, LN15T, LN16 and LN17T) representing three novel methylotrophic yeast species were isolated from the external surface of plant leaves by three-consecutive enrichments. On the basis of morphological, biochemical, physiological and chemotaxonomic characteristics, the sequence analysis of the D1/D2 domain of the large subunit (LSU) rRNA gene and the phylogenetic analysis, the five strains were assigned to be one novel Ogataea species and two novel Candida species. Three strains (LN12, LN14T and LN16) represent a single novel species of the genus Ogataea, for which the name Ogataea phyllophila sp. nov. is proposed. The type strain is LN14T (= BCC 42666T = NBRC 107780T = CBS 12095T). Strain LN15T was assigned to be Candida chumphonensis sp. nov. (type strain LN15T = BCC 42667T = NBRC 107781T = CBS 12096T). Strain LN17T represented another novel species of Candida that was named Candida mattranensis sp. nov. (type strain LN17T = BCC 42668T = NBRC 107782T = CBS 12097T).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Methylotrophic yeasts can utilize methanol as a sole source of carbon and energy. They represent a relative small proportion of yeasts and belong to a limited number of yeast genera, including Ogataea (Yamada et al. 1994; Mikata and Yamada 1995; Suh et al. 2006), Komagataella (Yamada et al. 1995; Dlauchy et al. 2003; Kurtzman 2005), Kuraishia (Yamada et al. 1994; Peter et al. 2005) and Candida (Meyer et al. 1998). In the recent years member of Ogataea increased rapidly not only because of transferring many Pichia species to this genus after emendation of the genus description for nitrate assimilation, ascospores shaped and ascospores number but also from discovering many novel species in the past few years (Glushakova et al. 2010; Kurtzman and Robnett 2010; Limtong et al. 2008; Nagatsuka et al. 2008; Peter et al. 2008, 2009; Suh and Zhou 2010). At present more than 30 species are accepted in the genus Ogataea.
The external surface of plant leaves usually refer to the phylloplane or phyllosphere (Phaff and Starmer 1987; Fonseca and Inacio 2006). Phylloplane from various regions of the world are found to be colonized by members of both basidiomycetous and ascomycetous yeasts (Fonseca and Inacio 2006; Glushakova et al. 2007; Landell et al. 2010; Nakase et al. 2001; Slavikova et al. 2009). Although most common phylloplane yeasts are basidiomycetous species (Fonseca and Inacio 2006; Nakase et al. 2001) Peter et al. (2007) reported the occurrence of methylotrophic ascomycetous species on leaves in Hungary. It is suggested that the methylotrophic yeasts on phylloplane utilize methanol that produced by pectin demethylation inside the leaves and emitted to the surface through stomata during transpiration for their growth (Peter et al. 2007).
During investigation of methylotrophic yeasts on phylloplane in Mattra island, Chumphon Islands National Park, Chumphon province, Thailand, five strains of novel species were isolated. Detailed analysis demonstrated that they are belonged to Ogataea clade. In this report one novel Ogataea species and two novels Candida species are described.
Materials and methods
Yeast isolation
Leaves were collected from 21 different plants included banana (Musa sapientum), cassava (Manihot esculenta), coconut (Cocos nucifera), fig (Ficus racemosa), sugarcane (Saccharum officinarum) and the other 16 unknown plants in a small area of primary rainforest in Mattra island (10°04′N 99°35′E), Chumphon Islands National Park, Chumphon province, Thailand in May, 2009. Methylotrophic yeasts were isolated by a technique involving three consecutive methanol enrichments as described by Limtong et al. (2004) but using 0.5% (v/v) methanol-yeast nitrogen base (YNB) broth (0.67% Difco yeast nitrogen base and 0.5% (v/v) methanol) instead of 1% (v/v) methanol-YNB broth. Three grams of cut leaves, derived from cutting few leaves to the size that can be put into a 250 ml Erlenmeyer flask, was inoculated into 50 ml enrichment broth in the flask and incubated on a rotary shaker at room temperature for 4–5 days. After three consecutive enrichment cultivation a loopful of the enriched culture was streaked on 0.5% (v/v) methanol-YNB agar and incubated at room temperature until yeast colonies appeared. Yeast colonies of different morphologies were picked and purified by cross streaking on YM agar. Purified yeast strains were suspended in YM broth supplemented with 10% glycerol and maintained at −80°C.
DNA sequencing and phylogenetic analysis
Methods for DNA isolation and amplification of the D1/D2 domain of the LSU rRNA gene were as described previously by Limtong et al. (2007). The PCR product was checked by agarose gel electrophoresis and purified by using the QIA quick purification kit (Qiagen, Hilden, Germany). The purified product was sequenced commercially by Macrogen Inc. (Seoul, Korea) for sequencing with primers, NL1 and NL4. The sequences were compared pairwise using a BLASTN search (Altschul et al. 1997) and were aligned with the sequences of related species retrieved from GenBank using the multiple alignment program CLUSTAL_X version 1.81 (Thompson et al. 1997). A phylogenetic tree was constructed from the evolutionary distance data with Kimura’s two-parameter correction (Kimura 1980), using the neighbor-joining method (Saitou and Nei 1987). The other phylogenetic trees were also constructed with the neighbor-joining and maximum likelihood methods using MEGA 5.0 package (Tamura et al. 2007) and the aligned sequences obtained from MUSCLE version 3.8 alignment program (Edgar 2004). Confidence levels of the clades were estimated from bootstrap analysis (1,000 replicates) (Felsenstein 1985).
Examination of taxonomic characteristics
The strains were characterized morphologically, biochemically, and physiologically according to the standard methods described by Yarrow (1998). Mycelium formation was investigated on corn meal agar in slide culture at 25°C for up to 7 days. Ascospores formation was investigated on 5% malt extract agar, Gorodkowa agar, Fowell’s acetate agar and corn meal agar at 15 and 25°C for up to 4 weeks. Carbon assimilation tests were conducted in liquid medium as described by Yarrow (1998). Assimilation of nitrogen compounds was examined on solid media with starved inocula following the method of Nakase and Suzuki (1986). Growth at various temperatures was determined by cultivation in YM broth. Ubiquinones were extracted from cells cultivated in 500 ml Erlenmeyer flasks containing 250 ml of yeast extract peptone dextrose (YPD) broth (1% yeast extract, 2% peptone and 2% dextrose) on a rotary shaker at 28°C for 24–48 h and purified according to the method described by Yamada and Kondo (1973) and Kuraishi et al. (1985). Isoprenologues were identified by HPLC as described previously (Limtong et al. 2007).
Results and discussion
Yeasts isolation and identification
Six methylotrophic yeast strains could be isolated by the enrichment isolation using 0.5% (v/v) methanol-YNB broth from six samples of leaves from cassava and the other five unknown plants out of 21 samples used for isolation. Identification on the basis of similarities of the D1/D2 domain of the LSU rRNA gene sequences revealed that five strains (LN12, LN14T, LN15T, LN16 and LN17T) were found to represent three novel species and one strain was assigned to Ogataea polymorpha.
Member of the methylotrophic yeast have been found associated with plant materials including tree bark, tree exudate, flower, leaf, gall on leaf and rotten wood (Dlauchy et al. 2003; Glushakova et al. 2010; Limtong et al. 2004, 2008; Morais et al. 2004; Peter et al. 2003, 2007, 2008, 2009). The result of this investigation showed that methylotrophic yeasts were detected on leaves collected from the small island in Thailand though only a small number of samples were investigated. However, methylotrophic yeast could be detected on only 27% of the leaves which was much lower than 45% that was reported in Hungary (Peter et al. 2007) while 95% of leaves were colonized by the other yeasts.
Novel species delineation
Among the five strains (LN12, LN14T, LN15T, LN16 and LN17T), the sequences of the D1/D2 domain of the LSU rRNA gene of three strains (LN12, LN14T and LN16) were identical. In terms of pairwise sequence similarity the closest species to the three strains (LN12, LN14T and LN16) was Ogataea sp. NCAIM Y.01896 but with 1.4% nucleotide substitutions (9 nucleotide substitutions out of 568 nt) and the three strains differed from Ogataea minuta, their closest described relative, by 1.8% nucleotide substitutions (10 nucleotide substitutions and 1 indel out of 567 nt). Strain LN15T differed from Candida sp. ZIM 2322, it closest phylogenic relative, by 2.8% nucleotide substitutions (18 nucleotide substitutions and 2 gaps out of 570 nt) and from Ogataea dorogensis, the known species closest to it, by 3.2% nucleotide substitutions (18 nucleotide substitutions and 3 gaps out of 570 nt). Strain LN17T was closest to Candida sp. NRRL YB-2442 but with 3.5% nucleotide substitutions (20 nucleotide substitutions and 1 indel out of 566 nt) while it differed by 3.7% nucleotide substitutions (21 nucleotide substitutions and 1 indel out of 566 nt) from Ogataea cecidiorum, it closest known species. Therefore, the three strains (LN12, LN14T and LN16) were considered to represent a single novel species while strains LN15T and LN17T represented two other novel species (Kurtzman and Robnett 1998).
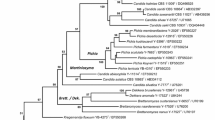
The phylogenetic trees based on the sequences of the D1/D2 domain of the LSU rRNA gene were constructed by neighbor-joining method using the aligned sequences obtained from CLUSTAL_X alignment program (Fig. 1) or MUSCLE alignment program (data not shown) and by maximum likelihood method using the aligned sequences obtained from either alignment program (data not shown). Though the trees constructed by both methods were not congruent completely, they demonstrated that the five strains were in the Ogataea clade. The neighbor-joining tree using aligned sequences obtained from CLUSTAL_X alignment program was selected to show the phylogenetic placement of the new species (Fig. 1). In this neighbor-joining tree the five strains were in the same cluster that separated from the so-called Pichia methanolica cluster proposed by Nagatsuka et al. (2008), which latter the Pichia species of this cluster were transferred to the genus Ogataea (Kurtzman and Robnett 2010). The three strains (LN12, LN14T and LN16) were located in the same position and formed a subcluster with Ogataea sp. NCAIM Y.01896 and O. minuta, there closest undescribed and described relatives in term of pairwise sequence similarity, but with low bootstrap support. Strain LN15T form a subcluster with O. dorogenis with low bootstrap support its closet described species. Strain LN17T was related to Candida sp. NRRL YB-2442 in a separate subcluster. Therefore, it is clear that the five strains were separated into three novel species belonging to the Ogataea clade but their correct phylogenetic position within the clade remains unclear which was also happened in the case of using single gene sequences for phylogenetic tree construction reported by Nagatsuka et al. (2008); Nakase et al. (2010) and Peter et al. (2010).
Phylogenetic tree based on the sequences of the D1/D2 domain of the LSU rRNA gene, showing positions of Ogataea phyllophila sp. nov. (LN12, LN14T and LN16), Candida chumphonensis sp. nov. (LN15T) and Candida mattranensis sp. nov. (LN17T) with respect to closely relate species. The phylogenetic tree was constructed from evolutionary distance data corrected by two-parameter transformation of Kimura (1980), using the neighbor-joining method. Numbers indicate percentages of bootstrap sampling, derived from 1,000 samples. Bar 0.01 Knuc
On the basis of the evidence of molecular and other taxonomic criteria here we propose three novel species, O. phyllophila sp. nov. (MycoBank no.: MB561002) for strains LN12, LN14T and LN16, C. chumphonensis sp. nov. (MycoBank no.: MB561003) for strain LN15T, and C. mattranensis sp. nov. (MycoBank no.: MB561004) for strain LN17T.
The three novel species O. phyllophila sp. nov., C. chumphonensis sp. nov. and C. mattranensis sp. nov. can be distinguished from each other and from their closest known species O. minuta, O. dorogensis and O. cecidiorum, respectively, not only on the basis of the sequences of the D1/D2 domain of the LSU rRNA gene and ascospores formation but also by several phenotypic characteristics as shown in Table 1.
Latin diagnosis of Ogataea phyllophila Koowadjanakul, Jindamorakot, Yongmanitchai et Limtong sp. nov.
In medio liquido ‘cum extracto levidins et extracto malti (YM)’, post dies 3 ad 25°C cellulae globosae aut ovoideae (2–3 × 2–4 μm), singulae aut binae, per germinationem multipolarem reproducentes. In agaro ‘YM’, post dies 3 ad 25°C, cultura butyrosa, cremea, glabra et margo glabra. Pseudohyphae et hyphae non formantur in agaro farina Zea mays post dies 7 ad 25°C. Ascosporae galeiformes aut pileiformes, 4 in ascum. Asci inconjugati, conjugati cellularum vel, conjugati cellularum gemmarum que oriumtur. Fermentatio nulla. d-Glucosum, l-sorbosum, d-ribosum, d-xylosum (lente), d-arabinosum (infirme), α-α-trehalosum, cellobiosum, salicinum, glycerolum, erythritolum, ribitolum, d-glucitolum, d-mannitolum, acidum 2-keto-d-gluconicum (infirme), acidum 5-keto-d-gluconicum, acidum d l-lacticum (infirme), acidum succinicum, acidum citricum, methanolum, ethanolum, natrium nitrosum, kalium nitricum, ethylaminum, l-lysinum et cadaverinum assimilantur at non d-galactosum, N-acetyl-d-glucosaminum, l-arabinosum, l-rhamnosum, sucrosum, maltosum, α-methyl-d-glucosidum, melibiosum, lactosum, raffinosum, melizitosum, inulinum, amylum solubile, galactitolum, inositolum, acidum d-gluconicum nec acidum d-glucuronicum. Vitamina externa ad crescentiam necessaria non sunt (infirme). Crescere potest in temperatura 25, 30 et 35°C at non crescit in temperatura 37, 40, 42 nec 45°C. Crescit in 50% glucosum, et 10% NaCl/5% glucosum. Non crescit in 60% glucosum, 16% NaCl/5% glucosum, 0. 1% cycloheximido et 0.01% cycloheximido. Amylum non formatur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur. Ubiquinonum majus: Q-7.
Holotypus
Stirps LN14T isolatus ex folio, in Chumphon provincia, Thailandia. Cultura et conservatus in Collectionie Culturarum in BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailandia ut BCC 42666T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japonia conservatus ut NBRC 107780T et Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands ut CBS 12095T.
Description of Ogataea phyllophila Koowadjanakul, Jindamorakot, Yongmanitchai and Limtong sp. nov.
Growth in yeast extract malt extract (YM) broth: After 3 days at 25°C, cells are spherical to ovoid (2–3 × 2–4 μm) and occur singly or in pairs (Fig. 2a). Budding is multilateral. Growth on YM agar: After 3 days at 25°C, the streak culture is butyrous, cream-coloured, with a smooth surface and has an entire margin. Pseudohyphae and true hyphae are not formed in slide culture on corn meal agar after 7 days at 25°C. Ascospores were produced on 5% malt extract agar and corn meal agar after 2 days at 15 and 25°C (Fig. 2b). Four hat-shaped ascospores are formed in a deliquescent ascus that may be produced unconjugation or conjugation between a parent cell and its bud or between independent cells and ascospores tend to agglutinate after liberation. The other phenotypic characteristics of the species are shown in Table 1. The MycoBank number is MB561002.
Holotype
LN14T is the holotype of O. phyllophila. The strain was isolated from leaves of an unknown tree on Mattra island (10°04′N 99°35′E) in Chumphon Islands National Park, Chumphon province, Thailand, collected in May, 2009. The living culture from type was deposited at the BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailand, as BCC 42666T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japan, as NBRC 107780T and Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands as CBS 12095T.
Etymology
The species epithet phyllophila refers to phylloplane where the three strains of this species were isolated.
Latin diagnosis of Candida chumphonensis Limtong, Koowadjanakul, Jindamorakot et Yongmanitchai sp. nov.
In medio liquido ‘YM’, post dies 3 ad 25°C cellulae globosae aut ovoideae (2–4 × 2–4 μm), singulae, binae, aut congregationibus, per germinationem multipolarem reproducentes. In agaro ‘YM’, post dies 3 ad 25°C, cultura butyrosa, cremea, glabra, umbonatus et margo undulata. Pseudohyphae et hyphae non formantur. Ascosporae non formantur. d-Glucosum (lente) fermentature at non d-galactosum, maltosum, sucrosum, α-α-trehalosum, lactosum nec raffinosum. d-Glucosum, d-ribosum (infirme), cellobiosum (infirme), salicinum (infirme), inulinum (infirme), glycerolum (infirme), erythritolum (infirme), ribitolum (infirme), d-glucitolum (infirme), d-mannitolum (infirme), acidum 2-keto-d-gluconicum (infirme), acidum 5-keto-d-gluconicum, acidum d-gluconicum (infirme), acidum succinicum (infirme), methanolum, ethanolum (infirme), ethylaminum et cadaverinum assimilantur at non d-galactosum, l-sorbosum, N-acetyl-d-glucosaminum, d-xylosum, l-arabinosum, d-arabinosum, l-rhamnosum, sucrosum, maltosum, α-α-trehalosum, α-methyl-d-glucosidum, melibiosum, lactosum, raffinosum, melizitosum, amylum solubile, galactitolum, inositolum, acidum d-glucuronicum, acidum d l-lacticum, acidum citricum, kalium nitricum, natrium nitrosum nec l-lysinum. Vitamina externa ad crescentiam necessaria non sunt (infirme). Crescere potest in temperatura 25, 30 et 35°C at non crescit in temperatura 37, 40, 42 nec 45°C. Non crescit in 50% glucosum, 60% glucosum, 10% NaCl/5% glucosum, 16% NaCl/5% glucosum, 0. 1% cycloheximido et 0.01% cycloheximido. Amylum non formatur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur. Ubiquinonum majus: Q-7.
Holotypus
Stirps LN15T isolatus ex folio, in Chumphon provincia, Thailandia. Cultura et conservatus in Collectionie Culturarum in BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailandia ut BCC 42667T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japonia conservatus ut NBRC 107781T et Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands ut CBS 12096T.
Description of Candida chumphonensis Limtong, Koowadjanakul, Jindamorakot and Yongmanitchai sp. nov.
Growth in YM broth: After 3 days at 25°C, cells are spherical to ovoid (2–4 × 2–4 μm) and occur singly or in pairs or in groups (Fig. 3a). Budding is multilateral. Growth on YM agar: After 3 days at 25°C, the streak culture is butyrous, cream-coloured, with an umbonate surface and has an undulate margin. Pseudohyphae and true hyphae are not formed in slide culture on corn meal agar after 7 days at 25°C. Ascospores were not produced on 5% malt extract agar, Fowell’s acetate agar, Gorodkowa agar and corn meal agar after 4 weeks at 15 and 25°C. The other phenotypic characteristics of the species are shown in Table 1. The MycoBank number is MB561003.
Holotype
LN15T is the holotype of C. chumphonensis. The strain was isolated from leaves of an unknown tree on Mattra island (10°04′N 99°35′E) in Chumphon Islands National Park, Chumphon province, Thailand, collected in May, 2009. The living culture from type was deposited at the BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailand, as BCC 42667T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japan, as NBRC 107781T and Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands as CBS 12096T.
Etymology
The species epithet chumphonensis refers to Chumphon province, Thailand, where the strain was isolated.
Latin diagnosis of Candida mattranensis Limtong, Koowadjanakul, Jindamorakot et Yongmanitchai sp. nov.
In medio liquido ‘YM’, post dies 3 ad 25°C cellulae subglobosae aut ovoideae (1–4 × 2–5 μm), singulae aut binae, per germinationem multipolarem reproducentes. In agaro ‘YM’, post dies 3 ad 25°C, cultura butyrosa, cremea, glabra et margo glabra. Pseudohyphae et hyphae non formantur. Ascosporae non formantur. d-Glucosum (infirme) fermentature at non d-galactosum, maltosum, sucrosum, α-α-trehalosum, lactosum nec raffinosum. d-Gucosum, d-galactosum (infirme), l-sorbosum (lente), d-ribosum (lente), d-xylosum (infirme), α-α-trehalosum, cellobiosum, salicinum, glycerolum, erythritolum, ribitolum, d-glucitolum, d-mannitolum, galactitolum, acidum d l-lacticum (infirme), acidum succinicum, acidum citricum, methanolum, ethanolum, kalium nitricum, natrium nitrosum, ethylaminum, l-lysinum et cadaverinum at non N-acetyl-d-glucosaminum, l-arabinosum, d-arabinosum, l-rhamnosum, sucrosum, maltosum, α-methyl-d-glucosidum, melibiosum, lactosum, raffinosum, melizitosum, inulinum, amylum solubile, inositolum, acidum 2-keto-d-gluconicum, acidum 5-keto-d-gluconicum, acidum d-gluconicum nec acidum d-glucuronicum. Vitamina externa ad crescentiam necessaria non sunt. Crescere potest in temperatura 25, 30, 35 et 37°C (lente) at non crescit in temperatura 40, 42 nec 45°C. Crescit in 50% glucosum, 60% glucosum et 10% NaCl/5% glucosum. Non crescit in 16% NaCl/5% glucosum, 0. 1% cycloheximido et 0.01% cycloheximido. Amylum non formatur. Diazonium caeruleum B non respondens. Ureum non hydrolysatur. Ubiquinonum majus: Q-7.
Holotypus
Stirps LN17T isolatus ex folio, in Chumphon provincia, Thailandia. Cultura et conservatus in Collectionie Culturarum in BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailandia ut BCC 42668T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japonia conservatus ut NBRC 107782T et Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands ut CBS 12097T.
Description of Candida mattranensis Limtong, Koowadjanakul, Jindamorakot and Yongmanitchai sp. nov.
Growth in YM broth: After 3 days at 25°C, cells are subspherical to ovoid (1–4 × 2–5 μm) and occur singly or in pairs (Fig. 3b). Budding is multilateral. Growth on YM agar: After 3 days at 25°C, the streak culture is butyrous, cream-coloured, with a smooth surface and has an entire margin. Pseudohyphae and true hyphae are not formed in slide culture on corn meal agar after 7 days at 25°C. Ascospores were not produced on 5% malt extract agar, Fowell’s acetate agar, Gorodkowa agar and corn meal agar after 4 weeks at 15 and 25°C. The other phenotypic characteristics of the species are shown in Table 1. The MycoBank number is MB561004.
Holotype
LN17T is the holotype of C. mattranensis. The strain was isolated from leaves of an unknown tree on Mattra island (10°04′N 99°35′E) in Chumphon Islands National Park, Chumphon province, Thailand, collected in May, 2009. The living culture from type was deposited at the BIOTEC Culture Collection (BCC), National Center for Genetic Engineering and Biotechnology (BIOTEC), Pathumthani, Thailand, as BCC 42668T; NITE Biological Resources Center (NBRC), Department of Biotechnology, National Institute of Technology and Evaluation, Chiba, Japan, as NBRC 107782T and Centraalbureau voor Schimmelcultures (CBS), Utrecht, the Netherlands as CBS 12097T.
Etymology
The species epithet mattranensis refers to Mattra island, Chumphon province, Thailand, where the strain was isolated.
References
Altschul SF, Madden TL, Schaffer JZ, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Barnett JA, Payne RW, Yarrow D (2000) Yeasts: characteristics and identification. Cambridge University Press, Cambridge
Dlauchy D, Tornai-Lehoczki J, Fulop L, Peter G (2003) Pichia (Komatagaella) pseudopastoris sp. nov., a new yeast species from Hungary. Antonie van Leeuwenhoek 83:327–332
Edgar PC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acid Res 32:1792–1797
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Fonseca A, Inacio J (2006) Phylloplane yeasts. In: Rosa C, Peter G (eds) Biodiversity and ecophysiology of yeasts. Springer-Verlag, Berlin, Heidelberg, Germany, pp 263–301
Glushakova AM, Yurkov AM, Chernov IY (2007) Massive isolation of anamorphous ascomycete yeasts Candida oleophila from plant phyllosphere. Microbiology 76:799–803
Glushakova AM, Maximova IA, Kachalkin AV, Yurkov AM (2010) Ogataea cecidiorum sp. nov., a methanaol-assimilating yeast isolated from galls on willow leaves. Antonie van Leeuwenhoek 98:93–101
Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kuraishi H, Katayama-Fujimura Y, Sugiyama J, Yokoyama T (1985) Ubiquinone systems in fungi. I. Distribution of ubiquinones in the major families of ascomycetes, basidiomycetes, and deuteromycetes, and their taxonomic implications. Trans Mycol Soc Jpn 26:383–395
Kurtzman CP (1998) Pichia E. C. Hansen emend Kurtzman. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 273–352
Kurtzman CP (2005) Description of Komagataella phaffii sp. nov. and the transfer of Pichia pseudopastoris to the methylotrophic yeast genus Komagataella. Int J Syst Evol Microbiol 55:973–976
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leewenhoek 73:331–371
Kurtzman CP, Robnett CJ (2010) Systematics of methanol assimilating yeasts and neighboring taxa from multigene sequence analysis and the proposal of Peterozyma gen. nov., a new member of the Saccharomycetales. FEMS Yeast Res 10:353–361
Landell MF, Billodre R, Ramos JP, Leoncini O, Vainstein MH, Valente P (2010) Candida aechmeae sp. nov. and Candida vrieseae sp. nov., novel yeast species isolated from the phylloplane of bromeliads in Southern Brazil. Int J Syst Evol Microbiol 60:244–248
Limtong S, Srisuk N, Yongmanitchai W, Kawasaki H, Yurimoto H, Nakase T, Kato N (2004) Three new thermotolerant methylotrophic yeasts, Candida krabiensis sp. nov., Candida sithepensis sp. nov., and Pichia siamensis sp. nov., isolate in Thailand. J Gen Appl Microbiol 50:119–127
Limtong S, Yongmanitchai W, Tun MM, Kawasaki H, Seki T (2007) Kazachstania siamensis sp. nov., an ascomycetous yeast species from forest soil in Thailand. Int J Syst Evol Microbiol 57:419–422
Limtong S, Srisuk N, Yongmanitchai W, Yurimoto H, Nakase T (2008) Ogataea chonburiensis sp. nov. and Ogataea nakhonphanomensis sp. nov., thermotolerant, methylotrophic yeast species isolated in Thailand, and transfer of Pichia siamensis and Pichia thermomethanolica to the genus Ogataea. Int J Syst Evol Microbiol 58:302–307
Meyer SA, Payne RW, Yarrow D (1998) Candida Berkhout. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 454–573
Mikata K, Yamada Y (1995) Ogataea kodamae, a new combination for a methanol-assimilating yeast species, Pichia kodamae van der Walt et Yarrow. Res Commun Inst Ferment 17:99–101
Morais PB, Teixeira LCRS, Bowles JM, Lachance M-A, Rosa CA (2004) Ogataea falcaomoraisii sp. nov., a sporogenous methylotrophic yeast from tree exudates. FEMS Yeast Res 5:81–85
Nagatsuka Y, Saito S, Sugiyama J (2008) Ogataea neopini sp. nov. and O. corticis sp. nov., with the emendation of the ascomycete yeast genus Ogataea, and transfer of Pichia zsoltii, P. dorogensis, and P. trehaloabstinens to it. J Gen Appl Microbiol 54:353–365
Nakase T, Suzuki M (1986) Bullera megalospora, a new species of yeast forming large ballistospores isolated from dead leaves of Oryza sativa, Miscanthus sinensis and Sasa sp. in Japan. J Gen Appl Microbiol 32:225–240
Nakase T, Takashima M, Itoh M, Fungsin B, Potacharoen W, Atthasampunna P, Komagata K (2001) Ballistoconidium-froming yeasts found in the phyllosphere of Thailand. Microbiol Cult Coll 17:23–33
Nakase T, Imanishi Y, Ninomiya S, Takashima M (2010) Candida rishirensis sp. nov., a novel methylotrophic anamorphic yeast species isolated from soil on Rishiri Island in Japan. J Gen Appl Microbiol 56:169–173
Peter G, Tornai-Lehoczki J, Fulop L, Dlauchy D (2003) Six new methanol assimilating yeast species from wood material. Antonie van Leeuwenhoek 84:147–159
Peter G, Dlauchy D, Tornai-Lehoczki J, Kurtzman CP (2005) Kuraishia molischiana sp. nov., the teleomorph of Candida molischiana. Antonie van Leeuwenhoek 88:241–247
Peter G, Tornai-Lehoczki J, Dlauchy D (2007) Ogataea allantospora sp. nov., an ascomycetous yeast species from phylloplane. Antonie van Leeuwenhoek 92:443–448
Peter G, Tornai-Lehoczki J, Dlauchy D (2008) Ogataea nitratroaversa sp. nov., a methylotrophic yeast species from temperate forest habitats. Antonie van Leewenhoek 94:217–222
Peter G, Tornai-Lehoczki J, Dlauchy D (2009) Ogataea populialbae sp. nov., a yeast species from white poplar. FEMS Yeast Res 9:936–941
Peter G, Tornai-Lehoczki J, Dlauchy D (2010) Ogataea pignaliae sp. nov., the teleomorph of Candida pignaliae. Int J Syst Evol Microbiol 60:2496–2500
Phaff HJ, Starmer WT (1987) Yeasts associated with plants, insects and soil. In: Rose AH, Harrison JS (eds) The yeasts, 2nd edn. Academic, London, pp 123–180
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Slavikova E, Vadkertiova R, Vranova D (2009) Yeasts colonizing the leaves of fruit trees. Ann Microbiol 59:419–424
Suh S-O, Zhou JJ (2010) Methylotrophic yeasts near Ogataea (Hansenula) polymorpha: a proposal of Ogataea angusta comb. nov. and Candida parapolymorpha sp. nov. FEMS Yeast Res 10:631–638
Suh SO, Blackwell M, Kurtzman CP, Lachance M-A (2006) Phylogenetics of Saccharomycetales, the ascomycete yeasts. Mycologia 98:1006–1017
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics (MEGA) soft-ware version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Yamada Y, Kondo K (1973) Coenzyme Q system in the classification of the yeast genera Rhodotorula and Cryptococcus, and the yeast-like genera Sporobolomyces and Rhodosporidium. J Gen Appl Microbiol 19:59–77
Yamada Y, Maeda K, Mikata K (1994) The phylogenetic relationships of the hat-shaped ascospore-forming, nitrate-assimilating Pichia species, formerly classified in the genus Hansenula Sydow et Sydow, based on the partial sequences of 18S and 26S ribosomal RNAs (Saccharomycetaceae): the proposal of three new genera, Ogataea, Kuraishia, and Nakazawaea. Biosci Biotechnol Biochem 58:1245–1257
Yamada Y, Matsuda M, Maeda K, Mikata K (1995) The phylogenetic relationships of methanol-assimilating yeasts based on the partial sequences of 18S and 26S ribosomal RNAs: the proposal of Komagataella gen. nov. (Saccharomycetaceae). Biosci Biotechnol Biochem 59:439–444
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Acknowledgments
This work was partially supported by the Thailand Graduate Institute of Science and Technology (TGIST) grant TGIST 01-52-030. The authors would like to thank Assoc Prof Dr Hiroya Yurimoto, Laboratory of Microbial Biotechnology, Graduate School of Agriculture, Kyoto University for allowing to use the microscope for taking yeast micrographs.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koowadjanakul, N., Jindamorakot, S., Yongmanitchai, W. et al. Ogataea phyllophila sp. nov., Candida chumphonensis sp. nov. and Candida mattranensis sp. nov., three methylotrophic yeast species from phylloplane in Thailand. Antonie van Leeuwenhoek 100, 207–217 (2011). https://doi.org/10.1007/s10482-011-9579-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-011-9579-9