Abstract
One of existing strategies to engineer active antibody is to link V H and V L domains via a linker peptide. How the composition, length, and conformation of the linker affect antibody activity, however, remains poorly understood. In this study, a dual approach that coordinates molecule modeling, biological measurements, and affinity evaluation was developed to quantify the binding activity of a novel stable miniaturized anti-CD20 antibody or single-chain fragment variable (scFv) with a linker peptide. Upon computer-guided homology modeling, distance geometry analysis, and molecular superimposition and optimization, three new linker peptides PT1, PT2, and PT3 with respective 7, 10, and 15 residues were proposed and three engineered antibodies were then constructed by linking the cloned V H and V L domains and fusing to a derivative of human IgG1. The binding stability and activity of scFv-Fc chimera to CD20 antigen was quantified using a micropipette adhesion frequency assay and a Scatchard analysis. Our data indicated that the binding affinity was similar for the chimera with PT2 or PT3 and ~24-fold higher than that for the chimera with PT1, supporting theoretical predictions in molecular modeling. These results further the understanding in the impact of linker peptide on antibody structure and activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Whole antibodies possess excellent antigen-binding specificity and efficiency. But they are limited in clinical imaging application in vivo because of their long circulating lives and associated effector functions. During the past decades numerous works have been focused on constructing the Fv fragment of antibodies, a hetero-dimer of variable region of heavy chain (V H) and light chain (V L) to form the antigen-binding site,6,17,18 in which the Fv portion of antibody is considered as the smallest fragment that consistently retains the binding specificity and affinity of the whole antibody. However, many native Fv antibody fragments have low yield, insufficient stability, and decreased avidity. Fv fragments are not linked by covalent bonds, and consequently the domains tend to dissociate at low protein concentrations. Accordingly, many different strategies have been developed to stabilize the binding of V H and V L domains. One of the approaches to improve the stability of Fv domain association involves the construction of single-chain antibody in which the two variable domains are linked via a short flexible linker peptide. Nowadays single-chain fragment variable (scFv)6 antibodies have become a popular format of engineered antibodies. They have gained substantial clinical interest by serving as immuno-therapeutic or as vehicles to deliver selectively diagnostic and therapeutic agents, such as radionuclides, enzymes, or toxin, to tumor cells.11,33
It has long been noticed that3,4,21,38 sufficient flexibility and suitable length for V H and V L domains are achieved by assembling them in the natural Fv orientation to form a monovalent antigen-binding site, which is comparable to the Fab fragment of parent antibody. It has also been indicated that the length and sequence of the linker peptide could significantly affect scFv’s expression and stability.9,32 A certain degree of flexibility in the linker is required for the functional cooperation of the two subunits. Thus, successful construction of scFv depends on the selection of a linker that neither interferes with the folding and association of V H and V L domains nor reduces the stability and recognition abilities of the Fv molecule. To satisfy these requirements, several design strategies have been developed. One approach is to use the flexible glycine-rich sequences (GGGGS)3 as tethers.14,24 Another set of useful linkers are derived from multidomain proteins or selected by phage display technique.1,42 There are no reasons to believe, however, that a linker suitable for one antibody will be optimal for others. Indeed, the expression level, solubility, and stability of scFv depend largely on linker length and sequence and vary significantly from one linker to another, which were attested in previous works.30,32
CD20 antigen is an integral transmembrane protein expressed by B-lineage cells from B-cell precursors through mature B cells but not plasma cells.26,34 This 33–37 kDa phosphoprotein was assumed to play a role in calcium conductance and B-cell activation, proliferation, and differentiation.12,35,36 Although the biological function of CD20 antigen has not been well understood, it was still considered as a candidate for monoclonal antibody (mAb) therapy of B-cell malignancies40 since anti-CD20 antibodies are effective on inducing regression of B-cell lymphomas.15,29 In a previous study, a novel neutralized murine immunoglobin (Ig) M (IgM)-type anti-human CD20 mAb 1-28 was obtained in one of our labs,23 which possessed excellent target-binding specificity. A chimeric antibody C1-28, which contained murine antibody 1-28 variable domain and human IgG1 constant region, was subsequently constructed but the antibody lost its binding activity.41 To address the issue, a new approach is required to design a scFv fragment of C1-28 antibody using a homology modeling rationale and to functionalize the binding activity of the engineered antibody using a quantitative binding kinetics assay.
Ig superfamily molecules contain one or more domains known as Ig fold. Each Ig fold has a disulfide bridge and proximately 100 amino acids.2 The highly conserved folding of the protein and the quickly increased quantity of Ig three-dimensional (3D) structures make the superfamily ideal for molecular design on a knowledge-based strategy.28 In the current study, the impact of linker peptide on binding activity of anti-CD20 scFv fragment to CD20 antigen was predicted theoretically using molecular modeling, and the binding affinity of engineered scFv-Fc chimeric antibody was quantified experimentally using a well-developed micropipette aspiration assay5,16,27,43 and a Scatchard analysis.5,16,43
Materials and Methods
Antibodies, Proteins, and Cells
Three anti-CD20 monoclonal antibodies (mAbs) containing a chemical linker with different length of 15 (termed as C1-28/GL3), 10 (C1-28/GL2), and 7 (C1-28/GL1) amino acids were reconstructed (as seen below). A reconstructed anti-CD20 mAb containing no linker (C1-28) was used as negative control while an anti-CD20 mAb used for clinical therapy of leukemia, rituxan, was purchased from Hoffmann-La Roche Inc. (Nutley, NJ) and used as positive control. Anti-CD58 mAb TS2/9 (mIgG1)19 was generous gift from Dr. Periasamy Selvaraj (Emory University School of Medicine, USA). FITC-labeled mouse anti-human IgG and goat anti-human IgG Fc polyclonal antibodies, goat anti-human IgG-HRP polyclonal antibody, and irrelevant control hIgG1 were from Sigma Chemical Co. (St. Louis, MO).
Human leukemia CD20+-Daudi cells or CD20−-Jurkat T cell from ATCC (Rockville, MD) were cultured in RPMI 1640 media supplemented with 2 mM l-glutamine, 100 U/mL penicillin, 10 μg/mL streptomycin, and 10% fetal bovine serum. Here Jurkat cells were used as a control cell line which does not express CD20 antigen. Expression of CD20 antigen in Daudi cells was regularly checked via flow cytometry (FACS Caliber, BD Biosciences, San Jose, CA). Chinese Hamster Ovary (CHO) cell line (CHO DG44) from ATCC (Rockville, MD) was used to transfect the scFv anti-CD 20 antibody. Fresh human red blood cells (RBCs) were isolated from healthy donors.
Homology Modeling
The sequences of V H and V L domains of murine anti-CD20 antibody 1-28 were compared with primary sequences of all Igs deposited in Brookhaven Protein Data Bank (PDB). Models of V H and V L domains were constructed by using four (PDB code: 1NDM, 1D9D, 1IC4, and 1J1X) and five (PDB code: 1EO8, 1MIM, 1RVF, 1QOK, and 1FOR) Fab crystal structures from PDB, respectively. The templates of V H and V L were superimposed by using main-chain atom coordinates (InsightII 2000 software, Biosym Technologies, San Diego, CA). To further identify the conserved structural region to build up the framework of the targets, the CLUSTALW programs37 were employed to perform multiple sequence alignment upon the selected templates. An in-house sequence-based structure superposition program with a rigid-body superposition algorithm was then used to determine the conserved regions of 1-28 V H and V L domains.
All atoms of the identical residues in the template were held fixed and residues requiring side-chain replacement were carried on using rotamer library to optimize the conformation. The atomic positions of the loop residues were constructed by sequentially attaching residues to the edged residues at both sides of the conserved regions with an arbitrary set of coordinates of amino acids. The modeling procedure was manipulated using the HOMOLOGY modules of InsightII 2000 software. The original structure of 1-28 V H and V L domains was optimized using Discover_3 models of InsightII 2000 software under CVFF and AMBER force field.
Linker Peptide and scFv Fragment Design
Considering the instability of 1-28 Fv fragment, a novel linker peptide was designed rationally upon the computer-guided design method. Using the optimized 3D structure of 1-28 Fv fragment, a precise definition of N- and C-terminal residues of V H and V L domains was determined, and the distance between V H C-terminal and V L N-terminal was measured. Since the linker peptide usually appears to be flexible and protease resistant, two glycine and serine residues were chosen to form a stable linker peptide. Upon the orientation of the linker peptide, 3D scFv conformation was obtained using computer homology modeling method. Under CVFF and AMBER force field, the conformation was optimized using steepest descent and conjugate gradient methods.
Structure and Optimization of scFv-CD20 Complex
Upon the structural conservation between scFv and rituximab antibodies, an existing structure of rituximab-CD20 complex (PDB code: 2OSL) was used as a template7 to construct 3D structure of a novel CD20-scFv complex using a rigid-body superimposition procedure. Analyses in intermolecular hydrogen bonds, solvent accessible surface area, and hydrophobicity were used to identify the interacting residues at the interface. The interacting residues were then subjected to molecular dynamics at a given temperature of 298 K to optimize the conformation of interacting interface when non-interacting residues were held fixed. Here the interacting residues were initially assigned at 100 K and heated to 300 K in an increment of 20 K and the dynamic equilibrium was done in a time step of 1 fs for 0.2 ps at each temperature. To further map the possible structural variations of CD20 conformations in the new complex, an additional molecular dynamics simulation combined with an annealing process was performed when the structure of scFv antibody was fixed. The complex was heated to 1000, 2000, 3000, and 4000 K in a rate of 50 K/ps, and at each temperature the interacting residues of the complex were subjected to a 5-ps dynamics simulation run followed by gradually cooling to 300 K with heat bath coupling.39 Energy minimization using a steepest descent method for 300 steps followed by a conjugate gradient method for 3000 steps was then performed. The energy landscape was analyzed and compared using interactive graphics in all the structures generated. The structures with minimum steric clashes and lowest interaction energy were finally determined.
Plasmid Construction, Transfection, and Production of Chimeric Antibodies
Three anti-CD20 mAbs consisting of different lengths of chemical linkers (15, 10, or 7 amino acid length) were constructed. The variable cDNAs were cloned from hybridoma cells producing anti-CD20 antibody 1-2823 with inserted chimeric antibody expression vector pCMV163, which contain different sequences encoding (GGGGS)3 (PT3), (GGGGS)2 (PT2), and GGGSAAA (PT1) linkers. Stock solutions of 1-28 scFv-Ig/pCMV163 was prepared in water at a final concentration of 1 mg/mL and stored frozen at −20 °C.
Suspension-adapted CHO DG44 cells were cultured in serum-free SFMCHO-II (Hyclone) with 0.68 g/L hypoxanthine and 0.194 g/L thymidine (Sigma Chemical. St Louis, MO). The cells were maintained in 5% CO2 at 37 °C and adjusted to 1 × 107/mL. An aliquot of 0.4 ml of cell suspension and 20 μg of chimeric antibody expression vector pCMV163 were added into the cuvettes (0.4 ml, BIO-RAD #2088), mixed, and then put on electroporator (BIO-RAD Gene Pulser Xcell) for shocking one time at 280 V and 20 ms. After electroporation, cell suspension were incubated on ice for 5 min and then in SFMCHO-II in 5% CO2 at 37 °C for 72 h to collect the chimeric antibody 1-28/GL3 (containing PT3), 1-28/GL2 (containing PT2), and 1-28/GL1 (containing PT1). Cell supernatants were harvested to detect chimeric antibody expression using a sandwich ELISA assay with goat anti-human IgG Fc and anti-human IgG-HRP.
Protein Purification and Identification
Supernatants containing the chimeric antibodies were purified by protein A-sephorose 4B column (GE), and the identity and purity of the antibodies were determined by 9% nonreducing SDS-PAGE electrophoresis without controlling the same amount of protein for each antibody. For western blotting analysis, supernatants containing the chimeric antibodies were resolved on SDS-PAGE under the same conditions. Gels were transferred to nitrocellulose membranes using a western semidry transfer apparatus at 100 V for 1 h. Two percent nonfat milk in PBS was used to block nonspecific interactions and then incubated with goat anti-human IgG-HRP and developed by ECL. For flow cytometry analysis, the purified antibody was added into 3 × 105 Daudi cells at a final concentration of 10 μg/mL and incubated for 40 min on ice. After washing three times with PBS, the cell pellet was incubated with FITC-labeled secondary mAbs at a concentration of 10 μg/mL in 200 μL of FACS buffer (RPMI/5 mM EDTA/1% BSA/0.02% sodium azide) for 40 min on ice. Following additional three washes, the cells were then analyzed by flow cytometry. An aliquot of 3 × 105 Daudi cells incubated with secondary FITC mAbs alone or with an irrelevant hIgG1 mAb prior to being incubated with secondary FITC mAbs was used as control. In some cases, the cells were preincubated with C1-28/GL1 before adding C1-28/GL2 or C1-28/GL3.
Protein Coupling and Site Density Determination
A modified chromium chloride (CrCl3) protocol5,8,16,27,43 was used to couple the anti-CD20 mAbs and isotype-matched irrelevant hIgG1 onto the surface of human RBCs. Briefly, collected RBCs from healthy donors were washed three times in PBS and then stored in EAS45+ solution20 at 4 °C. About 103 cells were washed three times with 0.85% phosphate-free saline and resuspended in 250 μL of saline solution. Antibody in phosphate-free saline was added to the cells, resulting in a final antibody concentration of 2 μg/mL. Equal volume of CrCl3 solution, prepared by diluting at 1:1000 ratios aged 1% CrCl3 in 0.02 M acetate buffer, was added dropwise to the cell suspension with continuous vortexing. After 5 min, PBS/5 mM EDTA/1% BSA at a volume equal to that of the reaction mix was added to terminate the reaction. Antibody-coupled cells were rinsed twice and stored in EAS45+ at 4 °C until use. Coupling efficiency of proteins was examined by flow cytometry, using CD58 that is constitutively expressed on RBC at a known density as a standard.31
Site densities of proteins coated on RBCs or expressed on Daudi cells were measured using flow cytometry. To determine site densities of antibodies coated via CrCl3 coupling or of CD20 constitutively expressed on Daudi cells, RBCs or Daudi cells were directly incubated with FITC-labeled mouse anti-human antibody and tested by flow cytometry analysis. The site densities were then calculated by comparing the fluorescence intensities of the cells with those of standard beads (Bangs Labs, Fishers, IN).5,16,27,43
Binding Affinity Measurements
Two assays of micropipette adhesion frequency assay and Scatchard analysis were used to quantify the binding affinity of anti-CD20 mAbs at nanogram- and microgram-level antibody, respectively. A micropipette adhesion frequency assay, used to measure the kinetics and affinity of surface-bound molecule pair, has been previously described.5,16,27,43 Briefly, RBC coated with anti-CD20 mAbs (or hIgG1) and Daudi cell bearing CD20 antigen were, respectively, aspirated by two micropipettes with respective inner diameters of ~2 and ~5 μm via a suction pressure of 1–4 mmH2O. The RBC was driven by a computer-controlled piezoelectric actuator to contact repeatedly with the Daudi cell kept stationary. The contact duration was kept constant in each 100 cycle for a RBC and Daudi cell pair but varied over a range (1–15 s) for different cell pairs. Adhesion between RBC and Daudi cell was staged by placing them onto controlled contact via micromanipulation. The presence of adhesion at the end of a given contact period was detected mechanically by observing microscopically the deflection of the flexible RBC membrane upon retracting it away from Daudi cell. This contact-retraction cycle was repeated a hundred times to estimate the adhesion probability, P a, at that contact duration, t. For each anti-CD20 mAb or hIgG1 examined, ~206 pairs of cells were used to obtain several P a vs. t curves that correspond to different protein densities, m r, and CD20 antigen density, m l. Each binding curve was fitted to a small system probabilistic kinetic model.5,16,27,43
to estimate a pair of parameters: the reverse rate, k r and effective binding affinity, A c K a, where A c is the contact area, which was kept constant in all experiments. Multiple pairs of (k r, A c K a) values were obtained for each anti-CD20 mAb or hIgG1 to allow evaluation of the mean and standard deviation. The statistical significance of the difference between the affinities of different surface-bound anti-CD20 mAbs was assessed by the Student t-test.
Scatchard analysis was used to determine the solution affinities of 125I-labeled anti-CD20 mAbs to CD20 antigens expressed on Daudi cells.5,16,43 Briefly, 200 μL of 0.1–6 μg/mL 125I-labeled anti-CD20 mAb was added to 5 × 106 CD20-expressing Daudi cells. After incubation at 4 °C for 30 min, 500 μL of a 2:8 ratio of GS-1 sealing oil (Sifang Oil Inc., Beijing):dibutyl phthalate (Sigma) oil mixture was added into each sample, which was then centrifuged to separate the free 125I-labeled anti-CD20 mAbs from the cells. Radioactivity associated with the cell pellets was measured using a gamma counter. Specific binding of anti-CD20 mAbs was calculated by subtracting the nonspecific binding, which was determined by adding the same volume 125I-labeled anti-CD20 mAbs to Jurkat cells, from total binding. All assays were performed in triplicate. The statistical significance of the difference between the solution affinities for different anti-CD20 mAbs was performed using the Student t-test.
Results
3D Structure of 1-28 Fv Fragment was Constructed from Homology Modeling
Upon computer-guided homology modeling procedure, the original 3D structure of murine anti-CD20 antibody 1-28 Fv fragment was obtained using HOMOLOGY program with multiple sequence alignment. The resulted 3D structure was optimized using Discover_3 models (Fig. 1a). A protein structure analysis program, Profile_3D available in the HOMOLOGY module of InsightII 2000, was also used for structural validation. It measured the bond distances, bond angles, and dihedral angles for a given protein structure, indicating that the modeling structure was reliable (data not shown). The backbone root means square distance (RMSD) between the optimized structure and the original modeling structure of 1-28 Fv fragment was 2.84 Å, suggesting that the energy minimization did not cause a substantial distortion in the structure with relieving steric clashes and close contacts. The free energy was calculated to be 471 kcal/mol for 1-28 Fv fragment.
Optimized 3D structures for murine anti-CD20 antibody Fv fragment involving the V H (green) and V L (red) domains (a), and for 1-28 scFv mAbs with different linker peptides of PT1 (b), PT2 (c), and PT3 (d) involving the V H (green) and V L (pink) domains, the linker peptide (yellow), as well as the CDRs of V H (red) and V L (blue) domains
1-28 scFv Antibodies were Designed to Contain Different Linker Peptides
A precise definition of N- and C-terminal residues of V H and V L domains is required when determining the length of linker peptide. Upon the 3D structure of 1-28 V H and V L domains shown in Fig. 1a, the linear distance of α-carbon atoms between V H C-terminal residue (i.e., Ala116) and V L N-terminal residue (i.e., Gln1) was measured to be 43.97 Å, which was then employed to measure roughly the quantity of amino acids used for linker peptide. Based on steric geometry distance matrix method, the glycine and serine residues were chosen to construct the linker peptide. No hydrogen bonds were found when Ala116 and Ser115 in V H C-terminal formed direct contact with other residues in V H domain whereas Thr114 and Val113 appeared to contact with framework residues Thr90, Ser10, and Val12 with intra-molecular hydrogen bonds, implying that the length of linker peptide should be designed to retain intra-molecular hydrogen bonds. Thus, Ser115 and Ala116 in V H conjugated to Gln1 in V L either directly or via additional (flexible) Gly/Ser residues served as a linker peptide with various lengths. However, the V L N-terminal residues Gln1-Ile2 could not act as defacto linker residues since their side-chain atoms made a contact with Tyr95 and Thr96 residues of CDR3 in V L. Upon the above theoretical analyses, a novel linker peptide Gly4SerGly4Ser (denominated as PT2) was designed as compared to a shorter linker peptide Gly3SerAla3 (denominated as PT1) and a common linker peptide Gly4SerGly4SerGly4Ser (denominated as PT3). The anti-CD20 scFv antibodies with three different linker peptides were then constructed and modeled (Figs. 1b–1d).
3D structure and conformation were analyzed using distance geometry matrix upon its optimized configuration as exemplified in Figs. 1a–1d. The core distance between the two CDR3s in V H and V L domains was measured (Table 1). The results indicated that the conformations of two anti-CD20 scFv antibodies with PT2 and PT3 were very similar to their parent antibody 1-28 whereas that of anti-CD20 scFv antibody with PT1 was very different from its parent antibody and the other two scFv antibodies. In fact, the pocket formed by CDR3 and the orientation of scFv with PT2 were most analog to those for the parent antibody (Fv fragment).
Energy differences were also observed between the scFv antibody and its parent antibody 1-28. Here the relevant stable energy, ΔE, was estimated using the formula,
where E L and E P are the free energy for scFv antibody and its parent antibody, respectively. It was indicated that the scFv with PT2 shared the lowest ΔE (Table 1), demonstrating that it presents the most proper conformation. Taken together, the stability and avidity order were proposed: PT2 > PT3 >> PT1, and the scFv antibodies with PT2 and PT3 would have similar affinities as its parent antibody 1-28 whereas the scFv with PT1 might lose its affinity.
Structure and Binding Energy of 3D Antibody–Antigen Complex were Evaluated
To further predict the interactions between the designed scFv antibodies and CD20 antigen, molecular superimposition procedure was performed using 3D structure of CD20-rituximab complex where murine antibody 1-28 identifies the similar epitope as rituximab does.26 Here 3D scFv-CD20 complex was modeled and the stable structures were constructed (Fig. 2). The binding energy between scFv antibody and CD20 fragment was estimated (Table 2). It was indicated that the scFv with PT2 possessed the strongest binding activity while the scFv with PT1 was the weakest antibody to bind with CD20 antigen.
Constructed 1-28 scFv-Ig Chimera Served as the Antibody to CD20
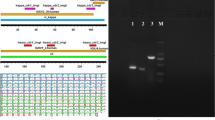
Upon the aforementioned molecular modeling, the primers were designed to amplify DNA fragments encoding the V H and V L regions of novel 1-28 scFv-Fc constructs by PCR. Isolated V H and V L DNA fragments were inserted one-by-one into the designed region of pCMV163 plasmid flanked by a sequence encoding a leader peptide at the N-terminus and a human IgG1 domain (hinge, CH2, and CH3) at the C-terminus. As exemplified in Fig. 3 for a C1-28/GL1/pCMV163 (PT1 scFv-Fc) vector, an open reading frame was created which encoded, from N- to C-terminus, the leader peptide, the V H and V L domains, and a derivative of human IgG1 Fc domain. The three vectors containing different linkers were then transfected into CHO DG44 cells. The secretion level was detected by sandwich ELISA using collected supernatants, and the yields of three proteins were ~0.2 μg/mL. The purity of three constructed scFv-Fc chimera, C1-28/GL3 with linker peptide PT3, C1-28/GL2 with PT2, and C1-28/GL1 with PT1, was tested using SDS-PAGE electrophoresis and western blotting. Nonreducing SDS-GAGE analysis indicated that the bulk of these three proteins were comprised of ~110 kDa disulfide-linked scFv-Fc, which is referred as scFv-Fc monomer (Fig. 4a). Western blotting analysis (nonreducing) demonstrated that productions of these proteins were reactive with anti-human Fc antibody with a migration consistent with the predicted molecular weight of 110 kDa for scFv-Fc monomer (Fig. 4b).
Characterization of engineered scFv mAbs. (a) SDS-PAGE analysis was carried out on 9% polyacrylamide gels and stained with Coomassie Brilliant Blue R250. (b) Western blotting analysis was performed for 1-28 scFv mAb from supernatants using HRP-conjugated GAH after separating the proteins on nonreducing SDS–PAGE gels
Binding of 1-28 scFv-Fc Construct to CD20 Antigen was Specific
CD20 binding activity of three novel 1-28 scFv-Fc constructs to Daudi cells was tested using flow cytometry. To exclude the background bindings, two measurements were done by incubating the cells with secondary FITC mAbs in the absence of the chimera or by coupling the cells with an irrelevant hIgG1 mAb prior to being incubated with secondary FITC mAbs. This turned out to be the mean fluorescent intensities (MFIs) of 8.9 and 9.4 for the former and the latter, respectively (Figs. 5a and 5b). By contrast, the MFI yielded 70.1, 133.1, and 155.3 (Figs. 5e–5g) when the cells were coupled by C1-28/GL1, C1-28/GL2, and C1-28/GL3, respectively, before being incubated with secondary FITC mAbs. To further identify if the low MFI for C1-28/GL1 is attributed to the conformation that impedes the recognition by secondary FITC mAbs, one more test was performed by pre-incubating the cells with C1-28/GL1 before adding C1-28/GL2 or C1-28/GL3. The measured MFI was 65.8 and 73.1 (Figs. 5c and 5d), very close to that for C1-28/GL1 (70.1). These data indicated that all of CD20 antigens were occupied by GL1 antibody and no free sites were available for GL2 or GL3 antibody. Thus, the reduction in MFI for GL1 (Figs. 5e–5g), was presumably attributed to its unfavorable conformation for binding to FITC antibody, even though the three 1-28 scFv-Fc constructs were recognized specifically by CD20 antigens on Daudi cells.
Flow cytometry analysis for binding of three 1-28 scFv constructs to Daudi cells. An aliquot of 3 × 105 Daudi cells was used for each case. The cells incubated with FITC-labeled GAH alone (a) or with an irrelevant hIgG1 mAb followed by the incubation of FITC-labeled GAH (b) were used as control. Blocking tests were done when the cells were pre-incubated with C1-28/GL1 before adding C1-28/GL2 (c) or C1-28/GL3 (d). Mean fluorescent intensities were measured for the three 1-28 scFv mAbs when the cells were coupled by 10 μg/mL of C1-28/GL1 (e), C1-28/GL2 (f), or C1-28/GL3 (g) in 100 μL for 30 min at 4 °C followed by incubating with FITC-labeled GAH for 20 min at 4 °C. All measurements were done in triplet
Binding specificity was also quantified, using micropipette adhesion frequency assay,5,16,27,43 by the adhesion frequency at sufficiently long contact time (t → ∞). As exemplified in Fig. 6, adhesions were mediated by specific anti-CD20 mAb-CD20 interactions, because they were present when the RBCs were coated with appropriate anti-CD20 mAbs (labeled P in four panels from the left) but were abolished when Jurkat cells or isotype-matched irrelevant mAbs (hIgG1) were used. In addition, binding was also blocked by adding soluble C1-28/GL3 construct against CD20 (labeled N in four panels from the left). The adhesion for anti-CD20 mAb C1-28 without chemical linker was indifferent with that for irrelevant hIgG1 (the first panel from the right).
Binding specificity of engineered scFv mAbs. The adhesion was present when an anti-CD20 mAb (rituxan, C1-28/GL3, C1-28/GL2, or C1-28/GL1) coated on RBC and bound to CD20 antigen expressed on Daudi cell (labeled P in four panels from the left), but was blocked when a C1-28-coated RBC bound to a Jurkat cell, a hIgG1-coated RBC bound to a Daudi cell, or an anti-CD20-coated RBC bound to a Daudi cell pre-incubated with the soluble anti-CD20 antibody (labeled N in four panels from the left). Binding of a C1-28-coated RBC to a Daudi cell was lowered to a nonspecific level when isotype-matched irrelevant hIgG1 was used (the first panel from right). Adhesion frequency for each condition was obtained by fitting the P a vs. t curves to Eq. (1) and then setting t → ∞, where the curves were obtained from 66 cell pairs with 100 contacts each cell pair. Data are presented as the mean ± standard deviation (SD)
Binding Curves Followed Simple Binding Kinetics
Dependence of adhesion frequency on contact duration was measured using the micropipette assay at contact times ranging from 1 to 15 s (Fig. 7). The adhesion probability, P a, was obtained by removing the nonspecific adhesion frequency, P n (dashed lines in Figs. 7a–7d, obtained by fitting the measured nonspecific adhesion frequency to Eq. 1), from the measured total adhesion frequency, P t, according to P a = (P t − P n)/(1 − P n).5,16,27,43 The adhesion for three scFv-Fc constructs with different lengths of linker peptides exhibited a transition phase when P a increased with contact duration, t, and then reached an equilibrium when P a keeps constant (Figs. 7a–7d). Experimental measurements were predicted by a simple kinetics (Eq. 1), which agreed well with the data (solid lines in Fig. 7).
Binding curves of engineered scFv mAbs. Adhesion probability was plotted against contact duration for C1-28/GL3 mAb (a) at two different site densities of m r = 6.5 (squares) and 1.2 (cycles) μm−2, for C1-28/GL2 mAb (b) at m r = 5.7 (squares) and 2.72 (cycles) μm−2, for C1-28/GL1 mAb (c) at m r = 22.9 (squares) and 7.9 (cycles) μm−2, and for a therapeutic anti-CD20 mAb rituxan (d) at m r = 3.9 (squares) and 1.3 (cycles) μm−2, respectively. The site density of CD20 antigen on Daudi cells was m l = 71.7 μm−2. Experimental data (points), presented as the mean ± SD at each contact time and obtained from 66 cell pairs for each curve, were compared to the predictions (solid lines) calculated from Eq. (1) using the averaged best-fit kinetic parameters and the corresponding m r and m l values. The dashed line represents nonspecific binding, obtained by fitting Eq. (1) to the nonspecific data (data not shown for clarity)
Linker Length Affected Binding Affinity
Kinetic parameters of k r and A c K a for each surface-bound anti-CD20 mAb were predicted from best fitting the data to kinetic model (Eq. 1). The mean A c K a value was obtained by averaging the values for different densities of anti-CD20 mAbs, m r, measured from independent experiments. As exemplified in Fig. 8 (right panel), the binding affinity A c K a was similar for C1-28/GL2 and C1-28/GL3 [(5.53 ± 0.61) × 10−3 and (5.25 ± 0.50) × 10−3 μm4, respectively; p > 0.90], which was ~24-fold higher than that for C1-28/GL1 [(0.22 ± 0.01) × 10−3 μm4; p < 0.01]. These values for C1-28/GL2 and C1-28/GL3 were also comparable with that for a therapeutic anti-CD20 antibody, rituxan [(7.04 ± 0.49) × 10−3 μm4; p > 0.06]. These data indicated that linker length in scFv-Fc chimera affected their binding affinities to CD20 antigen. By comparison, the reverse rates for four anti-CD20 mAbs varied slightly (k r = 0.33 ± 0.04, 0.16 ± 0.04, 0.22 ± 0.06, and 0.30 ± 0.01 s−1 for rituxan, C1-28/GL3, C1-28/GL2, and C1-28/GL1; p > 0.05).
Binding kinetics and affinities for surface-bound scFv mAbs. Reverse rate k r and binding affinity A c K a for each anti-CD20 mAb were obtained from fitting the data to kinetic model (Eq. 1). The mean k r (left panel) and A c K a (right panel) values were acquired by averaging the two values for two densities of anti-CD20 mAbs, m r, and the corresponding density of CD20 antigen, m l, measured from independent measurements. Data are presented as mean ± SD
To further test this prediction of linker length, the solution affinities were compared when soluble 1-28 scFv-Fc chimera or rituximab mAb bound to Daudi cells. The affinity obtained from the negative slope of the linear fit to the Scatchard plot was similar for C1-28/GL2, C1-28/GL3, and rituximab (0.10 ± 0.03, 0.13 ± 0.04, and 0.11 ± 0.03 nM−1, respectively; p > 0.50), which was ~3.3–4.3-fold higher than that for C1-28/GL1 (0.03 ± 0.02, p < 0.06) (Fig. 9). Taken together, these results provided the quantitative estimations to further support that the length of linker peptide affected the binding activity and affinity of 1-28 scFv-Fc chimera. The higher binding affinity of C1-28/GL2 or C1-28/GL3 constructs also supported the theoretical prediction that the binding activity of C1-28/GL2 or C1-28/GL3 to CD20 was higher than that of C1-28/GL1.
Discussion
The goal of this study was to characterize the novel anti-CD20 scFv mAbs and to quantify the impact of linker peptide on binding affinity. Using computer-guided homology and orientation alignment schemes, three anti-CD20 scFv mAbs with different linker peptide were proposed upon binding energy estimation and core distance analysis. The binding affinities of the engineered scFv-Fc mAbs were quantified using micropipette aspiration technique and Scatchard analysis (Figs. 8 and 9), supporting the prediction that the antibody with longer linker peptide is more stable with higher affinity than that with shorter linker. The advantage of the new approach is to combine the computer-aided engineered antibody design with the binding avidity and affinity quantification at nanogram- and microgram-level antibody, which is important to understand the structural bases of engineered antibody and the binding functionality to its target antigen in pharmaceutical industries.
Recombinant single-chain Fv antibody is able to be engineered in a stable form of high avidity that retains parent IgG specificity to target antigens and haptens.31 The stable form and multimeric size of a single-chain Fv unit are associated with the linker peptide length, the antibody orientation, and the core distance between V H C-terminal and V L N-terminal. To quantify the impact of linker peptide in stability and avidity of scFv mAbs, three linker peptides with different lengths were proposed and characterized in silico. With PT1 linker (GGGSAAA), V H domain was unable to associate with its attached V L domain to generate a functional orientation. With PT2 ((G4S)2) or PT3 ((G4S)3) linker, V H domain was able to bind to its attached V L in a stable form of scFv fragment and retained the bioactivity as its parent antibody (Fig. 1). Molecular mechanics procedure, computer-guided homology modeling, and 3D-QSAR analysis were coordinated in this study to predict the kinetics of scFv fragments with different linker peptides. Upon the constructed structures, three novel anit-CD20 scFv-Fc chimeric mAbs were proposed. Our data indicated that the novel mAbs so constructed bound specifically to CD20 antigens (Figs. 4–6). It should be pointed out that no direct comparisons were available between the therapeutic rituxan and scFv-Fc chimeric mAbs since the former is a whole antibody.
How the novel scFv mAbs accomplish their biological functionalities depends on their binding affinity. Physiologically a soluble antibody binds to its antigen, which seems not to be the case illustrated in the current adhesion frequency measurements where both molecules are surface-bound. One of the reasons to employ such an assay is because it enables us to quantify the binding affinity of a scFv-coated RBC to a CD20-expressing Daudi cell, as compared to semi-quantify the relative binding activity measured by ELISA assay. Even though coating scFv mAbs via CrCl3 on RBCs induced the random orientations of proteins presenting on the surface, the coupled proteins were still functional to bind to their counterpart molecules (Figs. 6 and 7; also seen in the work of Huang et al.16 and Wu et al.43). Moreover, the same effects should be expected even if the procedure of antibody coupling modifies the conformation of antigen recognition, which would not be against the conclusion that the affinity is quite lower for C1-28/GL1 than those for C1-28/GL2 and C1-28/GL3 (Figs. 8 and 9). Another line of reasoning comes from the technical advantages since only several or several-tens nanograms of proteins were required in micropipette adhesion frequency assay while at least several-tenths micrograms of proteins were required for Scatchard analysis in solution,5,16,27,43 which makes a sense for candidate antibody screening in labs. Since the former is based on a probabilistic model of small system kinetics and the latter is upon kinetics of bulk chemistry,5,16,27,43 the measured affinities were unavailable to be compared with each other. Nevertheless, both set of data indicated that the affinity for scFv mAbs with longer linker peptide was similar to that for a therapeutic anti-CD20 rituxan mAb and was much higher than that for scFv mAb with shorter peptide. This finding was in excellent agreement with that predicted from molecule modeling and theoretical analyses.
It should also be pointed out that there are several ways, such as a real-time flow cytometry and a BIAcore analysis, to quantify the binding affinity of a soluble antibody to an immobilized antigen. On one hand, a flow cytometry analysis was applicable to estimate the solution affinity between a soluble antibody and a surface-bound antigen by fitting the sigmoidal binding curve through nonlinear regression.13 However, either high affinity of the antibody or high expression level of surface-bound antigen or both was required presumably due to the low sensitivity of the analysis.10,25 Moreover, both solution affinity and reverse rate could only be determined from two independent sets of measurements,10,25 while they were able to be determined simultaneously from a single set of adhesion frequency measurements in the current study. On the other hand, BIAcore analysis was widely used in the determination of antibody affinity when the surface-bound antigen was purified out and subsequently immobilized onto the chip surface,10,25 which is not available in the case that the antigen of interest is constitutively expressed on cell membrane (e.g., CD20 antigens onto Daudi cell surface). Estimation of kinetic rates and affinity of surface-bound antigens from BIAcore analysis requires the new theoretical framework, since the impact of cell size, microtopology, deformability, as well as cell concentration, play the important roles in the process, and the perfusing flow also regulates the adhesion to and detachment from the chip. As the first step to address the issue, we recently developed a simplified cell kinetic model, which was used to quantify the kinetic rates per cell when the antigen of interest is coupled on a deformable, smooth surface.22 But it is still required to develop the new model(s) to correlate the kinetic parameters per cell to the intrinsic rates per molecule under flow, which is our on-going project.
It should also be pointed out that the impact of linker length on the activity and affinity of engineered antibody depends strongly on the distance between N- and C-terminal of V H domain. This is probably why another IF5 antibody with shorter linker peptide appeared to be a better candidate to target CD20 antigen as compared to its analog with longer linker peptide.32 Thus, the dual approach that coordinates the in-house molecular modeling and linker peptide design of engineered antibody with quantitative determinations of antibody activity and affinity is required to optimize the construction of functional engineered antibody. This study provided not only the rationale for designing the novel engineered antibodies upon molecular modeling, but also the new insight into quantifying the binding affinity of the antibodies, especially at a low level proteins.
References
Alfthan, K., K. Takkinen, D. Sizmann, H. Soderlund, and T. T. Teeri. Properties of a single-chain antibody containing different linker peptides. Protein Eng. 8:725–731, 1995.
Alzari, P. M., M. B. Lascombe, and R. J. Poljak. Three-dimensional structure of antibodies. Ann. Rev. Immunol. 6:555–580, 1988.
Atwell, J. L., L. A. Pearce, M. Lah, L. C. Gruen, A. A. Kortt, and P. J. Hudson. Design and expression of a stable bispecific scFv dimer with affinity for both glycophorin and N9 neuraminidase. Mol. Immunol. 33:1301–1312, 1996.
Bird, R. E., J. W. Jacobson, S. Johnson, B. M. Kaufman, S. M. Lee, T. Lee, S. H. Pope, G. S. Riordan, and M. Whitlow. Single-chain antigen-binding proteins. Science 242:423–426, 1988.
Chesla, S. E., P. Selvaraj, and C. Zhu. Measuring two-dimensional receptor-ligand binding kinetics by micropipette. Biophys. J. 75:1553–1572, 1998.
Davies, G. M., S. Bosze, F. Hudecz, M. R. Price, and S. J. Tendler. Characterisation of a recombinant Fv fragment of anti-MUC1 antibody HMFG1. Cancer Lett 82:179–184, 1994.
Du, J., H. Wang, C. Zhong, B. Peng, M. Zhang, B. Li, S. Huo, Y. Guo, and J. Ding. Structural basis for recognition of CD20 by therapeutic antibody rituximab. J. Biol. Chem. 282:15073–15080, 2007.
Dumaswala, U. J., M. J. Wilson, T. Jose, and D. L. Daleke. Glutamine- and phosphate-containing hypotonic storage media better maintain erythrocyte membrane physical properties. Blood 88:697–704, 1996.
Feng, J. N., Z. G. Xie, N. Guo, and B. F. Shen. Design and assembly of anti-CD16 ScFv antibody with two different linker peptides. J. Immunol. Meth. 282:33–43, 2003.
Geuijen, C. A. W., M. Clijsters-van der Horst, F. Cox, P. M. L. Rood, M. Throsby, H. H. J. Backus, E. van Deventer, A. M. Kruisbeek, J. Goudsmit, and J. de Kruif. Affinity ranking of antibodies using flow cytometry: application in antibody phage display-based target discovery. J. Immunol. Meth. 302:68–77, 2005.
Glockshuber, R., M. Malia, I. Pfitzinger, and A. Pluckthun. A comparison of strategies to stabilize immunoglobulin Fv-fragments. Biochemistry 29:1362–1367, 1990.
Golay, J. T., E. A. Clark, and P. C. Beverley. The CD20 (Bp35) antigen is involved in activation of B cells from the G0 to the G1 phase of the cell cycle. J. Immunol. 135:3795–3801, 1985.
Green, C. E., D. N. Pearson, N. B. Christensen, and S. I. Simon. Topographic requirements and dynamics of signaling via L-selectin on neutrophils. Am. J. Physiol. Cell Physiol. 284:C705–C717, 2003.
Hoedemaeker, F. J., T. Signorelli, K. Johns, D. A. Kuntz, and D. R. Rose. A single chain Fv fragment of P-glycoprotein-specific monoclonal antibody C219. Design, expression, and crystal structure at 2.4 A resolution. J. Biol. Chem. 272:29784–29789, 1997.
Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61–68, 1989.
Huang, J., J. Chen, S. E. Chesla, T. Yago, P. Mehta, R. P. McEver, C. Zhu, and M. Long. Quantifying the effects of molecular orientation and length on two-dimensional receptor–ligand binding kinetics. J. Biol. Chem. 279:44915–44923, 2004.
Huston, J. S., D. Levinson, M. Mudgett-Hunter, M. S. Tai, J. Novotny, M. N. Margolies, R. J. Ridge, R. E. Bruccoleri, E. Haber, and R. Crea. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA. 85:5879–5883, 1988.
Huston, J. S., J. McCartney, M. S. Tai, C. Mottola-Hartshorn, D. Jin, F. Warren, P. Keck, and H. Oppermann. Medical applications of single-chain antibodies. Int. Rev. Immunol. 10:195–217, 1993.
Kofler, R., and G. Wick. Some methodologic aspects of the chromium chloride method for coupling antigen to erythrocytes. J. Immunol. Meth. 16:201–209, 1977.
Kortt, A. A., O. Dolezal, B. E. Power, and P. J. Hudson. Dimeric and trimeric antibodies: high avidity scFvs for cancer targeting. Biomol. Eng. 18:95–108, 2001.
Kortt, A. A., R. L. Malby, J. B. Caldwell, L. C. Gruen, N. Ivancic, M. C. Lawrence, G. J. Howlett, R. G. Webster, P. J. Hudson, and P. M. Colman. Recombinant anti-sialidase single-chain variable fragment antibody. Characterization, formation of dimer and higher-molecular-mass multimers and the solution of the crystal structure of the single-chain variable fragment/sialidase complex. Eur. J. Biochem. 221:151–157, 1994.
Li, B., J. Chen, and M. Long. Measuring binding kinetics of surface-bound molecules using surface plasmon resonance technique. Anal. Biochem. 377:195–201, 2008.
Li, A. L., Y. Li, Y. X. Sun, M. Yu, J. S. Guo, and B. F. Shen. Studies on apoptosis of Daudi cells induced by monoclonal antibodies against human CD20. Immunol. J. 18:292–295, 2002.
Lilley, G. G., O. Dolezal, C. J. Hillyard, C. Bernard, and P. J. Hudson. Recombinant single-chain antibody peptide conjugates expressed in Escherichia coli for the rapid diagnosis of HIV. J. Immunol. Meth. 171:211–226, 1994.
Lofblöm, J., J. Sandberg, H. Wernérus, and S. Ståhl. Evaluation of staphylococcal cell surface display and flow cytometry for postselectional characterization of affinity proteins in combinatorial protein engineering applications. Appl. Environ. Microbiol. 73:6714–6721, 2007.
Loken, M. R., V. O. Shah, K. L. Datillio, and C. I. Civin. Flow cytometric analysis of human bone marrow. II. Normal B lymphocyte development. Blood 70:1316–1324, 1987.
Long, M., H. Zhao, K. S. Huang, and C. Zhu. Kinetic measurements of cell surface E-selectin/carbohydrate ligand interactions. Ann. Biomed. Eng. 29:935–946, 2001.
Petit, S., F. Brard, G. Coquerel, G. Perez, and F. Tron. Structural models of antibody variable fragments: a method for investigating binding mechanisms. J. Comput. Aid Mol. Des. 12:147–163, 1998.
Press, O. W., F. Appelbaum, J. A. Ledbetter, P. J. Martin, J. Zarling, P. Kidd, and E. D. Thomas. Monoclonal antibody 1F5 (anti-CD20) serotherapy of human B cell lymphomas. Blood 69:584–591, 1987.
Schmiedl, A., J. Zimmermann, J. E. Scherberich, P. Fischer, and S. Dubel. Recombinant variants of antibody 138H11 against human gamma-glutamyltransferase for targeting renal cell carcinoma. Hum. Antibodies 15:81–94, 2006.
Selvaraj, P., M. L. Plunkett, M. Dustin, M. E. Sanders, S. Shaw, and T. A. Springer. The T lymphocyte glycoprotein CD2 binds the cell surface ligand LFA-3. Nature 326:400–403, 1987.
Shan, D. M., O. W. Press, T. T. Tsu, M. S. Hayden, and J. A. Ledbetter. Characterization of scFv-Ig constructs generated from the anti-CD20 mAb 1F5 using linker peptides of varying lengths. J. Immunol. 162:6589–6595, 1999.
Sheikholvaezin, A., P. Sandstrom, D. Eriksson, N. Norgren, K. Riklund, and T. Stigbrand. Optimizing the generation of recombinant single-chain antibodies against placental alkaline phosphatase. Hybridoma (Larchmt) 25:181–192, 2006.
Stashenko, P., L. M. Naler, R. Hardy, and S. F. Schlossman. Characterization of a human B lymphocyte-specific antigen. J. Immunol. 125:1678–1685, 1980.
Tedder, T. F., A. W. Boyd, A. S. Freedman, L. M. Nadler, and S. F. Schlossman. The B cell surface molecule B1 is functionally linked with B cell activation and differentiation. J. Immunol. 135:973–979, 1985.
Tedder, T. F., and P. Engel. CD20: a regulator of cell-cycle progression of B lymphocytes. Immunol. Today 15:450–454, 1994.
Thompson, J. D., D. G. Higgins, and T. Gibson. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. J. Nucleic Acids Res. 22:4673–4680, 1994.
Turner, D. J., M. A. Ritter, and A. J. George. Importance of the linker in expression of single-chain Fv antibody fragments: optimisation of peptide sequence using phage display technology. J. Immunol. Meth. 205:43–54, 1997.
van Gunsteren, W. F., and H. J. Berendsen. Computer simulation as a tool for tracing the conformational differences between proteins in solution and in the crystalline state. J. Mol. Biol. 176:559–564, 1984.
von Schilling, C. Immunotherapy with anti-CD20 compounds. Semin. Cancer Biol. 13:211–222, 2003.
Wang, Y. G., Y. Huang, X. Gu, M. Yu, J. N. Feng, Y. X. Sun, Y. Li, and B. F. Shen. Construction and expression of chimeric anti-human CD20 monoclonal antibody. J. Cell. Mol. Immunol. 22:363–367, 2006.
Wang, S. H., J. B. Zhang, Z. P. Zhang, Y. F. Zhou, R. F. Yang, J. Chen, Y. C. Guo, F. You, and X. E. Zhang. Construction of single chain variable fragment (ScFv) and BiscFv-alkaline phosphatase fusion protein for detection of Bacillus anthracis. Anal. Chem. 78:997–1004, 2006.
Wu, L., B. T. Xiao, X. L. Jia, Y. Zhang, S. Q. Lü, J. Chen, and M. Long. Impacts of carrier stiffness and microtopology on 2D kinetics of P-selectin-PSGL-1 interactions. J. Biol. Chem. 282:9846–9854, 2007.
Acknowledgments
We thank Periasamy Selvaraj of Emory University School of Medicine for generous gifts of TS 2/9 mAb. This work was supported by National High Technology Research and Development Program of China Grant 2007AA02Z306, National Key Basic Research Foundation of China Grant 2006CB910303, National Natural Science Foundation of China Grants 30730032, 30771982 and 10332060, and Knowledge Innovation Program of Chinese Academy of Sciences Grant KJCX2-YW-L08 (M.L.), as well as National High Technology Research and Development Program of China Grant 2006AA02A254 and Beijing Grant 5062037 (Y.L.).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Xin Gu, Xiaoling Jia, and Jiannan Feng contributed equally to this work.
Rights and permissions
About this article
Cite this article
Gu, X., Jia, X., Feng, J. et al. Molecular Modeling and Affinity Determination of scFv Antibody: Proper Linker Peptide Enhances Its Activity. Ann Biomed Eng 38, 537–549 (2010). https://doi.org/10.1007/s10439-009-9810-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-009-9810-2