Abstract
Yeast populations in the Shark River Slough of the Florida Everglades, USA, were examined during a 3-year period (2002–2005) at six locations ranging from fresh water marshes to marine mangroves. Seventy-four described species (33 ascomycetes and 41 basidiomycetes) and an approximately equal number of undescribed species were isolated during the course of the investigation. Serious human pathogens, such as Candida tropicalis, were not observed, which indicates that their presence in coastal waters is due to sources of pollution. Some of the observed species were widespread throughout the fresh water and marine habitats, whereas others appeared to be habitat restricted. Species occurrence ranged from prevalent to rare. Five representative unknown species were selected for formal description. The five species comprise two ascomycetes: Candida sharkiensis sp. nov. (CBS 11368T) and Candida rhizophoriensis sp. nov. (CBS 11402T) (Saccharomycetales, Metschnikowiaceae), and three basidiomycetes: Rhodotorula cladiensis sp. nov. (CBS 10878T) in the Sakaguchia clade (Cystobasidiomycetes), Rhodotorula evergladiensis sp. nov. (CBS 10880T) in the Rhodosporidium toruloides clade (Microbotryomycetes, Sporidiobolales) and Cryptococcus mangaliensis sp. nov. (CBS 10870T) in the Bulleromyces clade (Agaricomycotina, Tremellales).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Everglades, which is the largest subtropical wetland in the United States, occurs in the southeastern state of Florida. The system spans from the fresh water Lake Okeechobee south to Florida Bay, which is a lagoonal estuary. Water moves from north to south as a slow river that is 97 km wide and over 160 km long. Over the past 100 years, the water flow has been channeled and diverted as South Florida has undergone changes to accommodate urban growth and agricultural practices. With national attention to loss of natural environments, attempts are being made to restore parts of the Everglades ecosystem. Concurrently, environmental studies are in progress to examine the ecological changes that occur during the restoration process.
One of these studies, the Florida Coastal Everglades Long Term Ecological Research (LTER) is monitoring (http://fcelter.fiu.edu/) the Shark River Slough (SRS, Fig. 1), which is a major distributor of fresh water within the Everglades. The upper reaches of the SRS are a fresh water habitat dominated by a sawgrass community, whereas the lower reaches are characterized by an estuarine mangrove habitat. The water sheet flow is seasonally driven with greater intrusions of estuarine waters during the dry season. As part of the LTER studies, we are examining the yeast populations and communities associated with the different habitats in the SRS. One of the characteristics of the SRS yeast communities is the presence of novel species (Statzell-Tallman and Belloch 2008). In the following report, a formal description of five new species from the Florida Everglades ecosystem and their relationships to ecological conditions and other members of the yeast community are presented.
Methods
Sample collection and isolation
Sampling sites were in the Shark River Slough (SRS, Fig. 1), which is on the southwest corner of the Florida Everglades. The entire region is located in a subtropical moist environment with distinct wet (June–Nov) and dry (Dec–May) seasons and a seasonally driven freshwater sheet flow. The Shark River Slough initiates in a fresh water marsh and terminates in a mangrove habitat in Florida Bay and the Gulf of Mexico. Collections were made at six sites (Fig. 1) that are maintained and routinely sampled for physical and biological data by the Florida International University (FIU) National Science Foundation (NSF) Long Term Ecological Research (LTER) program. Detailed information for each site can be obtained at the Florida Coastal Everglades LTER web site: http://fcelter.fiu.edu/.
Stations SRS 1a, 2 and 3 are located in freshwater wetlands dominated by sawgrass (Cladium jamaicense) interspersed with spike rushes (Eleocharis cellulosa) and maidencane (Panicum hemitomon). Stations SRS 4, 5 and 6 are in the slough with tidally driven oceanic inputs within a habitat of red (Rhizophora mangle) and black (Avicennia germinanus) mangrove trees. At SRS 4 the trees have a dwarf stature (~2–4 m high), at SRS 5 an intermediate stature (~6 m), whereas at SRS 6 the trees are tall (~12 m). The tidal influence is apparent from the variations in salinity at the different times of collection. Specifically, the salinities at the stations over times of our collections were; SRS 1a: 0 ppt, SRS 2 and SRS 3: 0–2 ppt, SRS 4: 0–17 ppt, SRS 5: 0–30 ppt and SRS 6: 12–33 ppt.
Eight collections were taken in the Shark River Slough. All stations were sampled during five collections: Collection (C) 1-July 2002, C2-November 2002, C3-February 2003, C4-June 2003 and C8-September 2004. The remaining three collections were: C5-October 2003 at station SRS 1a, 2, 3, 4 and 6, C7-April 2004 at stations SRS 1a and 4, and C9-March 2005 at stations SRS 3, 4, 5 and 6. The samples from inland stations 1a, 2 and 3 were collected from a motorboat or from an airboat during periods of low water. Within a few days, stations 4, 5 and 6, which were consistently in deeper water, were sampled from a motorboat. Surface waters (30 cm below the surface) in the SRS were collected in sterile, one liter Nalgene bottles. Three samples were collected at each station to increase the accuracy of the estimate of population variability. The samples were kept at 5–12°C until processed, usually within 24 h. Preliminary sampling results showed widely variable total numbers of cfu at each station. Consequently, a series of aliquots (10, 25, 50 and 100 ml) was filtered through 0.45 μm pore size cellulose acetate Millipore filters for each sample. The filters were placed on enrichment agar in 50 mm Millipore petri dishes. The enrichment medium (5.0 ml) consisted of 2% glucose, 1% peptone and 0.5% yeast extract agar, with 0.02% chloramphenicol to retard bacterial growth. The plates were incubated at 17°C. Incubation of samples from high nutrient waters require temperatures below ambient to slow the growth of filamentous fungi, which are prevalent in these environments. The resulting colonies were counted and streaked onto enrichment agar for purity within 7–10 days of sample filtration. The isolates were grown in duplicate in 1 ml of glucose peptone yeast extract (GPY) broth in 2 ml microcentrifuge tubes on a roller drum for 24–48 h. A separate collection was made north of the SRS in the mangroves of the Florida Everglades National Park at Chatham Bend (25°41′N, 85°1′W) on 10/23/1996.
Morphological and physiological analyses
The morphological descriptions and physiological tests followed modified methods of Yarrow (1998). The carbon and nitrogen assimilation assays were tested in 2-ml microcentrifuge tubes with 1 ml of liquid media. Culture tube lids were tight to avoid contamination. Based on multiple controls with cells inoculated in tubes with glucose media, neither oxygen depletion (as indicated by growth rates) nor gas build up, were recorded. The tubes were placed on a roller drum modified to accept the microcentrifuge tubes. Readings were taken after 3 days, 1, 2 and 3 weeks.
Phylogenetic analysis
DNA extraction followed the methods of Fell et al. (2000). Alternatively, DNA was directly amplified from a light cell suspension in water, as the DNA template, in 50 ml PCR reactions with a HotMaster Kit (Eppendorf North America, Westbury, NY). Molecular sequence analysis of the D1/D2 domains of the LSU and the ITS1, 5.8S and ITS2 rRNA gene regions followed the procedures of Fell et al. (2000) and Scorzetti et al. (2002). Trees, based on the D1/D2 LSU rDNA region were constructed based on likelihood analysis (heuristic search, stepwise addition) with bootstrap values calculated with 1000 replicates.
Results and discussion
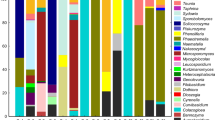
Yeasts were isolated from eight collections at six stations in the Shark River Slough (SRS) from July 2002 to June 2005. Seventy-four previously described species, and an approximately equal number of undescribed species, were isolated during the course of the investigation. The described species, including the five previously undescribed species that we selected to describe in this study, consisted of 35 ascomycetes and 44 basidiomycetes (Table 1). The habitat distribution of these yeast species included sawgrass stations (Stas. 1a–3): ascomycetes seven species, basidiomycetes 17 spp; mangrove stations (Stas 4–6) ascomycetes 15 spp, basidiomycetes 8 spp; mixed sawgrass and mangrove stations ascomycetes 13 spp. and basidiomycetes 19 spp.
Many of the species, particularly those species, which were restricted to sawgrass or mangrove habitats, were only encountered during one or two collections, e.g., Candida dendronema and Bullera sinensis (sawgrass stations) and Candida choctaworum and Auriculibuller fuscus (mangrove stations). There were species that were isolated at multiple times or stations within the individual habitats, for example Kregervanrija fluxuum and Sporobolomyces beijingensis in the sawgrass and Candida thaimueangensis and Kwoniella mangroviensis in the mangroves. There were also species that were widespread throughout both habitats and occurred at multiple collections, e.g., Candida silvae and Cryptococcus laurentii.
The role of these species in the Everglades system is difficult to pinpoint, due largely to our meager knowledge of yeast ecology. A common theme of the majority of these species is that their habitats, as known from published observations, are plant material or invertebrates associated with plants. Of particular note are: Candida amphixiae, which has only one known strain that was isolated from the gut of a fungus beetle in Panama (Suh et al. 2005) and Candida carpophila, which has close associations with insects (Lachance et al. 2010). Some of these species are only known from mangrove, marsh or other marine environments for example: Candida thaimueangensis from mangrove forests in Thailand (Limtong et al. 2007), Candida maris from the great barrier reef in Australia (van Uden and Zobell 1962), Kluyveromyces aestuarii, which occurs widely in North and South America in mangrove sediments and in association with specific species of mangrove crabs (de Araujo et al. 1995; Lachance et al. 2010). Similarly, Lachancea meyersii (Fell et al. 2004), Kwoniella mangroviensis (Statzell-Tallman and Belloch 2008) and Rhodosporidium paludigenum (Sampaio 2010) are mangrove-associated yeasts. Additionally, the SRS list includes species reported from both terrestrial and marine sources: Rhodosporidium diobovatum, Rhodosporidium sphaerocarpum (Sampaio 2010), Pseudozyma aphidis (Boekhout 2010) and C. silvae, which is abundant in invertebrates in Brazilian mangroves (de Araujo et al. 1995) and has been isolated from terrestrial sources such as a horse intestine (Lachance et al. 2010).
A few of the SRS isolated species (Cryptococcus liquefaciens, Candida glabrata, C. parapsilosis and C. silvae) have been isolated from humans. However, there did not appear to be an occurrence of the more important clinically related yeasts, Candida tropicalis, Candida albicans and Trichosporon asahii, which were reported at bathing beaches in south Florida (Vogel et al. 2007). Similarly, Papadakis et al. (1997) reported C. albicans, C. tropicalis and Trichosporon spp at bathing beaches in Greece. In a study along the coast of China, Yan et al. (2010), indicated that C. tropicalis has a wide distribution in marine environments, where the species may play an important role. In addition, we isolated C. tropicalis in the vicinity of human inhabitations from waters of the reef off Rendezvous Cay, Belize and from mangrove habitats of the Monkey River and Middle Long Cay, Belize (Fell and Statzell-Tallman, unpublished data). In contrast, Lazarus and Koburger (1974) in a study of the Suwannee River, Florida estuary, did not report the presence of these human pathogens. In concurrence with the SRS data, Cryptococcus laurentii was a prevalent species. The absence of C. tropicalis, C. albicans and Trichosporon spp in the natural environments of SRS and the Suwannee River estuary, suggests that their occurrence in marine habitats may indicate the presence of pollution.
Our SRS data demonstrated that there were as many undescribed as described species in the mangrove and sawgrass ecosytems. As part of the examination of mangrove fungi, our research provided the formal descriptions of species from the SRS and from Bahamian mangrove habitats (Fell et al. 2004, Statzell-Tallman and Belloch 2008, Statzell-Tallman et al. 2010). The present report presents the formal descriptions of five representative species, which have similar distribution patterns to the described species (Table 1) that range from abundant and widely distributed to rarely isolated. Two of the selected species are ascomycetes (Candida sharkiensis sp. nov. and Candida rhizophoriensis sp. nov.) and three are basidiomycetes (Cryptococcus mangaliensis sp. nov., Rhodotorula cladiensis sp. nov. and Rhodotorula evergladiensis sp. nov.). The species showed some indication of habitat preference. Candida sharkiensis was an abundant species in the SRS; 73 isolates were recorded from a broad salinity range (0–30 ppt). The majority of the strains were at stations 1a–4, with diminishing numbers at stations 5 and 6. Rhodotorula evergladiensis was repeatedly isolated at station 1a, with lesser numbers of recordings at stations 2, 3 and 6, which suggests that R. evergladiensis originated at Sta 1a and was carried downstream to the other locations. Similarly, the occurrence of C. sharkiensis at Stas 4, 5 and 6 may also be the result of southerly water flow. Among the less abundant species, Rhodotorula cladiensis (seven strains) was isolated at stations 1a, 2, 3 and 4. In contrast to the other species, which may have their origin in fresh water habitats, Candida rhizophoriensis (four strains) was isolated in the mangroves at Stations 4 and 6. Another mangrove habitat species, Cryptococcus mangaliensis did not appear to inhabit SRS, rather the source of isolation was at a separate location (Chatham Bend) in the Florida Everglades. The role of these species in the Everglades ecosystem is open to speculation. An in-depth study of the yeasts associated with the diversity of micro and macro-plant and insect species is required to identify the specific land- or water-based habitats or hosts.
Phylogenetic relationships
Phylogenetic analysis of D1/D2 domains of the LSU rRNA gene positioned Candida sharkiensis and C. rhizophoriensis in the Metschnikowia clade, which is comprised of 23 Candida species in four divergent subclades (Lachance et al. 2010). The neighbor-joining tree in Fig. 2 shows the position of C. sharkiensis and C. rhizophoriensis within one of these subclades and their phylogenetic relationship with other closely related species in the genera Clavispora and Metschnikowia. C. sharkiensis is in a cluster (bootstrap value <50%) with C. tsuchyiae, C. akabanensis, C. flosculorum, C. intermedia and C. pseudointermedia, which are, in general, plant associated species. For example, C. flosculorum was isolated from tropical flowers in Brazil (Rosa et al. 2007), C. tsuchiyae was collected from moss in Japan (Nakase and Suzuki 1985), C. akabanensis was isolated from frass of insects infesting the bark of a grape vine in Japan (Nakase et al. 1994) and C. intermedia has been isolated from various plants and soils (Lachance et al. 2010). In contrast, C. pseudointermedia was found in a traditional fish paste product in Japan (Nakase et al. 1976). The possibility exists that C. sharkiensis is associated with specific plants in the freshwater portion of the Everglades mangrove ecosystem.
Phylogenetic tree (likelihood analysis with heuristic search and stepwise addition) based on the LSU D1/D2 rRNA gene sequences, which shows the placement of Candida sharkiensis and Candida rhizophoriensis with related species. Bootstrap values were calculated with 1,000 replicates and reported on branches when higher than 50%
Candida rhizophoriensis is shown, in Fig. 2, as related to C. melibiosica (72% bootstrap value) with a low (86%) D1/D2 sequence identity. C. melibiosica occurred in both the sawgrass and mangrove habitats of the SRS; in addition, it has been isolated from environments such as wine, soil, insect tunnels in wood and clinical isolates (Lachance et al. 2010). Lazarus and Koburger (1974) reported the presence of C. melibiosica in low salinity areas of Suwannee River estuary in Florida, but with a low frequency of isolation.
Rhodotorulacladiensis is an urediniomycetous species, which belongs to the Sakaguchia group in the Erythrobasidiales of the Cystobasidiomycetes (Fig. 3). Bootstrap value in the D1/D2 domains is 100% for a branch cluster that includes Sakaguchia dacryoidea, Rhodotorula cladiensis and R. oryzae. The teleomorphic member of this cluster, S. dacryoidea, has a bifactorial mating system (Fell et al. 1973) and there appears to be intraspecific sequence variation within mating strains. Three mating strains were sequenced in the D1/D2 and ITS domains: the type strain CBS 6353 (mating type A1B1), CBS 6356 (mt A1B2) and CBS 7999 (mt A2B1). The latter strain mates with strain CBS 6356 although their sequences differ by 3 bp in D1/D2 and 14 bp in ITS (Scorzetti et al. 2002). Sequence analysis and fertility studies of mating types of multiple strains of S. dacryoidea from diverse geographical regions are required to understand the genetic variability in this species. The original mating studies of S. dacryoidea used strains from marine sources in the Indian and Pacific Oceans. In contrast, CBS 7999 was isolated from a brewery in France (http://www.cbs.knaw.nl/yeast/biolomics). Fertility studies between the marine and brewery strains have not been reported. Based on our observations, mating did not occur between strains of S. dacryoidea and R. cladiensis, which indicates the presence of genetically separate taxa, although a mating system in R. cladiensis has not been discovered. Sequence differences between the two species are significant. The D1/D2 base pair differences for R. cladiensis are 6 bp from S. dacryoidea strain CBS 6353 and 5 bp from strain CBS 7999.The ITS region differences for C. cladiensis are more than 20 bp from CBS 7999 and more than 30 bp from strains CBS 6356 and CBS 6353.
Phylogenetic tree (likelihood analysis with heuristic search and stepwise addition) based on the LSU D1/D2 rRNA gene sequences, which shows the placement of Rhodotorula cladiensis with related species. Bootstrap values were calculated with 1,000 replicates and reported on branches when higher than 50%
R. cladiensis also differs from S. dacryoidea in ribitol, DL lactate, succinate, citrate, hexadecane and 2-keto d-gluconate assimilation. Sakaguchia dacryoidea is a widespread species in the Antarctic waters (Fell et al. 1973), and was also found in more temperate waters along the coast of Portugal (Gadanho et al. 2003) as well as from field grasses in Georgia, USA (Allen et al. 2004). Rhodotorula oryzae was isolated as a single strain from paddy rice in Japan (Bai et al. 2004).
Rhodotorula evergladiensis is a urediniomycetous species, which clusters within the Order Sporidiobolales in a group of species, which includes Rhodotorula graminis, Rhodotorula glutinis, Rhodosporidium babjevae, Rhodosporidium diobovatum and strain CBS 9072 (Fig. 4). The group has a 97% support in the D1/D2 domains that decreases to 62% in the ITS regions (data not shown). R. evergladiensis differs from all the species in the branch cluster by at least 9 bp in the D1/D2 and 24 bp in the ITS region.
Phylogenetic tree (likelihood analysis with heuristic search and stepwise addition) based on the LSU D1/D2 rRNA gene sequences, which shows the placement of Rhodotorula evergladiensis with related species. Bootstrap values were calculated with 1,000 replicates and reported on branches when higher than 50%
Cryptococcusmangaliensis is a member of the Tremellomycetes and clusters with Bullera pseudoalba and Bullera hoabinhensis. Nucleotide differences from these two species are 4 and 2 bp, respectively, in the D1/D2 region (Fig. 5) with 100% bootstrap value. In the ITS regions the differences increase to 14 bp from B. pseudoalba and to 13 from B. hoabinhensis, while bootstrap values are 98% for the three species and 90% for the pair C. mangaliensis/B. pseudoalba (data not shown). Cryptococcusmangaliensis does not produce ballistoconidia, unlike the two Bullera spp. The species is positive for lactose assimilation and negative for d-arabinose assimilation. B. hoabinhensis was found as a single isolate in a National Park in Japan from a leaf of Anadendrum montanum, which is a flowering species in the Araceae family (Luong et al. 2005). All the strains of B. pseudoalba and the synonym, Cryptococcus cellulolyticus, were found on dead leaves or decaying wood in Japan (Nakase et al. 1996).
Phylogenetic tree (likelihood analysis with heuristic search and stepwise addition) based on the LSU D1/D2 rRNA gene sequences, which shows the placement of Cryptococcus mangaliensis with related species. Bootstrap values were calculated with 1,000 replicates and reported on branches when higher than 50%
Formal descriptions of the new species
Latin diagnosis of Candida sharkiensis Fell, Gutiérrez, Statzell-Tallman et Scorzetti sp. nov.
Post dies 3 ad 22°C in medio liquido (ME) cellulae ovoideae (1.3–5.2 × 1.7–10.2 μm), singulae et binatae. Post mensem unum sedimentum apparet at non pellicula, nec anulus. Post dies 7 ad 22°C in medio ME cum agaro cultura alba, lucida levisque. Post mensem unum ad 22°C in medio ME cum agaro ascosporae et hyphae non formantur. Post mensem unum ad 22°C in medio CM cum agaro mycelium non formatur.
Glucosum, galactosum, sucrosum, maltosum (lente) et trehalosum (lente) fermentantur. Lactosum et raffinosum non fermentantur. Glucosum, galactosum, l-sorbosum (lente), sucrosum, maltosum, cellobiosum, trehalosum, lactosum, melezitosum, d-xylosum, l-rhamnosum, d-glucosaminum (varie), ethanolum (lente), glycerolum (lente), d-mannitolum, d-glucitolum (lente), methyl-α-d-glucosidum (lente), salicinum, d-gluconicum (exigue), acidum 2-keto-d-gluconicum (exigue), N-acetyl-d-glucosaminum, acidum succinicum (lente), acidum citricum (lente), hexadecanum, creatininum et creatinum (lente) assimilantur. Melibiosum, raffinosum, inulinum, amylum solubile, l-arabinosum, d-arabinosum, d-ribosum, erythritolum, ribitolum, galactitolum, acidum 5-keto-d-gluconicum, dl-acidum lacticum, methanolum, inositolum, nitratum, nitritum, acidum saccharicum et acidum d-glucuronicum non assimilantur. Materia amyloidea non formatur. Crescit ad 30°C et 37°C et in medio cum 10% sodii chloridio/5% glucoso, at non in medio cum 0.1 g et 1 g cycloheximido per litrum nec in medio cum 50% glucoso. Ureum non finditur. Ad crescentiam vitaminae additae non necessariae sunt.
Typus CBS 11368, isolatus in Shark River Slough, Florida Everglades, USA.
Standard description of Candida sharkiensis Fell, Gutiérrez, Statzell-Tallman and Scorzetti sp. nov.
Etymology: The species name sharkiensis refers to the Shark River Slough, where the strains were collected.
After 3 days on 5% malt extract (ME) at 22°C, the cells are ovoid, measure 1.3–5.2 × 1.7–10.2 μm and occur singly or in pairs (Fig. 6). Budding is multilateral with one or two buds per cell and inflated spherical cells. After 1 month white sediment is formed and no ring or pellicle is present. Colonies growing on 5% ME agar after 7 days at 22°C, are white, glistening and the surface is smooth. After 1 month the cells have a “peeling” outer wall and abundant oil drops are observed (Fig. 6a). No hyphae or pseudohyphae were observed. Cultures growing under cover glass on corn meal (CM) agar (Dalmau plate culture) after 1 month at 22°C did not produce either hyphae or pseudohyphae. Ascospores were not observed in individual colonies growing in 5% ME agar after 1 month at 22°C. Mixing of strains on 5% ME agar, produced neither ascospores, nor conjugated cells after 1 month at 22°C. See Table 2 for physiological tests.
Type strain: NRRL Y-48380, CBS 11368 (EY8-093), which was obtained at Station 3 (25°47′N, 80°85′W), Collection 8 (8 Sept 2004). Additional strains submitted to culture collections include EY8-094 (NRRL Y-48381, CBS 11369) from the same collection, station and date as the type strain, EY0972 (NRR Y-48463, CBS 10856) and EY0984 (NRRL Y-48464, CBS 10857), which were obtained from Station 3, Collection 5, 19 June 2003.
Collection sites: A total of 73 strains were collected: Station (Sta) 1a: Collection (C) 1 (1 strain), C 2 (2), C3 (5), C4 (6) C7 (9). Sta 2: C3 (1), C4 (1). Sta 3: C2 (1), C3 (3), C4 (4), C5 (11), C7 (3), C8 (9), C 9 (7). Sta 4: C2 (2), C3 (1), C9 (1). Sta 5 C2 (1), C8 (1). Sta 6 (2). (For dates and geographical positions see “Methods” section).
GenBank accession numbers: CBS 11368—D1/D2 region of the large subunit rDNA = GU592922.
Latin diagnosis of Candida rhizophoriensis Fell, Gutiérrez, Statzell-Tallman et Scorzetti sp. nov.
Post dies 3 ad 22°C in medio liquido (ME) cellulae ovoideae (1.7–3.9 × 3.0–6.0 μm), singulae, binatae vel catenis ramosis cohaerentes. Post mensem unum copiosum album sedimentum et conspicuus anulus apparent. Post dies 7 ad 22°C in medio ME cum agaro cultura alba, hebes levisque. Post mensem unum ad 22°C in medio ME cum agaro ascosporae non formantur; pseudohyphae apparent. Post mensem unum ad 22°C in medio CM cum agaro pseudomycelium formatur.
Glucosum et galactosum fermentantur; sucrosum, lactosum, maltosum, trehalosum et raffinosum non fermentantur. Glucosum, galactosum, l-sorbosum (lente), sucrosum, maltosum, cellobiosum, trehalosum, raffinosum (lente), melezitosum, ethanolum (lente), ribitolum (lente), galactitolum (lente), d-mannitolum, d-glucitolum, methyl-α-d-glucosidum, salicinum, d-gluconicum (lente), acidum 2-keto-d-gluconicum, N-acetyl-d-glucosaminum, acidum succinicum, acidum citricum, hexadecanum (lente) et creatinum (lente) assimilantur. Lactosum, melibiosum, inulinum, amylum solubile, d-xylosum, l-arabinosum, d-arabinosum, d-ribosum, l-rhamnosum, d-glucosaminum, glycerolum, erythritolum, acidum 5-keto-d-gluconicum, dl-acidum lacticum, methanolum, inositolum, nitratum, nitritum, acidum saccharicum, acidum d-glucuronicum et creatininum non assimilantur. Materia amyloidea non formatur. Crescit ad 30°C at non ad 37°C. Crescit in medio cum 10% sodii chloridio/5% glucoso, et in medio cum 0.1 g cycloheximido per litrum (exigue) at non in medio cum 1 g cycloheximido per litrum nec in medio cum 50% glucoso. Ureum non finditur. Ad crescentiam vitaminae additae non necessariae sunt.
Typus CBS 11402, isolatus in Shark River Slough, Florida Everglades, USA.
Standard description of Candida rhizophoriensis Fell, Gutierrez, Statzell-Tallman and Scorzetti sp. nov.
Etymology: The species name rhizophoriensis refers to collection sites dominated by red mangrove trees (Rhizophora mangle) in Shark River Slough.
After 3 days at 22°C on 5% ME, the cells are ovoid, measuring 1.7–3.9 × 3.0–6.0 μm (Fig. 7). They occur singly, in pairs and branched chains, elongate cells are present and form chains. Budding is multilateral with 1 or 2 buds per cell. After 1 month, an abundant white sediment and definite ring are present. After 7 days at 22°C on 5% ME agar, the colony color is white, dull and the surface is smooth, pseudohyphae were observed. Cultures growing under a cover glass on CM agar (Dalmau plate culture) after 1 month at 22°C showed short pseudohyphae. Ascospores are not present after 1 month of growth on 5% ME agar at 22°C. See Table 2 for the results of the physiological tests.
Type strain: NRRL Y 48382, CBS 11402 (EY0384), which was isolated from a water sample at Station 4 (25°41′N, 80°96′W), Collection 3, 18 Feb 2003.
Collection sites: Four strains were obtained, three strains at the type locale collection and one strain at Station 6, Collection 2, 20 Nov 2002.
GenBank accession numbers: CBS 11402—D1/D2 region of the large subunit rDNA = GU592921.
Latin diagnosis of Rhodotorula cladiensis Fell, Statzell-Tallman et Scorzetti sp. nov.
In medio liquido (ME) post 3 dies ad 22°C, cellulae ellipsoidae aut elongatae (1.3–3.4 × 2.1–7.3 μm), singulae aut binae aut breviter catenatae.Tenue sedimentum formatur. Post unum mensem ad 25°C in medio ME cum agaro, cultura laevigata, madida et rubra/aurantiaca. In medio CM cum agaro post 7 dies hyphae non formantur.
Fermentatio nulla. Glucosum, galactosum, sucrosum, maltosum, cellobiosum (varie), trehalosum, raffinosum (varie), melezitosum, d-arabinosum (varie), l-rhamnosum (lente), glycerolum, ribitolum, d-mannitolum (lente), methyl-α-d-glucosidum (varie), salicinum (lente), acidum d-gluconicum, hexadecanum, acidum saccharicum (varie) et acidum d-glucoronicum assimilantur, at non l-sorbosum, lactosum, melibiosum, inulinum, amylum solubile, d-xylosum, l-arabinosum, d-ribosum, d-glucosaminum, N-acetyl d-glucosaminum, ethanolum, methanolum, erythritolum, galactitolum, d-glucitolum, acidum dl-lacticum, acidum succinicum, acidum citricum, inositolum, acidum 2-keto d-gluconicum, acidum 5-keto d-gluconicum neque nitratum. Creatininum creatinumque assimilantur (lente). Crescit in medio cum 10% sodii chloridio-5% d-glucoso (varie) et in medio cum 0.1 g cycloheximido per litrum (lente) at non in medio cum 1 g cycloheximido per litrum, neque in in medio cum 50% glucoso. Materia amyloidea non formatur. Ureum finditur. Crescit ad 30°C at non ad 37°C. Ad crescentiam vitaminae additae non necessariae sunt.
Typus CBS 10878 isolatus ex aqua fluviatilis, depositus in collectione zymotica Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
Standard description of Rhodotorula cladiensis Fell, Statzell-Tallman and Scorzetti sp. nov.
Etymology: The species is named after the type locale, which is a sawgrass (Cladium jamaicense) marsh.
After 3 days at 22°C in 5% ME the cells are elliptical or elongate measuring 1.3–3.4 × 2.1–7.3 μm (Fig. 8). Cells occur singly, in pairs or in short chains. Light sediment is present. The growth on 5% ME agar after 1 month at 25°C: the surface of the colony is smooth, moist and red/orange. No hyphae could be detected after 7 days at 17°C in Dalmau plate culture on CM agar. Mating studies between strains and with the related strains of Sakaguchia dacryoidea (CBS 7999 and CBS 6365) were negative. For results of physiological tests see Table 2.
Type strain: CBS 10878 (NRRL Y-48720), which was isolated from water at SRS Station 1a (25°76′N, 80°73′W), Collection 8, 8 September 2004. Additional strains submitted to CBS are CBS 10883, Station 1a Collection 4, 17 June 2003 and CBS 10879 from Station 3, Collection 8, 8 Sept 2004.
Collection sites: A total of seven strains were obtained: Station (Sta) 1a Collection (C) 4 (3 strains), C8 (1), Sta 2: C1 (1), Sta 3: C8 (1), Sta 4: C4 (1).
GenBank accession numbers for CBS 10878 are FJ008055 in the ITS regions and FJ008049 in the D1/D2 domains.
Latin diagnosis of Rhodotorula evergladiensis Fell, Statzell-Tallman et Scorzetti sp. nov.
In medio liquido (ME) post 3 dies ad 25°C, cellulae globosae aut ellipsoidae (2.1–4.2 × 3.4–7.2 μm), singulae aut binae aut breviter catenatae. Tenue sedimentum formatur. Post unum mensem moderatum sedimentum annulusque apparent. Post unum mensem ad 25°C in medio ME cum agaro, cultura rubra, mucosa expressaque. In medio CM cum agaro post 7 dies hyphae non formantur.
Fermentatio nulla. Glucosum, galactosum, l-sorbosum, sucrosum, maltosum, cellobiosum (lente), trehalosum (lente), melezitosum, raffinosum, l-rhamnosum (lente), ethanolum (lente), glycerolum (lente), ribitolum (lente), d-mannitolum, d-glucitolum, methyl-α-d-glucosidum, acidum d-gluconicum (lente), acidum succinicum, hexadecanum et acidum 2-keto d-gluconicum (lente) assimilantur, at non lactosum, melibiosum, inulinum, amylum solubile, d-xylosum, l-arabinosum, d-arabinosum, d-ribosum, d-glucosaminum, N-acetyl d-glucosaminum, methanolum, erythritolum, galactitolum, salicinum, acidum DL lacticum, acidum citricum, inositolum, acidum 5-keto d-gluconicum, acidum saccharicum, acidum d-glucuronicum neque nitratum. Creatininum creatinumque assimilantur. Ad crescentiam vitaminae additae necessariae sunt. Crescit in medio cum 10% sodii chloridio -5% d-glucoso (lente) at non in medio cum 0.1 g cycloheximido per litrum neque in medio cum 1 g cycloheximido per litrum Materia amyloidea non formatur. Crescit ad 30°C et 37°C (varie). Ureum finditur.
Typus CBS 10880 isolatus ex aqua fluviatili, depositus in collectione zymotica Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
Standard description of Rhodotorula evergladiensis Fell, Statzell-Tallman and Scorzetti sp. nov.
Etymology: The species, evergladiensis, is named after the Everglades region where the strains were isolated.
After 3 days of growth in 5% ME at 25°C, the cells are round to elliptical measuring 2.1–4.2 × 3.4–7.2 μm (Fig. 9). Cells occur singly, in pairs or in short chains. Light sediment is present. After a month, a moderate ring and a moderate sediment are present. After 1 month at 25°C on 5% ME agar, the colony is red, runny, mucoid and raised. No hyphae could be detected on Dalmau plate culture on CM agar after 7 days at 17°C. Mating studies between strains were negative. See Table 2 for the results of the physiological tests.
The Type strain CBS 10880 (NRRL Y-48721) was isolated from water in the sawgrass community at SRS Station 3 (25°47′N, 80°85′W) along the Shark River in the Everglades, Florida, Collection 8, 8 September 2004. Additional strains submitted to CBS are CBS 10881 and CBS 10882, which were collected from the same location and date as the type strain.
GenBank accession numbers for CBS 10880 are FJ008054 in the ITS regions and FJ008048 in the D1/D2 domains.
Collection sites: A total of 58 strains were isolated. Sta 1a: C2 (6), C3 (2), C4 (8), C8 (5), Sta 2: C2 (2), C8 (2). Sta 3: C2 (3), C3 (1), C8 (25), C9 (2). Sta 6: C3 (1), C9 (1)
Latin diagnosis of Cryptococcus mangaliensis Fell, Statzell-Tallman et Scorzetti sp. nov.
In medio liquido (ME) post 3 dies ad 25°C, cellulae ellipsoideae vel elongatae (2.0–3.4 × 3.4–5.4 μm) aut globosae. Sterigmata cellulas ferunt. Post unum mensem ad 25°C in medio ME cum agaro, cultura lucida, cremea, butyracea, mucosa, plana vel leniter expressa, glabraque, margine integro. In medio CM cum agaro post 7 dies hyphae aut conidia non formantur.
Fermentatio nulla. Glucosum, galactosum, l-sorbosum (lente), sucrosum, cellobiosum, maltosum, trehalosum, lactosum, raffinosum, melezitosum (lente), d-xylosum, l-arabinosum, d-ribosum (lente), l-rhamnosum (lente), d-glucosaminum (lente), N-acetyl d-glucosaminum, ethanolum, glycerolum, erythritolum (vel lente et exigue) ribitolum, galactitolum (lente), d-mannitolum, d-glucitolum, acidum d-gluconicum, salicinum, acidum succinicum, acidum citricum (exigue), inositolum (vel lente et exigue), acidum 2-keto d-gluconicum, acidum 5-keto d-gluconicum, acidum d-glucuronicumque assimilantur, at non melibiosum, inulinum, amylum solubile, d-arabinosum, methanolum, methyl-α-d-glucosidum, acidum DL lacticum, hexadecanum, acidum saccharicum, neque nitratum. Creatininum non assimilatur. Ad crescentiam vitaminae additae non necessariae sunt. Crescit in medio cum 10% sodii chloridio-5% d-glucoso. Materia amyloidea non formatur. Non crescit ad 37°C.
Typus CBS 10870 isolatus in Chatham Bend, Everglades National Park, in collectione zymotica Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands.
Description of Cryptococcus mangaliensis Fell, Statzell-Tallman and Scorzetti sp. nov.
Etymology: The name for the species (mangaliensis) is derived from the mangrove (mangal) habitat.
After 3 days of growth at 25°C in 5% ME the cells are ellipsoidal to elongate or globose, 2.0–3.4 × 3.4–5.4 μm (Fig. 10). Cells may be budding on short pedicels and a blunt apical bud scar may be present on the single cells. The streak culture is glossy, cream-colored, butyrous, mucoid, smooth, flat or slightly raised with entire margins after 1 month growth on 5% ME agar at 25°C. Pseudohyphae are not present on Dalmau plate culture on CM agar after 1 week at 17°C. Ballistoconidia were not observed. For the results of the physiological tests, see Table 2.
Type strain, CBS 10870 (NRRL Y-48462) was isolated in the mangroves in Chatman Bend (25°41′N, 85°1′W) in the Florida Everglades National Park on 10/23/1996.
Genbank accession numbers for CBS 10870 are FJ008052 in the ITS and FJ008046 in the D1/D2 domains.
References
Allen TW, Quayyum HA, Burpee LL, Buck JW (2004) Effect of foliar disease on the epiphytic yeast communities of creeping bent grass and tall fescue. Can J Microbiol 50:853–860
Bai FY, Yimin C, Qi-Ming W, Ohkubo H (2004) Rhodotorula oryzae sp nov., a novel basidiomycetous yeast species isolated from paddy rice. Antonie van Leeuwenhoek 86:295–299
Boekhout T (2010) The genus Pseudozyma. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam
de Araujo FV, Soares CA, Hagler AN, Mendonça-Hagler LC (1995) Ascomycetous yeast communities of marine invertebrates in a Southeast Brazilian mangrove ecosystem. Antonie van Leeuwenhoek 68:91–99
Fell JW, Hunter I, Tallman AS (1973) Marine basidiomycetous yeasts (Rhodosporidium spp. n.) with tetrapolar and multiple allelic bipolar mating systems. Can J Microbiol 19:643–657
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Fell JW, Statzell-Tallman A, Kurtzman CP (2004) Lachancea meyersii sp nov., an ascosporogenous yeast from mangrove regions in the Bahama Islands. Stud Mycol 50:359–363
Gadanho M, Almeida JM, Sampaio JP (2003) Assessment of yeast diversity in a marine environment in the south of Portugal by microsatellite-primed PCR. Antonie van Leeuwenhoek 84:217–227
Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP (2010) The Genus Candida. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam
Lazarus CR, Koburger JA (1974) Identification of yeasts from the Suwannee River Florida Estuary. Appl Environ Microbiol 27:1108–1111
Limtong S, Yongmanitchai W, Kawasaki H, Seki T (2007) Candida thaimueangensis sp. nov., an anamorphic yeast species from estuarine water in a mangrove forest in Thailand. Int J Syst Evol Microbiol 57:650–653
Luong DT, Takashima M, Van Ty P, Lang Dung N, Nakase T (2005) Bullera hoabinhensis sp. nov., a new ballistoconidiogenus yeast isolated from a plant leaf collected in Vietnam. J Gen Appl Microbiol 51:335–342
Nakase T, Suzuki M (1985) Candida tsuchiyae, sp. nov., isolated from moss collected in Japan. J Gen Appl Microbiol 31:507–512
Nakase T, Komagata K, Fukazawa Y (1976) Candida pseudointermedia sp. nov. isolated from “Kamaboko”, a traditional fish-paste product in Japan. J Gen Appl Microbiol 22:177–182
Nakase T, Suzuki M, Takashima M, Rosadi D, Hermosillo AM, Komagata K (1994) Candida akabanensis, a new species of yeast isolated from insect frass in bark of a grape-vine. Microbiol Cult Collect 10:35–43
Nakase T, Suzuki M, Hamamoto M, Takashima M, Hatano T, Fukui S (1996) A taxonomic study on cellulolytic yeasts and yeast-like microoganisms isolated in Japan II. The genus Cryptococcus. J Gen Appl Microbio 42:7–15
Papadakis JS, Mavridou A, Richardson SC, Lamprini M, Marcelou U (1997) Bather-related microbial and yeast populations in sand and seawater. Water Res 31:799–804
Rosa CA, Pagnocca FC, Lachance MA, Ruivo CC, Medeiros AO, Pimentel MR, Fontenelle JC, Martins RP (2007) Candida flosculorum sp. nov. and Candida floris sp. nov., two yeast species associated with tropical flowers. Int J Syst Evol Microbiol 57:2970–2974
Sampaio JP (2010) The genus Rhodosporidium. In: Kurtzman CP, Fell JW, Boekhout T (eds) The yeasts, a taxonomic study, 5th edn. Elsevier, Amsterdam
Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A (2002) Systematics of basidiomycetous yeasts: a comparison of large sub-unit D1/D2 and internal transcribed spacer rDNA regions. FEMS Yeast Res 2:495–517
Statzell-Tallman A, Belloch C, Fell JW (2008) Kwoniella mangroviensis gen nov., sp. nov. a tremellaceous yeast from mangrove habitats in the Florida Everglades and Bahamas. FEMS Yeast Res 8:103–113
Statzell-Tallman A, Scorzetti G, Fell JW (2010) Candida spencermartinsiae sp. nov., Candida taylorii sp. nov. and Pseudozyma abaconensis sp. nov., novel yeasts from mangrove and coral reef ecosystems. Int J Syst Evol Microbiol 60:1978–1984
Suh S-O, Nguyen NH, Blackwell M (2005) Nine new Candida species near C membranifaciens isolated from insects. Mycol Res 109:1045–1056
Van Uden N, Zobell C (1962) Candida marina nov. spec. Torulopsis torresii nov. spec. and T. maris nov. spec. three yeasts from the Torres Strait. Antonie van Leeuwenhoek 28:275–283
Vogel C, Rogerson A, Schatz S, Laubach H, Tallman A, Fell JW (2007) Prevalence of yeasts in beach sand at three bathing beaches in South Florida. Water Res 41:1915–1920
Yan K, Zhang Y, Chi Z (2010) Distribution and diversity of Candida tropicalis strains in different marine environments. J Ocean Univ China 9:139–144
Yarrow D (1998) Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW (eds) The yeasts, a taxonomic study, 4th edn. Elsevier, Amsterdam, pp 77–100
Acknowledgments
The continued and enthusiastic cooperation by the FIU/LTER personnel was of utmost importance to success of this project. The research was supported by the National Science Foundation through Biotic Surveys and Inventories Grant DEB 0206521 and the Florida Coastal Everglades Long-Term Ecological Research program under Grants DBI-0620409 and DEB-9910514. M. H. Gutiérrez, a visiting student at RSMAS, was sponsored by Fundacion Andes (Chile) through a WHOI/UDEC Agreement. Figure 1 is a map created with the Interactive Everglades Mapping tool available at http://fcelter.fiu.edu/data/GIS/interactive_map/.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Fell, J.W., Statzell-Tallman, A., Scorzetti, G. et al. Five new species of yeasts from fresh water and marine habitats in the Florida Everglades. Antonie van Leeuwenhoek 99, 533–549 (2011). https://doi.org/10.1007/s10482-010-9521-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-010-9521-6