Abstract
Root resorption is a common complication during orthodontic treatment. Microcracks occur on the root surface after an orthodontic force is applied and may be related to the root resorption caused by the orthodontic process. However, the mechanisms underlying root resorption induced by microcracks remain unclear. In this study, a rat orthodontic model was used to investigate the biological mechanisms of root resorption caused by microcracks. First, the first molar was loaded with 0.5-N orthodontic force for 7 days, and microcracks were observed on the root apex surface using a scanning electron microscope. Second, to describe the mechanical principle resulting in microcracks, a finite element model of rat orthodontics was established, which showed that a maximum stress on the root apex can cause microcrack extension. Third, after 7 days of loading in vivo, histological observation revealed that root resorption occurred in the stress concentration area and cementoclasts appeared in the resorption cavity. Finally, proteomics analysis of the root apex area, excluding the periodontal ligament, revealed that the NOX2, Aifm1, and MAPK signaling pathways were involved in the root resorption process. Microcrack extension on the root surface increases calcium ion concentrations, alters the proteins related to root resorption, and promotes cementoclast formation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
External apical root resorption is a common orthodontic complication.23,62 Root resorption can reduce the root surface area, structural support, and mechanical resistance of the periodontal tissue;42 in severe cases, root resorption can even lead to tooth loss.59 Root resorption can negatively affect orthodontic treatment and patient satisfaction. However, the mechanisms underlying root resorption remain unclear. Schwarz47 proposed that if the force exerted on the teeth exceeds capillary blood pressure, the capillaries will rupture, leading to hyalinization of the periodontal tissue, recruitment of multiple phagocytes, and subsequent root resorption.4,22,34 Current research suggests that root resorption is related to excessive local periodontal ligament (PDL) pressure caused by orthodontic force-loading. This results in a sterile process of pyknosis and necrosis.5,59 Most previous physiological experiments have analyzed the PDL but not the physiological and mechanical properties of the root tissue.
Microcracks have previously been found to occur on the root surface of the first molar of rabbits after an orthodontic force is applied. Finite element (FE) analysis shows that stress concentrated at the rabbit root apex can induce microcrack extension on the root surface.61 Moreover, clinical observations indicate that intrusion force applied to the crown is more likely to cause stress concentration at the root apex and cause root resorption. On the other hand, extrusion force applied to the crown does not easily produce stress concentration at the root apex, nor does it necessarily cause root resorption.42 Therefore, it can be speculated that the stress concentration at the root apex can easily cause microcracks on the root surface, and these microcracks may be a key root resorption indicator;61 however, the rabbit differs from the human tooth as the root is flat with no closed apical hole.21 Compared to other small animal models, rat teeth more closely resemble human teeth as they have multiple roots and a closed apical hole, so further study on the formation of microcracks in rats would better reflect human physiology. Furthermore, excluding PDL tissue in a study might help to identify a signaling pathway underlying root resorption in simple root tissue.
Bone tissue research has shown that microcracks can lead to tissue resorption; however, the underlying mechanisms remain controversial. Tami et al.54 demonstrated that linear microcracks disrupt the fluid transport of the lacunar-canalicular system between osteocytes. Herman et al.19 observed that microcracks directly transect osteocytes, leading to necrosis that further promotes adjacent osteocyte apoptosis and results in osteoclast formation. Cardoso et al.6 found that linear microcracks can induce osteocyte apoptosis by disrupting the osteocyte network. Vasquez-Sancho et al.56 proposed that cracks form when the pressured bone produces flexoelectricity, inducing bone cell apoptosis and promoting osteoclast activation. Although their respective mechanisms differ, previous studies have shown that microcracks can induce osteocyte apoptosis, affecting both the differentiation and formation of osteoclasts; however, whether microcracks on the root surface can cause cementoclast activation and the related molecular resorption mechanism remains unclear.
This study aimed to determine the microstructural changes and regional location of the root apex through scanning electron microscopy (SEM); analyze the stress and microcrack extension of the root apex region through FE simulation; and identify the sites of maximum stress. Through histological and proteomic analyses of the local key areas of the tooth root tissue after application of stress, this study explored the signaling pathways underlying microcrack-induced root resorption.
Materials and Methods
Animal protocols were approved by the Animal Care and Use Committee of the School of Biological Science and Medical Engineering, Beihang University. All sections adhered to the ARRIVE Guidelines27 for reporting animal research. Rats were obtained from the Department of Laboratory Animal Science, Peking University Health Science Center, Beijing, China.
Orthodontic Force-Loading and SEM Observation of the Isolated Rat Maxilla
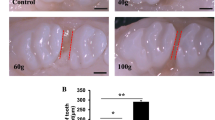
First, sixteen 8-week-old male Wistar rats (250 ± 5 g) were euthanized by intraperitoneal administration of an overdose of sodium pentobarbital. (Fig. 1a) After euthanasia, the heads of the rats were disinfected with 75% ethanol, and the fur was shaved. Thereafter, the maxilla of each rat was removed and washed in an ice-cold solution of penicillin-streptomycin-gentamicin (25 U/mL, 0.025 mg/mL, and 0.0125 mg/mL, respectively; Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) to remove the blood. Then, a 0.5-N load was immediately applied between the first molar and the anterior teeth on the right maxilla with a nickel-titanium spring (Sentalloy, Tomy Inc., Fukushima, Japan) and maintained for 7 days. The first molars and anterior teeth of the left maxilla, without any load, were used as controls (Fig. 2a). While the spring was still attached, the rat maxilla was soaked in a solution of penicillin-streptomycin-gentamicin (5 U/mL, 0.005 mg/mL, and 0.0025 mg/mL, respectively) and stored at 4 °C. The soaking solution was replaced daily. The spring was removed after 7 days of application of force to the maxillary bone. Then the maxillary first molar area bones were carefully dissected and separated, and a 2,940-nm Er:YAG laser (Fotona, San Ramon, CA) was used to separate the mesial roots of the first molars (Fig. 2b). The treated maxillary bone was placed in a 10-mL centrifuge tube, soaked in 0.4-mg/mL neutrase (200,000 U/g, Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) prepared in phosphate-buffered saline (PBS), and placed in a constant-temperature shaker maintained at 40 °C, with a speed of 50 revolutions per min. The solution was replaced every 12 h after it was left to cool to room temperature. A pH paper was used to ensure that the solution was pH-neutral, to avoid decalcification and brittleness. The PDL was allowed to dissolve until the root could be easily removed from the alveolar bone (48 h), after which the mesial roots of the first molars were carefully removed. The separated mesial roots were soaked in sodium hypochlorite with 0.25% available chlorine for 5 min to remove any residual PDL (Fig. 2c), and then rinsed in deionized water to remove residual sodium hypochlorite.9 Then, each of the separated mesial roots were dehydrated using gradient alcohol dehydration (25, 50, 75, 85, 90, 95, and 100%); each mesial root was left in each concentration of alcohol for 15 min each. After dehydrated, the alcohol was replaced with pure tert-butanol three times for 2 h each time. Lastly, they were randomly divided into four groups for different surface observation. The distal, mesial, buccal, and lingual surfaces of the roots were placed on an aluminum observation platform, fixed with conductive glue, and placed in a freeze dryer for vacuum drying. Gold spraying (Au 20 nm) (EMS 150TS; Quorum Technologies Ltd, United Kingdom) was then performed. The sample was observed under an SEM (JSM-6700F, JEOL, Tokyo, Japan) at 5 kV in the high-vacuum mode; all root apex areas were observed (Fig. 2d). The number of microcracks in the main microcrack concentrated area of each root was counted using ImageJ software (1.52i, U. S. National Institutes of Health, Bethesda, MD, USA).
A flow diagram of the study design. (a) The ex vivo force-loading model of 16 rats for SEM observation of microcracks. (b) Micro-CT scanning the maxilla of a normal rat for building the FE model. (c) The in vivo force-loading model of 4 rats for histological observation. (d) The in vivo force-loading model of 30 rats for proteomics analysis.
Orthodontic force-loading and SEM observation of isolated rat maxilla. (a) The rat maxilla is loaded ex vivo. The red circle represents the tooth loaded with orthodontic force. (b) An Er:YAG laser is used to separate the first molar’s mesial root after dissecting the maxillary first molar area bones. The red circle represents the first molar after laser separation. (c) The mesial root from the alveolar socket is removed after PDL dissolution. The red rectangle represents the apex of the mesial tooth observed by SEM. (d) The mesial, distal, buccal, and lingual surfaces of the mesial apex of the first molar are observed using an SEM on the force-loading and control groups under 0.5-N loading conditions. Yellow arrows represent microcracks. (e) Analysis of number of microcracks differences between force-loading and control groups. *p value < 0.05. Data are presented as the mean ± SD (n = 16). (f) Analysis of the number of microcracks difference between the mesial, distal, buccal and lingual observation groups in the force-loading group. *p < 0.05 and ***p < 0.001. Data are presented as the mean ± SD (n = 4).
Micro-computed Tomography Scanning
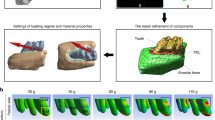
Micro-computed tomography (CT) scanning data of one rat was obtained. One 8-week-old male Wistar rat was sacrificed and its maxilla was dissected (Fig. 1b). The SkyScan 1076 (Bruker micro-CT; Kontich, Belgium) micro-CT system was used to scan the entire maxilla at a resolution of 9-µm/pixel (Fig. 3a). The first molar and nearby bones were then dissected; the SkyScan 1272 (Bruker micro-CT; Kontich, Belgium) micro-CT was used to perform a finer, 1-µm/pixel scan of the first molar area (Fig. 3b). See supplementary material for the detailed scanning parameters.
FE model analysis method and process. (a) Micro-CT with a 9-µm/pixel scan of the rat maxilla. The inserted picture is a physical representation of the rat’s maxilla. (b) The first molar and surrounding bone are dissected, and a 1-µm/pixel scan is performed. The inserted picture is a physical representation of the first molar and nearby bones. (c) Fine mesh of the mesial root of the first molar with 0.01 mm. (d) The FE model in the Abaqus software; the red arrow indicates the force-loading direction. (e) According to force analysis results, the 1-µm/pixel hexahedral mesh is refined on the rectangular area of the first molar mesial root apex. (f) One of the hexahedral elements is deleted and the adjacent surface of the element is set on the root apex surface as a defect to simulate crack extension.
Building the FE Analysis Model
Three-dimensional (3D) models were developed based on micro-CT images at resolutions of 1 and 9 µm/pixel using Mimics 19.0 (Materialise Inc., Leuven, Belgium), with which the thresholding, region growing, and 3D calculations were performed.31 Teeth, PDL, and cortical and cancellous bones were segmented and converted into non-uniform rational B-splines surface models using Geomagic Studio 12 (Raindrop Geomagic Inc., Research Triangle Park, NC, USA). The mesial roots of the first, second, and third molars, and the other parts were meshed with 0.01-mm C3D4, 0.2-mm C3D4, and 0.05-mm C3D4 tetrahedral elements, respectively, using an advancing-front algorithm software, HyperMesh 13.0 (Altair Engineering Inc., Troy, MI, USA) (Fig. 3c).46 Subsequently, the number of teeth, PDL, cortical, and cancellous bone elements were calculated to be 1,118,087, 105,999, 893,055, and 420,711, respectively. The entire model constituted 525,418 nodes and 2,537,852 elements. The model of each part was imported into FE software Abaqus 6.14 (Simulia Inc., Providence, RI, USA). The teeth, cortical bone, and cancellous bone were assumed to behave as linear isotropic elastic materials with a Poisson ratio of 0.3 and with Young’s moduli of 20 GPa, 20 GPa and 3 GPa, respectively.26,58 PDL behave as linear isotropic elastic materials with Young’s moduli of 0.7 MPa and with a Poisson ratio of 0.49.14,44 The cortical bone was set to be fixed, and the tie constraint was set to the following: root-PDL, PDL-cortical bone, PDL-cancellous bone, and cortical bone-cancellous bone. A 0.5-N load was applied on the orthodontic spring direction at the first molar, and the root apex force was analyzed (Fig. 3d).
Crack Extension Simulation
To simulate crack extension, the area of maximum principal stress root apex concentration was refined with a 1-µm hexahedral mesh (Fig. 3e). For the first molar, the number of C3D4 tetrahedral and C3D8 hexahedral elements was 350,489 and 97,842, respectively. A force analysis of the refined hexahedral mesh model was carried out. The area with the highest maximum principal stress value was selected, deleting one of the hexahedral elements and simulating the dent structure on the root surface.11,24,28 One surface of the element adjacent to the deleted hexahedral element was set as a crack defect to simulate crack extension (Fig. 3f). The maximum principal stress criterion was used with the extended FE method.33,61 Crack extensions of 0.25, 0.5, 0.8, and 1 N were simulated.
To establish the sensitivity of element deletion, a 0.5-N load was used to simulate crack extension in the model with no element deletion. To confirm the validity of the FE model in comparison with previously published results, a 0.1-N load was applied on the orthodontic spring direction at the first molar.26 The cortical bone was set to be fixed, and the tie constraint was set as: root-PDL, PDL-cortical bone, PDL-cancellous bone, and cortical bone-cancellous bone. The Von Mises strain of the PDL and the displacement of the first molar were analyzed.
Histological Observation
Four 8-week-old Wistar male rats (250 ± 5 g) were loaded with 0.5 N on the right maxillary first molars using a nickel-titanium spring; the left side with no loading was used as the control (Figs. 1c and 6a). Soft food was given, following an overdose of chloral hydrate anesthetics injection after 1 week, incurring their death. The maxillary bones were dissected, fixed with 4% paraformaldehyde for 48 h, and decalcified with 10% ethylenediaminetetraacetic acid for 45 days (pH 7.4). Following dehydration, the bones were embedded in paraffin and sliced continuously every 4 μm. Every fifth slice was stained with hematoxylin-eosin (HE) to find the area where the root was resorbed. TRAP staining was performed on 2 slices before and after the resorbed area. The control group was also subjected to TRAP staining of the corresponding sections. See supplementary material for the detailed process of TRAP staining. The root tissue resorption and the TRAP-positive osteoclast position in the root apex were observed by automatic digital section scanning (KF-PRO-120, KFBIO, Zhejiang, PR China). The number of TRAP+ cells in the slices of the force-loading and control groups was manually counted using the ImageJ software.
Proteomic Analysis
Sample Preparation
A total of 30 8-week-old male Wistar rats (250 ± 5 g) were included for proteomic analysis (Fig. 1d). The rats were randomly divided into three groups, each with 10 rats. A 0.5-N force was applied on the right side of the maxillary first molar using a nickel-titanium spring, and the left side with no loading was used as the control (Fig. 6a). Rats were given soft food. After 1 week, they were euthanized with an overdose of pentobarbital. The mesial root apex of the first molar of the loading and blank groups were removed (Fig. 6b). The PDL tissue was peeled off under a stereomicroscope and the molar was rinsed with PBS. The first molars were fixed with paraformaldehyde for 48 h and decalcified with ethylenediaminetetraacetic acid for 2 weeks. Three millimeters of the root apex of each group were cut off and freeze-dried (Fig. 6c). A data-independent acquisition (DIA) proteomic analysis was performed on each group.
Proteomic Analysis
The protein extraction method used in previous studies was performed;2 the Bradford method was used to quantify protein concentration after extraction. All samples were trypsinized using the filter-aided sample preparation method60 for 16 h. The partial, enzymatically hydrolyzed peptides were removed using the high-pH reverse phase method10 for classification. Orbitrap Fusion Lumos (Thermo Fisher, San Jose, CA) was used to collect mass spectrometry data. Using Proteome Discoverer 2.1 (Thermo Fisher), the data collected with data-dependent acquisition (DDA) were used for protein identification and analysis. The R. norvegicus protein database downloaded from Uniprot was used for analysis. The Percolator algorithm52 was employed to filter the identification results. The peptide false discovery rate was ≤ 1%, and the protein q-value was ≤ 0.01. Skyline 4.1 (University of Washington) was used to build a spectral library with high-confidence peptide results in the identification result file of Proteome Discoverer 2.1. Nanodrop 2000 (Thermo Fisher) was employed to quantify the peptide concentration. The amount of peptide detected in each sample was kept constant. The DIA collection method was used to collect three mass spectrometric data readings for each sample. Skyline 4.1 software (University of Washington) was employed to analyze the data. Differentially expressed proteins (filter with protein abundance ratio ≥ 1.2 or ≤ 0.83, and the t-test p value of ≤ 0.05 as the differential protein screening condition) were shown using the Blast2Go 5.0 software (http://www.blast2go.com)8,16 to conduct bioinformatics analysis. For detailed steps of the proteomics analysis, see the supplementary material.
Statistical Analysis
The results are expressed as the mean ± standard deviation (SD). One-way analysis of variance (ANOVA) was used to compare statistical differences between groups using IBM SPSS Statistics (version 19; IBM Corp., Armonk, NY, USA). Differences were considered significant when p < 0.05. Power analysis was calculated with G*Power software (v3.1.9.2, UCLA, Los Angeles, CA, USA). The value of the two-sided significance level (α) and a statistical power were 0.05 and 0.8, respectively.
Results
Orthodontic Force-Loading Promotes Microcracks on the Root Surface
The flow diagram of the study design is shown in Fig. 1a. The morphology of the force-loading and control groups on the root surface was observed under an electron microscope. There were almost no microcrack in the control group, and nor on the lingual and buccal sides in the force-loading group. However, a large number of microcracks had appeared on the surface of the third of the root apex in the mesial and distal sides of the force-loading groups (Figs. 2d and 2e). Compared with the mesial side, the number of microcracks in the distal side was increased significantly, by 63% (p < 0.01) (Fig. 2f).
FE Simulates Root Stress Concentration Area and Microcrack Extension
The results of Micro-CT scanning are shown in Figs. 3a and 3b.The FE analysis of stress distributions of 0.25, 0.5, and 1 N in the apical region of the rat model is shown in Fig. 4a. The maximum principal stress was high in one-third of the area of the root apex. Stress concentration and applied load values were positively correlated; however, the stress concentration area did not change (Fig. 4a). One of the hexahedral elements was removed to simulate the dent structure on the root surface. One element surface adjacent to the deleted hexahedral element was set as a crack defect to simulate its extension. The simulation result showed that the crack extension occurred at the nearby defect. The crack extension simulation process and PHILSM (a signed distance function to describe the microcrack surface)64 results under 0.1, 0.25, 0.5, 0.8, and 1 N loads are shown in Fig. 4b. The crack extension and load positively correlated, maintaining the crack extension path. After the 0.5-N loading in the model with no element deletion, crack extension was observed (Supplementary Fig. 1). Under the 0.1 N loading conditions, the FE analysis results showed that the maximum Von Mises stress of the PDL was 0.094 MPa, and the maximum displacement of the first molar was 12 µm, which is similar to the results of the maximum Von Mises stress of the PDL of 0.064 MPa and the maximum displacement of the first molar of 18 µm in published literatures.26,43
FE simulates the root stress concentration area and microcrack extension. (a) The stress distribution of the root apex under 0.25, 0.5, and 1 N loading conditions. (b) Root apex microcrack extension simulation image of mesial loading under 0.1, 0.25, 0.5, 0.8, and 1 N. Microcrack extension occurred at the defect, which was set to simulate microcrack extension near the deleted element. PHILSM is a signed distance function to describe the microcrack surface in the Abaqus software.
Orthodontic Force-Loading Promotes the Production of Resorption Cavities and Cementoclast with Histological Observation
The tissue sections of the first molar area of the rat were observed using HE staining. Resorption cavities (Fig. 5a) appeared on the apical surface of the force-loading group, and no cavities were observed in the control group (Fig. 5c). Purplish-red TRAP-positive particles appeared in the force-loading group near the resorption cavities (Fig. 5b), indicating that cementoclasts exist in this area. However, no resorption cavity was found in the control group, and the TRAP staining results were negative (Fig. 5d). There are significantly different (p < 0.001) between the ratio of the number of TRAP+ cells in the root apex area to the number of TRAP+ cells in the first molar area of the force-loading group (17.24 ± 1.37%) and the control group (3.69 ± 0.39%) (Fig. 5e). The number of TRAP+ cells per unit in the root apex area of the force-loading group (11.23 ± 1.66) and control group (2.73 ± 0.45) also exhibits significantly different (p < 0.001) (Fig. 5f).
Orthodontic force-loading promotes the production of resorption cavities and cementoclasts. (a) High-power microscopy results of the HE staining in the force-loading group (right), marked in the rectangle—resorption cavities appeared on the root surface; low-power microscopy results of the HE staining in the force-loading group (left). (b) TRAP staining results in the force-loading group; the black arrow indicates the purple-red cells as TRAP-positive (cementoclasts). (c) High-power microscopy of HE staining in the control group (right) indicates no resorption filling nest; low-power microscopy of the experimental group’s HE staining is shown (left). (d) The control group’s TRAP staining results were negative. (e) Analysis on the ratio of the number of TRAP+ cells in the root apex area to the number of TRAP+ cells in the first molar area between 2 groups. ***p < 0.001. Data are presented as the mean ± SD (n = 16). (f) Analysis on the number of TRAP+ cells per unit in the root apex area between 2 groups. ***p < 0.001. Data are presented as the mean ± SD (n = 16). B, bone; R, root; P, PDL.
High Expression of Orthodontic Force-Mediated of Root Resorption-Related Proteins Revealed by Proteomic Analysis
After strict quantitative peptide and product ion screening, 1969 proteins were reliably and quantitatively analyzed. The quality stability test is shown in Supplementary Fig. 2. Differential protein screening was performed for each sample group; 132 differentially expressed proteins were identified (with an abundance ratio ≥ 1.2 or ≤ 0.83, and p value ≤ 0.05 as the screening conditions). The force-loading group had 68 upregulated and 64 downregulated proteins, as shown in the volcano plot (Fig. 7a). Differential protein gene ontology function analysis was further performed. Among the differentially expressed proteins, 60 emergency response, 28 cell death-related, and 27 immune response-related proteins were detected (Fig. 7b). The analysis results of each sample group are shown in the heat map (Fig. 7c). Differential protein enzyme code-matching analysis showed that the differentially expressed proteins were mainly related to the functional activities of oxidoreductases, transferases, hydrolases, lyases, ligases, and other enzymes (Supplementary Fig. 3). Analysis of differentially expressed proteins with the Kyoto Encyclopedia of Genes and Genomes (KEGG) biological pathway (Supplementary Fig. 4). The results indicate that different proteins at the root apex, including nicotinamide adenine dinucleotide phosphate oxidase 2 (NOX2), Ca2+/calmodulin-dependent protein kinase II (CamKII), adenine nucleotide translocase (ANT), apoptosis-inducing factor mitochondrion-associated-1 (Aifm1), mitogen-activated protein kinase (MAPK), and signal transducer and activator of transcription 2 (STAT2), were involved in root resorption mediated by orthodontic force; they were also involved in the signaling pathways of osteocyte/cementocyte apoptosis and osteoclast/cementoclast differentiation (Fig. 8).
Discussion
In this study, the mesial root apex of the rat tooth was chosen as the research target, as it is the largest. However, surface microcracks are too small to be observed in the rat orthodontic model in vivo. The microcrack self-repair and phagocyte destruction affect microcrack observation.55Ex vivo model evaluations can reduce in vivo inherent biological interference, such as excessive inflammatory response, and tissue repair. The presence of masticatory loads in living rats also affects the distribution of orthodontic force.20,37 Hence, the ex vivo model was used in this study to observe microcracks on the root surface. The influence of orthodontic force on microcrack extension can be simply analyzed. Therefore, the obtained results decouple the force from the biochemical factors, which is more conducive to the micocracks observation by SEM. Previous studies had shown that a 0.5-N orthodontic force could cause significant rat root apex resorption; as such, a 0.5-N orthodontic force was used in this experiment.23,53
SEM results showed microcracks on the mesial and distal, but not on the buccal and lingual sides of the root. Microcracks were linearly distributed on the root surface. Previous studies found that, during bone reconstruction, linearly distributed microcracks induce osteocyte apoptosis through related physical injury, subsequently activate osteoclasts, and finally promote bone resorption.19,57 Therefore, our results confirmed the hypothesis that the microcracks at the root may cause cell apoptosis and resorption of the root apex. SEM results showed non-uniform structures on the root surface, which can easily form a stress concentration area.41 The stress value of this area is usually larger than its surrounding average, which is conducive to crack source formation. The increase and extension of microcracks were positively correlated with loading time; subsequently, small microcracks interconnected and eventually formed larger microcracks.61
This study established an FE model of rat orthodontic tooth loading. After the orthodontic force was loaded, the maximum principal stress value at the root apex was the largest (Fig. 4a). The location of stress concentration areas did not correlate with force magnitude (Fig. 4a). A hexahedral element was deleted to simulate the sunken structure of the root surface in an area with a large stress concentration, resulting both in obvious stress concentration and crack extension. The greater the loading force can generate the more obvious crack extension. The force-loading did not affect the crack extension’s direction (Fig. 4b).
Observation of the HE-stained slices after in vivo loading of rats revealed that resorption cavities appeared at the mesial root apex (Fig. 5a), consistent with the SEM crack and stress concentration areas of the FE analysis results, potentially indicating a close relationship between stress concentration and the root resorption of microcracks. The TRAP staining near the resorption cavity was positive, indicating the presence of cementoclasts (Fig. 5b). Consistent with our results, a previous study found that the location of root resorption was evident at one-third of the apex under 0.5-N orthodontic force.53 Therefore, this experiment focused on the root apex area, cutting off a 3-mm segment from the root apex, peeling off the peripheral tissue, and performing a proteomic analysis (Fig. 6c).
Proteomics analysis sample preparation and process. (a) Rat first molars loaded with orthodontic force in vivo. The crown enamel of the mesial root of the first molar was drilled off. (b) After the mesial roots of the first molars were separated, they were extracted. (c) The mesial apex’s PDL was peeled off, and the apex was washed, fixed, and decalcified, and a 3-mm segment was cut off from the mesial apex of the first molar for proteomic analysis.
Previous studies indicated that root resorption is related to the rupture and necrosis of PDL. To eliminate the interference of PDL, the PDL tissue was removed and proteomic analysis on only the cementum and other hard tissues in the root apex was conducted. This study investigated whether there are relevant signaling pathways involved in root resorption in the hard tissue of the root apex. Proteomics results mainly showed responses to stress, immune system process, and cell death-related proteins (Fig. 7b); hydrolases7 related to the phagocytosis of phagocytes were significantly upregulated (Supplementary Fig. 3). Cementocytes and cementoclasts share similar biological functions with osteocytes and osteoclasts, respectively.22,48,65 A consideration of the signaling pathways of osteocytes and osteoclasts thus provides insight. Previous studies reported that microcracks could increase calcium ion concentrations25,50 and activate downstream inflammatory response-signaling pathways.15,49 Moreover, this study detected an increase in calcium ion pathway-related proteins, indicating that this may be caused by root microcracks. The expression of the calcium ion pathway upstream-related transmembrane proteins, NOX2 and CamKII, and ANT were significantly increased (Supplementary Fig. 4).12,49 The calcium pathway downstream-related protein Aifm1 increased, and MAPK decreased.30,63 Reactive oxygen species (ROS) can cause Janus kinase (JAK) to increase,3,51 and experimental results have detected that STAT2 was elevated in the JAK-STAT pathway. Moreover, it has been reported that MAPK, Aifm1, and STAT2 can cause cell apoptosis.38,51,63 Cell apoptosis can turn off the inhibition of osteoclasts/cementoclasts and induces their formation13,17 (Fig. 8). In addition to osteoclast/cementoclast activation caused by osteocyte/cementocyte apoptosis, calcium ions can also promote osteoclast/cementoclast differentiation.35 Our results showed that orthodontic force-loading could change the expression of calcium ion-related signaling pathways. Previous studies showed that microcracks increase calcium release.50 Furthermore, in a rat orthodontic model, calcitonin, which can reduce local calcium ion concentration, can inhibit root resorption caused by orthodontic force.18 Hence, local calcium ion concentrations may be increased by microcrack generation and finally mediate the response of root resorption-related signaling pathways. Concomitantly, an increase in Htra1 was detected, which is capable of degrading osteoprotegerin in the bone microenvironment.40 Additionally, mechanics-related proteins, such as DNase and Pea15 may also be involved in the root resorption pathway.29,66 However, further verification and exploration of the relationship between these proteins and root resorption is warranted. In future work, it is worth analyzing the signaling pathways and mechanisms related to microcrack-mediated root resorption in order to improve the understanding of this process.
Orthodontic force-mediated high expression of root resorption-related proteins on proteomic analysis. (a) Volcano plot for differential protein screening; red dots indicate increased and green dots indicate decreased proteins. (b) Results of the biological function analysis of the differentially expressed proteins. (c) Heat-map of differentially expressed proteins.
The above results suggested that the excessive calcium ions released by the root surface microcracks can cause cell inflammation and apoptosis and activate the osteoclast/cementoclast cascade, which may be a key factor in causing root resorption. Normal root tissues are balanced between cementum resorption and formation.1 Therefore, when a continuous orthodontic force causes microcracks on the root surface, the balance between cement resorption and formation may be disrupted, resulting in clinically visible root resorption (Fig. 8).
This study has several limitations. First, there are still differences between rat teeth and human teeth in small animal models. The observation of human tooth microcracks would yield more meaningful insight. In the future, further studies on whether orthodontic force causes microcracks on the root surface of human teeth is necessary. Second, the accuracy of micro-CT scanning on the root surface is very low relative to the size resolution of microcracks. Therefore, the effect of finer microstructures on the microcrack extension should be studied in the future. Third, different orthodontic forces may cause different root stress concentration areas. Any part of root surface, not solely root apex, may suffer from great stress concentration, depending on the orthodontic force applied; however the majority of studies have targeted apical resorption due to its ease of examination on 2D X-ray images. In the future, with high-resolution CT detection, personalized treatment design can be carried out to adjust the magnitude and direction of orthodontic force and reduce the risk of root resorption caused by orthodontic force.
In conclusion, the present study investigated microcracks on the root apex surface after orthodontic force-loading and established an FE model of rat orthodontic loading to simulate crack extension on the root surface. The consistent relationship of the stress concentration area of the root tissue, the area of microcracks generation and extension, and the area of root resorption, were preliminarily explored. Finally, the signaling pathways causing root resorption in this area was analyzed by proteomics. The present results support the hypothesis that microcracks at the root apex may be one of the important factors that cause root resorption. Microcracks can serve as indicators of root resorption in the early stage of orthodontic treatment, and an ultra-high-resolution CT or ultrasound can be used for detection in the future. Previous studies have reported that high-frequency vibration, low-level laser therapy, and low-intensity pulsed ultrasound can reduce root resorption in clinical practice.23,32,36,39,45 To obtain the best treatment effect, these methods can be combined with the findings of this study on the generation of microcracks, the main root resorption area, and the high expression of local calcium ions. This combination may also further optimize the parameter selection, location position, or treatment time. Additionally, this study provides a theoretical basis for further research on biological mechanisms involved during orthodontic force-loading and yields a practical idea for the study of new drugs to prevent root resorption.
References
Abass, S. K., and J. K. Hartsfield. Orthodontics and external apical root resorption. Semin. Orthod. 13:246–256, 2007.
Alvares, K., C. J. DeHart, P. M. Thomas, N. L. Kelleher, and A. Veis. The unique biomineralization transcriptome and proteome of Lytechinus variegatus teeth. Connect. Tissue Res. 59:20–29, 2018.
Bhattacharjee, N., S. Barma, N. Konwar, S. Dewanjee, and P. Manna. Mechanistic insight of diabetic nephropathy and its pharmacotherapeutic targets: an update. Eur. J. Pharmacol. 791:8–24, 2016.
Brudvik, P., and P. Rygh. Multi-nucleated cells remove the main hyalinized tissue and start resorption of adjacent root surfaces. Eur. J. Orthod. 16:265–273, 1994.
Brudvik, P., and P. Rygh. Root resorption beneath the main hyalinized zone. Eur. J. Orthod. 16:249–263, 1994.
Cardoso, L., B. C. Herman, O. Verborgt, D. Laudier, R. J. Majeska, and M. B. Schaffler. Osteocyte apoptosis controls activation of intracortical resorption in response to bone fatigue. J Bone Miner Res 24:597–605, 2009.
Cohn, Z. A., and B. Benson. The in vitro differentiation of mononuclear phagocytes: II. The influence of serum on granule granule formation, hydrolase production, and pinocytosis. J. Exp. Med. 121:835–848, 1965.
Conesa, A., S. Götz, J. M. García-Gómez, J. Terol, M. Talón, and M. Robles. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676, 2005.
DeLaurier, A., A. Boyde, M. A. Horton, and J. S. Price. Analysis of the surface characteristics and mineralization status of feline teeth using scanning electron microscopy. J. Anat. 209:655–669, 2006.
Dimayacyac-Esleta, B. R. T., C.-F. Tsai, R. B. Kitata, P.-Y. Lin, W.-K. Choong, T.-D. Lin, Y.-T. Wang, S.-H. Weng, P.-C. Yang, S. D. Arco, T.-Y. Sung, and Y.-J. Chen. Rapid high-pH reverse phase StageTip for sensitive small-scale membrane proteomic profiling. Anal. Chem. 87:12016–12023, 2015.
Donaldson, F., D. Ruffoni, P. Schneider, A. Levchuk, A. Zwahlen, P. Pankaj, and R. Müller. Modeling microdamage behavior of cortical bone. Biomech. Model. Mechanobiol. 13:1227–1242, 2014.
El-Benna, J., P. M.-C. Dang, M.-A. Gougerot-Pocidalo, J.-C. Marie, and F. Braut-Boucher. p47phox, the phagocyte NADPH oxidase/NOX2 organizer: structure, phosphorylation and implication in diseases. Exp. Mol. Med. 41:217–225, 2009.
Giuliani, N., M. Ferretti, M. Bolzoni, P. Storti, M. Lazzaretti, B. Dalla Palma, S. Bonomini, E. Martella, L. Agnelli, A. Neri, F. Ceccarelli, and C. Palumbo. Increased osteocyte death in multiple myeloma patients: role in myeloma-induced osteoclast formation. Leukemia 26:1391–1401, 2012.
Gonzales, C., H. Hotokezaka, Y. Arai, T. Ninomiya, J. Tominaga, I. Jang, Y. Hotokezaka, M. Tanaka, and N. Yoshida. An in vivo 3D micro-CT evaluation of tooth movement after the application of different force magnitudes in rat molar. Angle Orthod. 79:703–714, 2009.
Görlach, A., K. Bertram, S. Hudecova, and O. Krizanova. Calcium and ROS: a mutual interplay. Redox Biol. 6:260–271, 2015.
Götz, S., J. M. García-Gómez, J. Terol, T. D. Williams, S. H. Nagaraj, M. J. Nueda, M. Robles, M. Talón, J. Dopazo, and A. Conesa. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36:3420–3435, 2008.
Gu, G., M. Mulari, Z. Peng, T. A. Hentunen, and H. K. Väänänen. Death of osteocytes turns off the inhibition of osteoclasts and triggers local bone resorption. Biochem. Biophys. Res. Commun. 335:1095–1101, 2005.
Guan, L., S. Lin, W. Yan, L. Chen, and X. Wang. Effects of calcitonin on orthodontic tooth movement and associated root resorption in rats. Acta Odontol. Scand. 75:595–602, 2017.
Herman, B. C., L. Cardoso, R. J. Majeska, K. J. Jepsen, and M. B. Schaffler. Activation of bone remodeling after fatigue: differential response to linear microcracks and diffuse damage. Bone 47:766–772, 2010.
Hichijo, N., E. Tanaka, N. Kawai, L. J. van Ruijven, and G. E. J. Langenbach. Effects of decreased occlusal loading during growth on the mandibular bone characteristics. PLoS ONE 10:e0129290, 2015.
Ibrahim, A. Y., S. Gudhimella, S. N. Pandruvada, and S. S. Huja. Resolving differences between animal models for expedited orthodontic tooth movement. Orthod. Craniofac. Res. 20:72–76, 2017.
Iglesias-Linares, A., and J. J. K. Hartsfield. Cellular and molecular pathways leading to external root resorption. J. Dent. Res. 96:145–152, 2017.
Inubushi, T., E. Tanaka, E. B. Rego, J. Ohtani, A. Kawazoe, K. Tanne, M. Miyauchi, and T. Takata. Ultrasound stimulation attenuates resorption of tooth root induced by experimental force application. Bone 53:497–506, 2013.
Jiang, H., Y. Ren, Z. Liu, and S. Zhang. Microscale finite element analysis for predicting effects of air voids on mechanical properties of single fiber bundle in composites. J. Mater. Sci. 54:1363–1381, 2019.
Jung, H., and O. Akkus. Activation of intracellular calcium signaling in osteoblasts colocalizes with the formation of post-yield diffuse microdamage in bone matrix. Bonekey Rep. 5:778, 2016.
Kawarizadeh, A., C. Bourauel, and A. Jäger. Experimental and numerical determination of initial tooth mobility and material properties of the periodontal ligament in rat molar specimens. Eur. J. Orthod. 25:569–578, 2003.
Kilkenny, C., W. J. Browne, I. C. Cuthill, M. Emerson, and D. G. Altman. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8:e1000412, 2010.
Lee, D. G., S.-H. Kim, S. Kim, J. H. Yu, and S. W. Cho. Prediction of material properties of ceramic composite material by porous structure and porosity using the finite element method. Int. J. Precis. Eng. Manuf. 20:805–814, 2019.
Levine, J., E. Kwon, P. Paez, W. Yan, G. Czerwieniec, J. A. Loo, M. V. Sofroniew, and I.-B. Wanner. Traumatically injured astrocytes release a proteomic signature modulated by STAT3-dependent cell survival. Glia 64:668–694, 2016.
Lim, J.-A., L. Li, O. Kakhlon, R. Myerowitz, and N. Raben. Defects in calcium homeostasis and mitochondria can be reversed in Pompe disease. Autophagy 11:385–402, 2015.
Lü, L.-W., G.-W. Meng, and Z.-H. Liu. Finite element analysis of multi-piece post-crown restoration using different types of adhesives. Int. J. Oral Sci. 5:162–166, 2013.
Lyu, C., L. Zhang, and S. Zou. The effectiveness of supplemental vibrational force on enhancing orthodontic treatment. A systematic review. Eur. J. Orthod. 41:502–512, 2019.
Marco, M., E. Giner, R. Larraínzar-Garijo, J. R. Caeiro, and M. H. Miguélez. Numerical modelling of femur fracture and experimental validation using bone simulant. Ann. Biomed. Eng. 45:2395–2408, 2017.
Matsumoto, Y., S. Sringkarnboriboon, and T. Ono. Proinflammatory mediators related to orthodontically induced periapical root resorption in rat mandibular molars. Eur. J. Orthod. 39:686–691, 2017.
Mentaverri, R., S. Yano, N. Chattopadhyay, L. Petit, O. Kifor, S. Kamel, E. F. Terwilliger, M. Brazier, and E. M. Brown. The calcium sensing receptor is directly involved in both osteoclast differentiation and apoptosis. FASEB J. 20:2562–2564, 2006.
Michelogiannakis, D., D. Al-Shammery, Z. Akram, P. E. Rossouw, F. Javed, and G. E. Romanos. Influence of low-level laser therapy on orthodontically-induced inflammatory root resorption. A systematic review. Arch. Oral Biol. 100:1–13, 2019.
Milne, T. J., I. Ichim, B. Patel, A. McNaughton, and M. C. Meikle. Induction of osteopenia during experimental tooth movement in the rat: alveolar bone remodelling and the mechanostat theory. Eur. J. Orthod. 31:221–231, 2009.
Miyake, N., N. I. Wolf, F. K. Cayami, J. Crawford, A. Bley, D. Bulas, A. Conant, S. J. Bent, K. W. Gripp, A. Hahn, S. Humphray, S. Kimura-Ohba, Z. Kingsbury, B. R. Lajoie, D. Lal, D. Micha, A. Pizzino, R. J. Sinke, D. Sival, I. Stolte-Dijkstra, A. Superti-Furga, N. Ulrick, R. J. Taft, T. Ogata, K. Ozono, N. Matsumoto, B. A. Neubauer, C. Simons, and A. Vanderver. X-linked hypomyelination with spondylometaphyseal dysplasia (H-SMD) associated with mutations in AIFM1. Neurogenetics 18:185–194, 2017.
Nayyer, N., T. Tripathi, P. Rai, and R. Gopal. Effect of photobiomodulation on external root resorption during orthodontic tooth movement—a scoping review. Lasers Dent. Sci. 3:219–226, 2019.
Ochiai, N., Y. Nakachi, T. Yokoo, T. Ichihara, T. Eriksson, Y. Yonemoto, T. Kato, H. Ogata, N. Fujimoto, Y. Kobayashi, N. Udagawa, S. Kaku, T. Ueki, Y. Okazaki, N. Takahashi, and T. Suda. Murine osteoclasts secrete serine protease HtrA1 capable of degrading osteoprotegerin in the bone microenvironment. Commun. Biol. 2:86, 2019.
Ouyang, S., Z. Deng, and X. Hou. Stress concentration in octagonal honeycombs due to defects. Compos. Struct. 204:814–821, 2018.
Parker, R. J., and E. F. Harris. Directions of orthodontic tooth movements associated with external apical root resorption of the maxillary central incisor. Am. J. Orthod. Dentofac. Orthop. 114:677–683, 1998.
Rahimi, A., L. Keilig, G. Bendels, R. Klein, T. M. Buzug, I. Abdelgader, M. Abboud, and C. Bourauel. 3D Reconstruction of dental specimens from 2D histological images and μCT-Scans. Comput. Methods Biomech. Biomed. Eng. 8:167–176, 2005.
Ru, N., S. S.-Y. Liu, Y. Bai, S. Li, Y. Liu, and X. Wei. BoneCeramic graft regenerates alveolar defects but slows orthodontic tooth movement with less root resorption. Am. J. Orthod. Dentofac. Orthop. 149:523–532, 2016.
Sato, M., K. Nagata, S. Kuroda, S. Horiuchi, T. Nakamura, M. Karima, T. Inubushi, and E. Tanaka. Low-intensity pulsed ultrasound activates integrin-mediated mechanotransduction pathway in synovial cells. Ann. Biomed. Eng. 42:2156–2163, 2014.
Schileo, E., F. Taddei, L. Cristofolini, and M. Viceconti. Subject-specific finite element models implementing a maximum principal strain criterion are able to estimate failure risk and fracture location on human femurs tested in vitro. J. Biomech. 41:356–367, 2008.
Schwarz, A. M. Tissue changes incidental to orthodontic tooth movement. Int. J. Orthod. Oral Surg. Radiogr. 18:331–352, 1932.
Shah, A., D. Lee, M. Song, S. Kim, M. K. Kang, and R. H. Kim. Clastic cells are absent around the root surface in pulp-exposed periapical periodontitis lesions in mice. Oral Dis. 24:57–62, 2018.
Shirokova, N., and E. Niggli. Cardiac phenotype of duchenne muscular dystrophy: insights from cellular studies. J. Mol. Cell. Cardiol. 58:217–224, 2013.
Shu, Y., M. J. Baumann, E. D. Case, R. K. Irwin, S. E. Meyer, C. S. Pearson, and L. R. McCabe. Surface microcracks signal osteoblasts to regulate alignment and bone formation. Mater. Sci. Eng. C 44:191–200, 2014.
Simon, A. R., U. Rai, B. L. Fanburg, and B. H. Cochran. Activation of the JAK-STAT pathway by reactive oxygen species. Am. J. Physiol. Physiol. 275:C1640–C1652, 1998.
Spivak, M., J. Weston, L. Bottou, L. Käll, and W. S. Noble. Improvements to the percolator algorithm for peptide identification from shotgun proteomics data sets. J. Proteome Res. 8:3737–3745, 2009.
Sringkarnboriboon, S., Y. Matsumoto, and K. Soma. Root resorption related to hypofunctional periodontium in experimental tooth movement. J. Dent. Res. 82:486–490, 2003.
Tami, A. E., P. Nasser, O. Verborgt, M. B. Schaffler, and M. L. K. Tate. The role of interstitial fluid flow in the remodeling response to fatigue loading. J. Bone Miner. Res. 17:2030–2037, 2002.
Taylor, D., J. G. Hazenberg, and T. C. Lee. Living with cracks: damage and repair in human bone. Nat. Mater. 6:263–268, 2007.
Vasquez-Sancho, F., A. Abdollahi, D. Damjanovic, and G. Catalan. Flexoelectricity in bones. Adv. Mater. 30:1705316, 2018.
Verborgt, O., G. J. Gibson, and M. B. Schaffler. Loss of osteocyte integrity in association with microdamage and bone remodeling after fatigue in vivo. J. Bone Miner. Res. 15:60–67, 2000.
Verna, C., P. M. Cattaneo, and M. Dalstra. Corticotomy affects both the modus and magnitude of orthodontic tooth movement. Eur. J. Orthod. 40:107–112, 2018.
Viecilli, R. F., M. H. Kar-kuri, J. Varriale, A. Budiman, and M. Janal. Effects of initial stresses and time on orthodontic external root resorption. J. Dent. Res. 92:346–351, 2013.
Wiśniewski, J. R., A. Zougman, N. Nagaraj, and M. Mann. Universal sample preparation method for proteome analysis. Nat. Methods 6:359–362, 2009.
Xiao, S., L. Li, L. Wang, Y. Wang, M. Zhang, J. Yao, and Y. Fan. Root surface microcracks induced by orthodontic force as a potential primary indicator of root resorption. J. Biomech. 110:109938, 2020.
Yamaguchi, M., N. Aihara, T. Kojima, and K. Kasai. RANKL increase in compressed periodontal ligament cells from root resorption. J. Dent. Res. 85:751–756, 2006.
Yu, C., D. Huang, K. Wang, B. Lin, Y. Liu, S. Liu, W. Wu, and H. Zhang. Advanced oxidation protein products induce apoptosis, and upregulate sclerostin and RANKL expression, in osteocytic MLO-Y4 cells via JNK/p38 MAPK activation. Mol. Med. Rep. 15:543–550, 2017.
Yuan, S., Z. Yang, and G. Chen. 3D microstructure model and thermal shock failure mechanism of a Si3N4-bonded SiC ceramic refractory with SiC high volume ratio particles. Ceram. Int. 45:4219–4229, 2019.
Zhao, N., B. L. Foster, and L. F. Bonewald. The cementocyte—an osteocyte relative? J. Dent. Res. 95:734–741, 2016.
Zhao, Y., A. Sarkar, and X. Wang. Peptide nucleic acid based tension sensor for cellular force imaging with strong DNase resistance. Biosens. Bioelectron. 150:111959, 2020.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos. U20A20390 and 11827803). We thank Dr. Peng Xu and Dr. Tianyun Jiang for their assistance in guiding the use of related finite element softwares. We thank the Beijing Fangtailun Medical Technology Co., Ltd. for providing the laser transmitter.
Conflict of interest
The authors declare no competing interests.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Associate Editor Eiji Tanaka oversaw the review of this article.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xiao, S., Li, L., Yao, J. et al. Microcracks on the Rat Root Surface Induced by Orthodontic Force, Crack Extension Simulation, and Proteomics Study. Ann Biomed Eng 49, 2228–2242 (2021). https://doi.org/10.1007/s10439-021-02733-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-021-02733-y