Abstract
A novel species in the genus Candida was obtained from deep-sea hydrothermal fields on the Mid-Atlantic Ridge. Strains Mo39, MARY089 and CBS 5307, respectively, isolated from an unidentified deep-sea coral collected near Rainbow hydrothermal vent, from water samples near Menez Gwen hydrothermal field and from the stomach of a marine fish are considered as a novel taxon. Sequence similarities in the D1/D2 region of the 26S rRNA gene indicated that strains Mo39, MARY089 and CBS 5307 have for closest neighbors Candida spencermartinsiae, Candida taylorii, Candida atmosphaerica and Candida atlantica. The strains, respectively, differ from C. spencermartinsiae, C. taylorii, C. atmosphaerica andCandida atlantica by 4, 4.3, 4.3 and 4.7% in the D1/D2 domain. Strains Mo39, MARY089 and CBS 5307 were differentiated from others by differences in the ability to assimilate d-Gluconate and in the ability to grow at relatively high temperature. Only strain Mo39 displays an optimal growth at 3% sea salts, indicating that this strain is clearly adapted to live in marine conditions. Sequence similarities between strains Mo39, MARY089 and CBS 5307 and related species and differences in the ability to utilize specific carbon compounds revealed that these strains represent a hitherto unknown species. Sexual reproduction was not observed in strains Mo39, MARY089 and CBS 5307. An anamorphic name Candida oceani sp. nov. is proposed for the type strain Mo39T (= CBS 11857T = DSM 23777T) and the two other strains MARY089 and CBS 5307. To our knowledge, this is the first description of a micro-eukaryotic organism including a strain isolated from a deep-sea coral near a hydrothermal ecosystem.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Yeasts are microorganisms that represent a significant part of the microbiota in all natural ecosystems. Many studies have reported the occurrence of yeasts in heterogeneous substrates such as soils, plants, animals and other organic matter in aquatic and marine environments including shallow-waters but also deep-sea waters and sediments (Nagahama 2005). Recently, a survey was realized in order to better characterize the yeast communities that occurred at deep-sea hydrothermal vents (Burgaud et al. 2010). This work led to the isolation of yeasts from different type of substrates, from seawater to endemic animals as Rimicaris exoculata shrimps, Bathymodiolus azoricus mussels and even deep-sea corals. Most of the positive cultures were obtained from samples collected at Rainbow hydrothermal site (Mid-Atlantic Ridge). More precisely, isolates were cultivated from deep-sea hydrothermal shrimps and mussels suggesting an association between yeast communities and hydrothermal fauna. The strain collection established mainly consists of commonly observed yeasts as Rhodotorula mucilaginosa and Debaryomyces hansenii, two of the most ubiquitous yeast species known (Lachance and Starmer 1998), but some correspond to already known marine endemic species as Rhodosporidium diobovatum and Candida atlantica. Previously, several new yeast species were isolated from deep-sea samples. These new deep-sea species were affiliated to the genera Kluyveromyces (Nagahama et al. 1999), Cryptococcus (Nagahama et al. 2003a), Rhodotorula (Nagahama et al. 2001, 2003b, 2006) and Dipodascus (Nagahama et al. 2008). Three yeast strains respectively identified as Mo39, MARY089 and CBS 5307 were isolated from an unidentified deep-sea coral near Rainbow hydrothermal vent, from water samples near Menez Gwen hydrothermal field and from the stomach of a marine fish. These strains showed more than 4% nucleotide divergence in the D1/D2 domain of the LSU rRNA from their closest relatives Candida spencermartinsiae, Candida taylorii, Candida atlantica and Candida atmosphaerica. These strains also differ morphologically and in several physiological tests from related species, suggesting that Mo39, MARY089 and CBS 5307 represent a new species in the Candida genus.
Materials and methods
Sample collection, isolation and conventional characterization
For strain Mo39, samples were collected from Rainbow hydrothermal site (36°08′N, 34°00′W, −2300 m) during MoMARDREAM-Naut oceanographic cruise (08/07/2007–19/07/2007) on the Mid-Atlantic Ridge. Sampling was processed using the manned submersible Nautile. To prevent any contamination by open water, we used sterilized insulated boxes filled with sterile seawater as described by Burgaud et al. (2009). On board, hydrothermal samples were rinsed several times with sterile seawater before being plated on culture media GYPS (Per liter: Glucose, 1 g; Yeast Extract, 1 g; Starch, 1 g; Peptone, 1 g and Sea salts, 30 g). Yeast isolates were then purified using the same culture medium. Long-term storage was performed in this culture medium with 5% DMSO (Dimethyl sulfoxyde) at −80°C. For strain MARY089, samples were collected from Menez Gwen hydrothermal site (37°50′N, 31°31′W, −825 m) during SEAHMA-1 oceanographic cruise. Yeast isolate was obtained on MYPssS (0.7% malt extract, 0.05% yeast extract, 0.25% peptone soytone, 3% sea salts (Sigma), 0.605% PIPES buffer (Sigma), 1% sulfur, and 1.5% agar supplemented with 0.05% chloramphenicol) as described by Gadanho and Sampaio (2005). The strain CBS 5307 was isolated from the stomach of a marine fish on GYPA culture medium composed of 4% glucose, 0.5% peptone, 0.5% yeast extract and 1.5% agar (unpublished data, CBS). Sporulation tests were performed for the three strains on GYPS, yeast malt extract agar (YM), potato-dextrose agar, corn meal agar, acetate agar and malt extract agar at 25°C for 6 weeks. Fermentation of carbohydrates and carbon assimilation tests were assessed according to standard methods (Yarrow 1998). Urea hydrolysis was tested on a solid medium using BBL urea agar slants (Difco) according to manufacturer’s recommendations.
Specific characterization
Adaptation to marine conditions was assessed using GYPS broth media with different sea salt concentrations: 0, 1.5, 3, 4.5 and 6%. The effect of temperature was also determined at 5, 15, 25, 35 and 40°C. Experiments were done in triplicate. Optical densities (OD) were measured at 600 nm with a Nanocolor 100D (Macherey–Nagel) at 17, 22, 25 and 28 h of growth under each conditions of temperature and salinity.
Sequence analysis
Cells were harvested from a 3-day-old culture on GYPS medium. DNA was extracted using FastDNA Spin Kit (MP Biomedicals) specific for fungi and yeasts. Amplifications of the D1/D2 region of 26S rRNA gene, small subunit and internal transcribed spacers including 5.8S rRNA genes were processed as described by Gadanho and Sampaio (2005) and White et al. (1990). All PCR reactions were performed as described earlier (Burgaud et al. 2010). Amplified DNA fragments were visualized by electrophoresis separation in 0.8% agarose gel (Promega) in 0.5× Tri-Borate-EDTA (TBE) Buffer at 90 V for 1 h and stained with ethidium bromide. A molecular size marker was used for reference (Lambda DNA/EcoR1 + Hind III Markers, Promega). DNA banding patterns were visualized under UV transillumination and picture files were generated using Gel-Doc 2000 (Biorad). Sequences were obtained by “Big Dye Terminator” technology (Applied Biosystems) realized at “Ouest Genopole” sequencing facility in the “Station Biologique de Roscoff” (www.sb-roscoff.fr). Chromatograms obtained were translated in nucleotidic sequences with Sequencher v 4.8 (Gene Codes). After cleaning, sequences were imported to MEGA 4.0 software (Tamura et al. 2007). Similarities between sequences were assessed using pairwise distance calculation with MEGA 4.0. Sequences were trimmed to ensure that all sequences had the same start and end-point. Alignments were processed using ClustalW v.1.83 (Thompson et al. 1994). After visual checking and manual curation, alignments were analysed using MODELTEST v.3.7 (Posada and Crandall 1998), in order to obtain the more realistic evolutionary model used for phylogenetic analyses. Phylogenies were then evaluated using three different methods: (i) Bayesian inference with MrBayes v.3.1.2 software (Ronquist and Huelsenbeck 2003) analysis using 2,000,000 generations and the mcmc method. The tree search included two mcmc searches with four chains (setting default temperature for heating the chains) and a sampling frequency of 100 generations. A ‘burnin’ of 5,000 (25% of the 2,000,000 generations/100 sample frequency) was set in order to exclude the first 5,000 trees generated. (ii) Maximum likelihood with 100 bootstrap iterations using PHYML (Guindon et al. 2005) and the parameters obtained with MODELTEST v.3.7. (iii) Neighbor-Joining with 2,000 bootstrap iterations using MEGA4.0 with Maximum Likelihood Composite model. Final phylogenetic tree topology was realized using MrBayes v.3.1.2 analysis results. Nodes in the tree show ML and NJ bootstraps and Bayesian posterior probabilities.
Results and discussion
Deep-sea coral samples were collected near Rainbow hydrothermal vents during the MoMARDREAM-Naut oceanographic cruise in 2007. Aseptically sampled deep-sea corals were used for yeast isolation on GYPS media. One yeast identified as Mo39 was isolated from an unidentified deep-sea coral sample. This isolate has grown directly on coral substrate and has been purified by streaking.
An initial megablast similarity search in the NCBI database using D1/D2 domain of the LSU rRNA gene allowed the identification of similar sequences. Interestingly, a strain harbouring a close sequence (one substitution and two indels) was previously isolated and identified as MARY089 (Gadanho and Sampaio 2005). These authors reported the occurrence and diversity of yeasts in the Mid-Atlantic hydrothermal fields. Strain MARY089 was isolated from a water sample collected at Menez Gwen hydrothermal site. Authors indicated that their isolate was a new phylotype but no further description has been published. Recently, Nguyen et al. (2009) suggested that Candida sp. MARY089 and the strain Candida sp. CBS 5307, previously described as Debaryomyces hansenii, were conspecific. CBS 5307 harbours a close sequence to Mo39 (one substitution). These two strains were integrated in our analyses to know whether they represent a distinctive species with Mo39. Regarding the megablast similarity search, the next most similar published sequences with valid species names were those of Candida atlantica (95.3% identity) and Candida atmosphaerica (95.8% identity) strains. According to Kurtzman and Robnett (1998), nucleotide substitutions >1% are indicative of separate species. Here, the >4% difference from the closest sequences suggests that these deep-sea (Mo39 and MARY089) and marine (CBS 5307) isolates represent a hitherto undescribed taxon. The chromosomal regions coding for the small subunit 18S rRNA gene, ITS1, 5.8S and ITS2 were also sequenced for strain Mo39. This strain differs from C. atlantica and C. atmosphaerica, respectively, by 1.2 and 1.3% in the SSU rRNA and 9.1 and 8.2% in the internal transcribed spacer region. Recently, two novel species close to Candida atlantica and Candida atmosphaerica were described (Statzell-Tallman et al. 2010). Candida spencermartinsiae and Candida taylorii were isolated from seawater at a water depth of 10 m adjacent to corals at Looe Key Reef, Florida, USA. Sequence similarities of D1/D2 region of the 26S rRNA gene between the three strains Mo39, MARY089 and CBS 5307 and C. spencermartinsiae and C. taylorii were, respectively, 96 and 95.6%. The chromosomal region coding for the internal transcribed spacers was also analyzed. Strain Mo39 differs from C. spencermartinsiae and C. taylorii, respectively, by 9.9 and 32.3%.
To determine the phylogenetic position, one phylogenetic tree was designed from D1/D2 region of the 26S rRNA gene sequences of the isolates Mo39 and MARY089, the most similar sequences and members of the Yamadazyma clade as defined by Kurtzman and Suzuki 2010 (Fig. 1). This phylogenetic tree shows that Mo39, MARY089 and the conspecific strain CBS 5307 are close to C. taylorii, C. spencermartinsiae, C. atlantica and C. atmosphaerica, supporting the fact that strains are members of the Candida genus. Significant bootstrap values indicate that strains Mo39, MARY089 and CBS 5307 form a distinct taxon in the Candida genus. All these phylogenetic results confirm that the isolates, respectively, obtained from an unidentified deep-sea coral (Mo39), from water sample (MARY089) near Rainbow and Menez Gwen hydrothermal vents and from the stomach of a marine fish (CBS 5307) represent a new species.
D1/D2 rRNA phylogenetic tree of Candida oceani sp. nov Mo39T and related reference taxa in the Yamadazyma clade (Kurtzman and Suzuki, 2010). Node support values are given in the following order: PHYML 100 bootstrap values/MrBayes posterior probabilities/Neighbor-Joining 2000 bootstrap values. Values below 50% are represented with a minus sign. GenBank accession numbers of all sequences are indicated after strain name. Schizosaccharomyces pombe was used as outgroup
As strains Mo39, MARY089 and CBS 5307 were isolated from marine samples, one aim was to know whether these isolates display specific adaptations to marine conditions. Clear establishment of endemism or ubiquity of marine yeasts is a difficult task. The physiological properties of yeasts generally do not give clues as to whether they are marine or terrestrial species. Almost all yeast species can grow well in media with NaCl concentrations exceeding those normally present in the sea (Kohlmeyer and Kohlmeyer 1979). Figure 2 clearly indicates that strain Mo39 displays an optimal growth at 3% sea salts indicating that this yeast is adapted to live in marine conditions. Moreover, to date, strain Mo39 and close isolates are confined to marine habitats as Mo39 and MARY089 were harvested only in deep-sea ecosystems and Candida sp. CBS 5307 was retrieved from the stomach of a fish. Candida taylorii CBS 8508 and Candida spencermartinsiae CBS 10894 were harvested in seawater near corals. All those results indicate that these yeasts are forming a cluster of marine species. However, only Mo39 displays a clear adaptation to marine conditions as strains MARY089 and CBS 5307 were able to grow with sea salts but growth rates were lower with increasing sea salt concentrations (data not shown). Based on the definition of obligate marine yeasts, that are those yeasts that, thus far, have never been collected anywhere but in the marine environment (Kohlmeyer and Kohlmeyer 1979), strains Mo39, MARY089 and CBS 5307 can be considered as obligate marine yeasts. Regarding Mo39, this is the first report of an obligate marine yeast with a physiological dependence on NaCl.
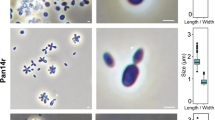
Microscopic observations of Mo39 (Fig. 3a), C. atlantica CBS 5263 (Fig. 3f) and C. atmosphaerica CBS 4547 (Fig. 3g) allowed clear distinction between the three strains. Cells of C. atlantica are defined as globose to ovoidal with multipolar buds (Meyer et al. 1998) but microscopic observations allowed to visualize elongated cells forming primitive pseudohyphae. Cells of C. atmosphaerica are globose to elongated and multilateral buds can also be visualized. Main differences between strain Mo39 and the two others are the cellular size and shape. Cell morphology of Mo39 is close to Candida sp. MARY089 (Fig. 3b), Candida spencermartinsiae (Fig. 3c) and Candida taylorii (Fig. 3d) except for the pseudohyphae formation. Cells of Candida sp. CBS 5307 (Fig. 3e) are globose and no ovoidal cells were visualized.
aCandida oceani sp. nov. Mo39T (= CBS 11857T = DSM 23777T). bCandida oceani sp. nov. MARY089, cCandida spencermartinisae CBS 10894 (Taken from Statzell-Tallman et al. 2010 with permission from Jack Fell), dCandida taylorii CBS 8508 (Taken from Statzell-Tallman et al. 2010 with permission from Jack Fell), eCandida oceani sp. nov. CBS 5307, fCandida atlantica CBS 5263, gCandida atmosphaerica CBS 4547. Cells visualized after 3 days at in GYPS medium a, b, e, f, g, after 3 days in malt extract broth (c) and after 7 days on cornmeal agar (d). (Bars 5 μm except c and d 10 μm)
Physiological data regarding Candida atlantica CBS 5263 and Candida atmosphaerica CBS 4547 indicate that these two yeasts are not able to grow at 35°C. Maximal growth were thus at temperatures comprised between 30 and 35°C unlike Mo39, MARY089 and CBS 5307 which are able to growth at 35°C (Table 1). Considering their physiological characteristics Candida sp. Mo39, Candida sp. MARY089 and Candida sp. CBS 5307 are rather close to Candida taylorii and Candida spencermartinsiae but Candida sp. Mo39, Candida sp. MARY089 and Candida sp. CBS 5307 are not able to assimilate d-gluconate.
The phenotypic and molecular data presented above suggest that the strains Mo39, MARY089 and CBS 5307 are divergent enough from all known species in the genus Candida. Therefore, we propose the name Candida oceani sp. nov. for these strains and designate strain Mo39T as the type strain of novel species.
Latin diagnosis of Candida oceani Burgaud and Barbier sp. nov
In medio liquido GYPS post 3 dies ad 35°C, cellulae exiguus (2–3 μm × 4–5 μm), ovoideae vel ellipsoidae, singulae aut binae. Flosculi sunt polares. Post 21 dies ad 25°C in agaro GYPS, exigua colonia (0.5-1 mm) albida vel cremea, rotundas et marginis integer. Pseudohyphae et hyphae non formantur. Ascosporae non fiunt.
Glucosum (lente) fermentantur, at non, galactosum, sucrosum, maltosum, lactosum, trehalosum raffinosum. d-glucosum, d-galactosum, l-sorbosum, sucrosum, maltosum, cellobiosum, trehalosum, melezitosum, d-xylosum, l-arabinosum, d-arabinosum (lente), d-ribosum (lente), l-rhamnosum, d-glucosaminum (lente), N-acetyl-glucosaminum, ethanolum, glycerolum, erythritolum, ribitolum, d-mannitolum, d-glucitolum, methyl-α-d-glucosidum, salicinum, acidum succinicum (lente et exigue), acidum citricum, acidum 2-keto-d-gluconicum (lente), acidum 5-keto-d-gluconicum, xylitolum, l-arabinitolum, arbutinum (lente et exigue) assimilantur at non lactosum, melibiosum, raffinosum, inulinum, methanolum, galactitolum, acidum d-gluconicum, acidum d l-lacticum, hexadecanum, nitratum, nitritum, acidum saccharicum. Ad crescentiam vitaminae additae necesseriae sunt. Creatininum non assimilantur. Crescit in 10% NaCl/5% d-glucoso. Non crescit in 0.01% et 0.1% cycloheximido. Diazonium caeruleum B non repondens. Ureum non hydrolysatur. Maris sales accretionem meliorem faciunt. Typus Mo39T (= CBS 11857T = DSM 23777T) in altissimo mari ab altis corallis divisa est. Isolatus depositus in collectione zymotica Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands et Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
Description of Candida oceani Burgaud and Barbier sp. nov
Candida oceani (o.ce.a’ni. L. gen. n. oceani, of an ocean, referring to its optimal growth under marine conditions).
In GYPS broth, after a 3 days culture at 35°C, cells are small (2–3 μm wide × 4–5 μm long), ovoid to ellipsoid and occur often singly or seldom in parent-bud pairs (Fig. 3a). Budding is lateral and only single bud per cell was observed. After 3 weeks on GYPS agar, colonies (0.5–1 mm) are creamy-white, smooth, round, convex and have an entire margin. No pseudomycelium or true hyphae are formed. No sporulation was observed. The physiological characteristics presented in Table 1 show that C. oceani and the most closely related species differ in numerous properties, which allow their differentiation by conventional taxonomic tests associated with one complementary test as the growth rate under increasing sea salt concentrations. Significant differences are the inability of C. oceani to assimilate d-gluconate and one strain displays an optimal growth at 3% sea salts in culture medium.
The type strain is Mo39T (= CBS 11857T = DSM 23777T), which was isolated from an unidentified deep-sea coral collected near Rainbow hydrothermal site at Mid-Atlantic Ridge (2300 meters depth), Atlantic Ocean, while two other strains (MARY089 and CBS 5307) of this species were found respectively in water samples from Rainbow hydrothermal site and from the stomach of a fish.
References
Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G (2009) Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol 11:1588–1600
Burgaud G, Arzur D, Durand L, Cambon-Bonavita MA, Barbier G (2010) Marine culturable yeasts in deep-sea hydrothermal vents: species richness and association with fauna. FEMS Microbiol Ecol 73:121–133
Gadanho M, Sampaio J (2005) Occurrence and diversity of yeasts in the Mid-Atlantic Ridge hydrothermal fields near the azores archipelago. Microb Ecol 50:408–417
Guindon S, Lethiec F, Duroux P, Gascuel O (2005) PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 33:557–559
Kohlmeyer J, Kohlmeyer E (1979) Marine Mycology: the Higher Fungi. Academic Press, New York, USA
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371
Kurtzman CP, Suzuki M (2010) Phylogenetic analysis of ascomycete yeasts that form coenzyme Q-9 and the proposal of the new genera Babjeviella, Meyerozyma, Millerozyma, Priceomyces, and Scheffersomyces. Mycoscience 51:2–14
Lachance MA, Starmer WT (1998) Ecology and yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier Science B.V, Amsterdam, pp 21–30
Meyer SA, Payne RW, Yarrow D (1998) Candida Berkhout. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 454–573
Nagahama T (2005) Yeast biodiversity in freshwater, marine and deep-sea environments. In: Rosa C and Peter G (eds) The yeasts handbook. Springer, Berlin, Heidelberg, New York, pp 241–262
Nagahama T, Hamamoto M, Nakase T (1999) Kluyveromyces nonfermentans sp. nov. a new yeast species isolated from the deep sea. Int J of Syst Evol Micr 49:1899–1905
Nagahama T, Hamamoto M, Nakase T, Horikoshi K (2001) Rhodotorula lamellibrachii sp. nov. a new yeast species from a tubeworm collected at the deep-sea floor in Sagami Bay and its phylogenetic analyses. Antonie Van Leeuwenhoek 80:317–323
Nagahama T, Hamamoto M, Nakase T, Takaki Y, Horikoshi K (2003a) Cryptococcus surugaensis sp. nov. a novel yeast species from sediment collected on the deep-sea floor of Suruga Bay. Int J of Syst Evol Micr 53:2095–2098
Nagahama T, Hamamoto M, Nakase T, Horikoshi K (2003b) Rhodotorula benthica sp. nov. and Rhodotorula calyptogenae sp. nov. novel yeast species from animals collected from the deep-sea floor and Rhodotorula lysiniphila sp. nov. which is related phylogenetically. Int J of Syst Evol Micr 53:897–903
Nagahama T, Hamamoto M, Horikoshi K (2006) Rhodotorula pacifica sp. nov. a novel yeast species from sediment collected on the deep-sea floor of the north-west Pacific Ocean. Int J of Syst Evol Micr 56:295–299
Nagahama T, Abdel-Wahab MA, Nogi Y, Miyazaki M, Uematsu K, Hamamoto M, Horikoshi K (2008) Dipodascus tetrasporeus sp. nov. an ascosporogenous yeast isolated from deep-sea sediments in the Japan Trench. Int J of Syst Evol Micr 58:1040–1046
Nguyen HV, Gaillardin C, Neuvéglise C (2009) Differentiation of Debaryomyces hansenii and Candida famata by rRNA gene intergenic spacer fingerprinting and reassessment of phylogenetic relationships among D. hansenii, C. famata, D. fabryi, C. flareri (= D. subglobosus) and D. prosopidis: description of D. vietnamensis sp. nov. closely related to D. nepalensis. FEMS Yeast Res 9:641–662
Posada D, Crandall K (1998) Applications note. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818
Ronquist F, Huelsenbeck J (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Statzell-Tallman A, Scorzetti G, Fell JW (2010) Candida spencermartinsiae sp. nov., Candida taylorii sp. nov. and Pseudozyma abaconensis sp. nov., novel yeasts from mangrove and coral reef ecosystems. Int J of Syst Evol Micr 60:1978–1984
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) Software version 4.0. Mol Biol Evol 24:1596
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Chap 38. PCR Protocols: a guide to methods and applications. Orlando, Florida: Academic Press. pp 315–322
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier B.V, Amsterdam, The Netherlands, pp 77–100
Acknowledgments
We thank the chief scientist of the MoMARDREAM-Naut cruise, pilots and support crews of oceanographic vessels and Deep Submergence Vehicles of Ifremer. We greatly acknowledge Justine Amborski for valuable help for latin diagnosis. We thank all members of the GDR Ecchis for discussions and suggestions. We would like to thank the editor and the anonymous reviewers who have provided helpful comments on the refinement of this manuscript. We also thank ANR Deep-Oases and French Research Ministry for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Burgaud, G., Arzur, D., Sampaio, J.P. et al. Candida oceani sp. nov., a novel yeast isolated from a Mid-Atlantic Ridge hydrothermal vent (−2300 meters). Antonie van Leeuwenhoek 100, 75–82 (2011). https://doi.org/10.1007/s10482-011-9566-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-011-9566-1