Abstract
The low membrane permeability of lipophilic drugs was resolved using liposomes as a solubilizing agent and the precise size control of them is a significant parameter in drug carrier technology. Here, we have established a microfluidic octanol-assisted liposome assembly method to produce a surfactant-assisted liposome which has merged by the cytoskeleton drug (Taxotere) encapsulation in a single process step, then a complete microfluidic cellular analysis was performed in trapping cell device with an optofluidic assay for quantifying drug permeability. The optimization of process variables resulted in the formation of liposomes with particle size 6.75 ± 0.5 µm and monodispersity 6.2%, representing encapsulation efficiency and loading capacity of 65.49 ± 3.08% and 10.16 ± 0.32%, respectively. Qualitative and quantitative studies of cellular uptake in MCF-7 cell line that was cultured in the cell trapping chip indicated a significant increase in cellular uptake of carboxyfluorescein-loaded liposomes, suggesting endocytic mechanisms. The drug-loaded liposomes with an IC50 value of 0.55 ± 0.04 μg mL−1 have shown a higher level of cellular inhibition and apoptosis in cells than free Taxotere (2.48 ± 0.01). Furthermore, real-time analysis of the dynamic labeling assay for live/dead cells was investigated. Our data revealed that lab-on-a-chip platforms for the time-lapse fluorescence imaging were applied for drug screening routines.
Graphic abstract
Cytoskeleton drugs encapsulated in liposome using microfluidic approaches and investigation of programmed cell death assay.

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Liposomes are spherical structures formed from microscopic aqueous compartments confined by a lipid bilayer. Many studies on membrane properties have been performed using liposomes as a model membrane as they mimic the self-assembled lipid matrix of the biological plasma membrane (Mijajlovic et al. 2013). Nowadays, liposomes are intensively used in different areas such as transport processes of the membrane (Becucci et al. 2015; Rezaei et al. 2020), in biophysics sciences (e.g., membrane fusion and membrane dynamics) or biochemistry (e.g., protein–lipid bilayer interactions and biomimetic membrane) (Bobone et al. 2016; Stella et al. 2016; Vaezi et al. 2019). Moreover, in the medical applications, liposomes are used as carriers for drug and gene delivery (Kuramoto et al. 2008; Mohammadi et al. 2019). Given the wide range of liposome-dependent disciplines, it is very useful to establish the methods for producing liposomes in a controlled and simple way. Traditionally, liposomes are produced in bulk with various relatively simple techniques; however, the conventional methods suffered from problems such as high and uncertain poly-dispersity, leading to variability in the amount of material encapsulated and adverse results with regard to encapsulation efficiency (van Swaay 2013).
In recent years, a number of microfluidic platforms have been introduced as a highly controlled and renewable environment for producing liposomes referred to as giant unilamellar vesicles (GUVs) in a microchip. The obtained double-emulsion droplets from water–organic solvent–water (W/O/W) are interesting in which the dissolved lipids form a bilayer by slowly removing the organic solvent (Shum et al. 2008). Giant liposomes, typically several microns in size, have attracted the attention of bio-analytical systems, especially in a microfluidic platform, due to easy imaging and monitoring of lipid membrane dynamics, permeability of the liposomal drug, and deformation of the treated cells (Deshpande et al. 2016; Edwards et al. 2012). Recently, Deshpande and Dekker (Deshpande et al. 2016; Deshpande and Dekker 2018) introduced a biocompatible organic solvent (1-octanol) that the fully assembled solvent-free liposomes were prepared by splitting off this solvent pocket within minutes. This solves a fundamental problem of the long-term presence of residual organic solvent in the bilayer of the liposome. To demonstrate drug delivery, a well-known anti-mitotic chemotherapy drug, Taxotere (TXT) (Cortes and Pazdur 1995), was encapsulated in liposomes by the microfluidic approach to improve solubility, reproducibility, and antitumor effects. This drug is highly lipophilic, practically insoluble in water, and extraordinarily toxic to normal tissues, which limit it for clinical applications. Therefore, to increase the solubility, high concentrations of the nonionic surfactant Tween 80 (polysorbate 80) and 13% (w/w, ethanol/water) were used in the commercial formulation of Taxotere. Unfortunately, there are serious side effects of their use, including febrile, fluid retention, and hypersensitivity reactions (Loos et al. 2003). However, the new micro-fabricated chip-based analytical technologies that support dynamic, real-time, cell-based analysis are necessary for uncovering the complexities of cell variability and supporting the accelerated anticancer drug discovery (He et al. 2011; Wlodkowic and Cooper 2010). The precise separation of cell-defined populations is very important for the biology of cancer cells and clinical diagnosis. Therefore, one of the essential parts of the discovery of mechanisms that regulate cell death and/or facilitate cell survival is the separation and analysis of living and dying cells, particularly, the cells in different stages of apoptosis (Wolbers et al. 2004). The goal of this study is to encapsulate Taxotere into POPC liposomes using the octanol-assisted microfluidics and then test its efficacy using a lab-on-a-chip apoptosis assay within a single cell in a trapping chip. A schematic presentation of the encapsulated a drug in liposome using microfluidic approaches and investigation of programmed cell death assay is shown in Scheme 1.
This lab-on-a-chip total analysis platform can be useful for developing liposome-based immunoassays.
2 Materials and methods
2.1 Materials
The powdered lipid of 1-palmitoyl, 2-oleoyl phosphatidylcholine (POPC) was obtained from Avanti Polar Lipids, Inc. Polyvinyl alcohol (PVA) (MW = 13,000–23,000 g mol−1, 87–88% hydrolyzed) and 1-octanol were purchased from Sigma-Aldrich. Pluronic® F-68 from ThermoFisher Scientific was used. Dulbecco’s Modified Eagle’s Medium (DMEM) and fetal bovine serum (FBS) were obtained from Gibco; MTT salt ([3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide]) (M5655), Nile Red, Calcein-AM (1 mg), Propidium iodide (P4864), and Fibronectin solution were from Sigma. Apoptosis detection kit containing PE Annexin V with 7-AAD was purchased from BioLegend.
2.2 Solution compositions
The different compositions of inner aqueous, lipid-carrying organic and outer aqueous channels are given in the electronic supplementary material (ESM) and Milli-Q water was used for the preparation of all solutions. To prevent the coalescence of liposomes and to increase their stability, a three-block copolymer surfactant, pluronic (F-68), was added to both inner and outer aqueous phases (Deng et al. 2016). The outer aqueous phase contained glycerol and PVA for improving the pinching-off process and stabilizing the liposomes. All solutions were passed through 0.22-μm filters (Millipore Corp., New Bedford, MA) before entering into the microfluidic device.
2.3 Soft lithography
Soft lithography technique was used for microfluidic devices and the fabrication of channels is completely explained in the ESM. For producing uniquely sized 10-µm liposomes, the microstructures were ~ 15 µm in height and the widths of the micro-channel at the junction were as follows: inner aqueous channel, 10 µm; lipid-carrying organic channel, 10 µm; outer aqueous channel, 20 µm; and post-junction channel, 300 µm. The height of the channels originated from spin-coater as a function of time and speed. Furthermore, there is a 15–20% fluctuation in width channels when they are copied on PDMS. For trapping single-cell device, consisting of trapping channel which was 200 μm × 700 μm (width × height) with U-shape structures (50 μm height) in series. The asymmetric U-shape rows were used for trapping because they were to be better at capturing cells throughout the chamber when compared to symmetrically rows (Zhao et al. 2013). Then after oxygen plasma treatment by Harrick Plasma 45 s at high power, the hydrophilic PDMS chip was attached to the glass coverslip, incubated in hotplate on 95 °C for 3 min to make hard binding between glass coverslip and PDMS. Before cell seeding, the device was filled with fibronectin solution (50 µg mL−1) for 1 h to allow the binding of fibronectin to the substrate to enhance cell adhesion. To remove residual fibronectin solution, the channels were rinsed by PBS.
2.4 Image acquisition and processing
An inverted microscope (Olympus IX81) equipped with fluorescence illumination and appropriate filter sets: × 10 LUPlanSApo (numerical aperture 0.40), × 20 LUCPlanFL N (numerical aperture 0.45), and × 40 LUCPlanFL N (numerical aperture 0.65) objectives (Olympus) was used to perform the experiments. For capturing the images, an Olympus DP72 digital camera was used and they were processed with AnalySIS LS professional® software. Image processing of liposomes formation was performed using MATLAB (R2019b). Circular Hough transform approach was applied to estimate the radius of liposomes.
2.5 Removal of residual solvent droplets
The density differences were utilized to remove the residual 1-octanol droplets in the formed liposome sample solution (see ESM). Briefly, the freshly prepared liposomes (density 1.017 g mL−1) and 1-octanol droplets (0.83 g mL−1) (Smallwood 1996) were collected from the outlet stream in a vial that contains an isotonic solution (as inside of liposome) whose density is between those of liposome and the 1-octanol droplets, such as sucrose or salt solution. For the demonstration, the sample solution was injected into 100 mM sucrose solution (density, 1.010 g mL−1) (Deng et al. 2016). As expected, 1-octanol droplets floated on the top of the sucrose solution within seconds, while liposomes settled down at the bottom of the vial. Lastly, the floating 1-octanol droplets were removed by pipetting, resulting in pure liposome samples in the vial and stored at 4 °C.
2.6 Liposome properties
2.6.1 Size dependence on lipid-carrying organic/inner water phase velocity
The flow rate ratio between lipid-carrying organic and inner water stream (FRR) was controlled by air pumps and varied from 1:1 to 1:3. The flow velocities for each stream of inner water and lipid-carrying organic were separately calculated by the relation
where is \(v\) and \(a\) are the flow velocity and the cross-sectional area of the channel; subsequently, the volumetric flow rate \(Q\) was calculated as
where Δp is the pressure difference across the channel (~ 40 mbar); w, h, and L are width, height, and the channel length (10 µm, 15 µm, 300 µm, respectively); η is the fluid viscosity (1-octanol droplets (0.83 g mL−1) (Bruus 2008).
2.6.2 Characterization of monodispersity of resultant liposomes
To determine the monodispersity of the resultant liposomes, coefficient of variation (CV) value was used which is defined as the ratio of the size standard deviation to its arithmetic mean with equation (Deng et al. 2014):
The diameter of the ith liposome is di, the arithmetic average diameter is \(\bar{d}\), and n is the number of liposomes. di was measured by ImageJ and n ≥ 100 in statistics.
2.6.3 Liposome production rate
Theoretical estimation of liposome production, assuming 100% yield with the known lipid content, is determined from (Hood and DeVoe 2015):
where the liposome production rate, R, (liposomes/second) resulted from Q the total flow rate (2.5e−9 m3 s−1), C the final lipid concentration (1 mg mL−1), NA Avogadro’s number, a the cross-sectional area of the lipid polar head group for POPC which is ≈ 0.6 nm2, rh the hydrodynamic radius of the liposome as obtained through circular Hough transform approach ≈ 3.3 µm, and e the thickness of the lipid bilayer (4 nm).
2.6.4 Physical stability of liposomes
This is the first report on octanol-assisted liposome assembly to encapsulate Taxotere, and time-course zeta potential (stability) for this preparation is needed and explained in the ESM.
2.7 Quantification of drug loading into liposomes and its concentration
Since the drug is very lipophilic, there might be a good chance that a large part of the drug was removed along with 1-octanol during the dewetting process. To check the concentration of the drug in the liposomes, the liposomal dispersion was centrifuged at 5000 rpm (2795×g) for 10 min at 4 °C to precipitate the un-entrapped TXT and in these conditions, the liposomes remained settled down at the bottom of the vial, above the precipitated drug. Finally, the TXT-loaded liposomes were collected and 10 µL of that was taken and dissolved in 900 mL of methanol. The encapsulation efficiency (EE) was calculated by
Moreover, the drug concentration in the bilayer was calculated by the drug loading efficiency (LE), according to the mass balance of the residual amount of drug in the liposome bilayer with a non-entrapped drug in the 1-octanol droplets:
where WTotal is the total weight of TXT added and Wunloading is the weight of the unloaded TXT in liposome; absorbance measurements were taken by a Perkin Elmer Lambda 25 UV–Visible spectrometer at 229 nm, and the purified samples as well as a serial dilution of Taxotere in 1-octanol were compared with the standard curves to determine the final drug concentration (Immordino et al. 2003).
2.8 Cell culture and 2D cell experiments in chip
2.8.1 Cell culture in the trapping cell device
For viability, cellular uptake, and apoptosis assay, the MCF-7 cells were expanded in a complete DMEM supplement with fetal bovine serum (FBS), 10% (v/v), a solution of penicillin: streptomycin (1%), and incubated in a humid environment (5% CO2, 37 °C). For effective filling of the micro-channels without any trapped air bubble, a vacuum chamber is required, which involves immersing the device in distilled water and setting it in the vacuum chamber. The air is then pumped out of the chamber to expand and escape the trapped air in the device. After the pressure is restored to the normal pressure, the remaining air is compressed and dissolved in the drawn water into the device. The cells were cultured in a T25 flask, centrifuged at 100g for 5 min, and then 10 μL of cell suspension containing 1 × 104 cells was added to the needle reservoir of the cell chamber. To remove unattached cells, after 12 h, the medium was replaced, and then to ensure that the cells are growing healthily, live/dead assay was performed. More details were explained in ESM.
2.8.2 Single-cell uptake assay
To study the qualitative cellular uptake, and assign cell internalization pathways, the MCF-7 cells were incubated with 6-carboxyfluorescein-loaded liposomes (model dye for cellular uptake analysis) that were fabricated in FRR 1:1 for 0.5, 1, 1.5, and 2 h. To visualize only the intracellular liposome, first, the treated cells were washed with PBS and then put under Olympus IX81 fluorescence microscope with a blue filter. The quantitative measurement of fluorescence intensity within the images has been carried out with Matlab 2019a software. To quantify the intensity of the green light (the cell regions), which results from cell uptake, we first segmented the green areas based on OTSU’s method (Otsu 1979) and then calculated the global statistics of the uptake areas.
2.8.3 Apoptosis study
The cytotoxicity potential of the cell was evaluated using an apoptotic assay based on Annexin V binding (Cummings et al. 2012). Briefly, MCF-7 cells were incubated with DMEM containing 0.5 μg mL−1 of free TXT and TXT-loaded liposomes incubated for 6 h. After incubation, the cells were washed by FBS (3 times) and stained with Annexin–Phycoerythrin conjugate (AnnPE) and 7-Amino-Actinomycin D (7-AAD) following the manufacturer’s protocol (Annexin V-PE Apoptosis Detection Kit, Sigma, USA) and visualized by Olympus IX81 under blue and green filters for AnnPE and 7-AAD, respectively. The necrotic and apoptotic processes can be distinguished from early apoptotic affairs in the cells using 7-AAD solution. The quantitative measure of fluorescence within the images has been carried out via Image J software (National Institutes of Health, Maryland, USA).
2.8.4 Kinetic assay of drug-induced cytotoxicity and apoptosis
Qualitative on-a-chip analysis of cancer cell death was performed using time-lapse photography. MCF-7 cells were loaded onto a trapping chip and allowed to attach to the PDMS surface overnight. Cells were then continuously perfused by 0.5 μg mL−1 liposomal drugs (less than IC50) in the presence of 2.0 µL 7-AAD. Then it was placed inside the incubator and imaged every hour over a period of 6 h.
2.9 Statistical analysis
The one-way ANOVA for multiple groups was performed for statistical analysis and **** referred to p value < 0.0001.
3 Results and discussion
3.1 On-a-chip synthesis of liposome drug
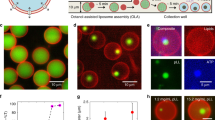
A microfluidic platform has recently been reported by Deshpande et al. in which the liposome assembly is designed with octanol-assist as lipid solvent to form cell-sized, monodispersible, and unilamellar liposomes (Deshpande et al. 2016). We used this double-emulsion solvent-extraction mechanism to produce liposomes in a designed microfluidic chip and then evaluated their ability as a carrier for a low-soluble drug; also, we developed a complete microfluidic cell analysis system for quantifying drug permeability and apoptosis assay (Figs. 1 and S1). POPC has been selected as the primary constituent of liposomes, whose properties are more similar to eukaryotic cellular membranes. The number of liposomes produced per second (the liposome production rate) is expected to increase in the microfluidic approach. The size and poly-dispersity of liposomes were measured at different flow rate ratios (FRR) and the liposome formation process has previously been reported using pressure-driven flow pumps from a single six-way junction; the inner water stream and the surrounding lipid–organic streams were hydro-dynamically focused, eventually with two outer water streams pinched off (Deshpande and Dekker 2018) (Fig. 1).
The microfluidic chip features of liposome production and the working principle on-chip using a outer water (OW), b lipid-carrying the organic phase (LO), c inner water (IW) and d downstream channel. The photo represents bright-field images of liposomes with the same diameters in the (1:1) ratio of (lipid phase: inner water phase) flow velocities, following the schematic diagram of the U-shaped cell trapping structure
Thus, the stable and monodispersible double-emulsion droplets were formed in a single step, similar to blowing the bubble (inset Fig. 1). The precise control over the expanding interfacial energies of W/O/W systems was the essential parameter throughout the liposome formation, which is obtained by adding surfactant in the system. To separate liposomes from 1-octanol droplets, it is necessary to identify them based on certain physical properties (Deshpande et al. 2017); density is one of the distinctive properties. As shown in Fig. S2, the unwanted residual 1-octanol droplets and possible free dye formed after dewetting can be easily removed from liposome samples by exploiting the density differences in the system (density: liposome ˃ sucrose ˃ 1-octanol droplet), see Sect. 2.5. Since water miscibility of 1-octanol is 0.05%, it will be very unlikely to affect the function of encapsulated drugs at a very low concentration of 1-octanol which was tested by toxicity assay; more details are given in ESM.
3.2 Simple geometric considerations and production rate of liposome
There is a linear relationship between the ratio of the two flow velocities (outer: inner water phase) and the size of the synthesized liposomes in this high-throughput manufacturing method, as reported in ref. (Deshpande et al. 2016). In this work, the ratio between the flow velocities of the inner water and lipid–organic streams was changed, and the flow velocities of these streams primarily qualify the liposome diameter as well. As shown in Fig. 2a, for production of smaller liposomes (around 6 μm), our microfluidic system optimally worked at relatively low pressure and in the (1:1) ratio of (lipid-organic: inner water) flow velocities, which this result was similar to the previous studies.
a The size distribution of liposome assembled on the microfluidic platform at increasing lipid–organic flow velocity: inner water flow velocity, b frequency histograms showing the biologically relevant size range of 1-octanol-based liposomes (~ 7 to 11 µm) and a high monodispersity for the population (n ≥ 100), solid red lines are Gaussian fits for individual histograms while arrows point out the mean diameters and corresponding SD. Note that each population was obtained under different experimental conditions, that is, in each case the distribution is unimodal (color figure online)
The inner water channel dimensions determined the minimum volume encapsulated in the liposomes and the lower limit of the liposome size. In this case, a single 10-µm size unilamellar liposome contained approximately 8.7 × 108 POPC molecules, which has calculated internal aqueous volume of 5.22 × 10−13 L (= 522 fL) (Walde et al. 2010). Although giant liposomes with ~ 50 µm diameter can easily be achieved in the microfluidic system (Kong et al. 2014; Shum et al. 2008), the lower size limit appeared around 10 µm (Matosevic and Paegel 2011) and in our fabricated device, the liposomes formed are as small as ~ 7 µm. Figures 2a and S3 represent liposomes of the same size confined within the microfluidic channels, and Fig. 2b shows a frequency histogram of three batches of liposomes formed in the size range of 6–11 µm. Solid curves are Gaussian fits to the individual histograms, while the arrows point out the mean diameters along with the corresponding standard division (SD). In the case of flow velocity ratio (0.98), both free and drug-encapsulated liposomes display a narrow, monodisperse size distribution (6.7 ± 0.2 µm) with the coefficient of variation 6.2% of the mean, while liposome size formulated with decreasing flow velocity ratio, increased around (8.9 ± 0.5 µm) with the coefficient of variation 18%. Maximum liposome production rates were also investigated by the particular flow conditions that were selected to yield liposome populations with diameters of 6.7 µm, microchannel dimensions, and concentration of lipid (1 mg mL−1). An estimated liposome production rate of 3.4 × 107 g s−1 lipid, corresponding to ≈ 100 liposomes per second was determined using initial lipid concentration and linear flow velocity, which is similar to other liposome-producing techniques (Hood and DeVoe 2015; Matosevic and Paegel 2011; Teh et al. 2011). Furthermore, drug encapsulation did not alter liposome size or polydispersity and it is independent of the loaded Taxotere concentration. The effective concentration of the drug (IC50) against MCF-7 cells was (1.0 µg mL−1) (Kushwah et al. 2018; Zhang et al. 2017), which encapsulated in liposomes (6.7 ± 0.2 µm) and suggested the saturation of the liposome system. Based on previous studies, the Pluronic® F-68 is a non-ionic, biocompatible surfactant often used in a drug delivery system and greatly improves the stability of liposomes by adjusting the interfacial energies of phospholipid-stabilized W/O/W emulsion drops, ensuring their complete dewetting and formation of oil-free monodispersible liposomes with multiple compartments (Deng et al. 2014, 2016; Teh et al. 2011). Consequently, the presence of F-68 outside the liposomes does not hinder future applications; it additionally provides certain advantages such as easier fabrication and better stability. Based on the results obtained from stability analysis, free liposome and TXT-loaded liposomes stored at 4 ± 1 °C were found to be comparatively stable and further subjected to in vitro cell uptake, cytotoxicity and acute toxicity studies (Fig. S4).
3.3 Pinching-off process and the concurrent encapsulation
To consider drug encapsulation profiles, two fluorescent dyes were encapsulated in liposomes. One inside the liposomes [6-carboxyfluorescein (CF), cell viability marker dye, 0.5 mM] which was dissolved exclusively in the inner water phase, and the other (Nile Red, hydrophobic dye, 0.1 mM) dissolved in the lipid–organic phase and located only in the bilayer (Fig. 3 and Supplementary Movie S1–3). As shown in Fig. 3a, the bright-field images of the pinching-off process highlighted in the junction at high temporal resolution and a single liposome were produced in a fraction of 30 ms. Moreover, the fluorescent of the inner water phase completely plunged into the resulting liposomes, without any visible leakage during pinching off. Thus, the excellent encapsulation yield was achieved and verified in the core of liposomes (Fig. 3b).
a High-temporal-resolution sequences showing the pinching-off process to form a liposome, b images showing the encapsulation of the fluorescent IW phase (CF) during the pinching-off process to form a double-emulsion droplet, c encapsulation of hydrophobic drug with hydrophobic dye (Nile Red), due to the very small size of liposome and high flow rate, the detection of the bilayer is difficult (color figure online)
We expected the hydrophobic drugs to be located predominantly in the hydrophobic portion of the lipid bilayer to protect the drugs from dissolution until they are taken by the cells (Fig. 3c). The encapsulation of low solubility drugs into the bilayer of liposomes allows not only their solubilization in an aqueous media, but, furthermore, can protect from degradation, improve therapeutic efficacy, and have control over the pharmacokinetic drug distribution. Additionally, the processing time requires the formation of liposome and the loading of the drug in the microfluidic device required around 3 min. Furthermore, the drug-to-lipid ratio improved five times as compared to conventional methods (Drummond et al. 1999). It should be noted that an ultra-sensitive digital camera is needed for capturing liposome production images with flow rate and this small size (~ 7 µm). Since Nile Red is a hydrophobic dye and completely embedded in the bilayer, particularly in the low concentration (0.1% lipid); therefore, upon stimulation with dye to induce TXT encapsulation, a red color liposome was observed, suggesting that the encapsulation of drugs in the bilayer can be sensitively visualized under this condition. The encapsulation efficiency was calculated to assess this microfluidic approach for a high-throughput liposome production as solubilizing agents.
The absorbance was measured at 229 nm and the concentration of loaded TXT was calculated from the calibration curve (Fig S5). The standard TXT curve was obtained through Taxotere 1-octanol solutions in a concentration gradient (y = 0.15654x + 0.06188, R2 = 0.99292), where y and x correspond to absorbance and Taxotere concentration (µg mL−1). The EE and LE for TXT-liposome were 65.49 ± 3.08% and 10.16 ± 0.32%, respectively, which is within the range of previous results in microfluidic systems (from 50 to 72%) (Hood et al. 2014; Kastner et al. 2015). Due to the low processing encapsulation time in a microfluidic device for poorly water-soluble drugs; this lower encapsulation efficiency rate is not surprising. However, it was significantly higher than drug loading via sonication in bulk scale (~ 40%) (Muthu et al. 2011). TXT was capsulated within liposomes at FRR (1:1) and total flow rate 1.0 mL min−1, with an effective concentration (50.0 µg mL−1 in the solvent stream), indicated that the solubility of TXT in water was significantly improved. It was represented a 2000-fold increase to the reported aqueous solubility of Taxotere, 0.025 μg mL−1, without (50:50, v/v) ethanol: Tween 80 (Sohail et al. 2018).
3.4 Cell culture studies
Viable cells on the trapping chip were stained with an intense green fluorescence in the cytosol by calcein-AM (a live cell indicator reagent), more details are in ESM (Bozyczko-Coyne et al. 1993). As shown in Fig. S6, similar cell morphology was observed between the control group and the microfluidic chamber during 2 days of cell culture. The quantitative cell scoring analysis (Eq. S1) exhibited high viability (~ 94%), based on cells staining positive for either calcein-AM (green fluorescence) or PI (red fluorescence). The cell culture chamber in the device is controlled by a hydrodynamic separation and sorting technique such as U-shaped trapping structures (Kazayama et al. 2015; Sajeesh and Sen 2014; Zhao et al. 2013). Using these trapping structures, cells can easily seed and immobilize to successfully increase the interaction of drug-loaded liposomes with cells. This allows a flexible strategy for testing different anticancer drugs in one single chip.
3.5 Evaluation of the microfluidic-formed liposome cytotoxicity
A significant part of post-genomic biomedical research is cell-based assays. The cytotoxicity of liposomes formed with the microfluidic method on the viability of MCF-7 human breast cancer cells was assessed by the MTT assay.
To perform this assay, cells were incubated with free TXT, free liposomes, both conventional and microfluidic form of TXT–liposomes, and 1-octanol at different concentration ranges 0.1–10 μg mL−1 for 24 and 48 h. The “dependency” of the cell viability to concentration is shown in Fig. 4a, b. According to the results, 0.05% (v/v) 1-octanol and TXT-free liposomes, as control, were used for the encapsulation which did not represent significant cell cytotoxicity in the main results. Both the TXT solution and TXT-loaded represented a clear dose-dependent cytotoxicity in which TXT-loaded liposomes had remarkable higher cytotoxicity than the free TXT in the same concentration. The differences between the toxicity effect of free TXT (Fig. 4a) and the TXT-loaded liposomes microfluidic form (Fig. 4b) were significant in all the concentrations tested for 48 h, see ESM for more details (Fig. S7). The increased cytotoxicity of liposomes may be attributed to relatively higher intracellular delivery of TXT due to higher permeability and more rapid molecularly absorption of the dissolved drug into the cancer cells by endocytosis, as evident from the results of cell uptake studies (Akhtartavan et al. 2019). The value of half-maximal inhibitory concentration (IC50) for the TXT-loaded liposomes was obtained 0.55 ± 0.04 μg mL−1 as compared to free TXT and bulk solution (Fig. 4c, Eq. S2). As shown in Fig. 4d, there is a significant difference between TXT-liposome microfluidic treated group, free TXT, and TXT–liposome bulk group. It strongly suggests that microfluidic produced liposomes have more efficient cellular uptake than bulk made liposomes, and one can see that free TXT has more cytotoxic effect in 24 h treatment due to the high amount of the available drug in the early hours of the treatment, in contrast, liposomal drug has a more gradual but stable cytotoxic effect besides they have a high amount of TXT uptake both in 24 h and 48 h treatments.
Cytotoxicity determination by the MTT assay and dependencies of the cell viability on the a tree TXT and b TXT-loaded in microfluidic liposome in the concentration range for 48 h, **** represents statistically significant difference (p < 0.0001) (mean ± SD, n = 3). c Different IC50 values for TXT against MCF-7 cell line at an incubation time of 24 and 48 h from Eq. S2. Sonication-formed liposome (bulk) and microfluidic-formed liposome (MF). d A multiple comparison test, the 24 and 48 h cell viability data (y-axis) for all the different test formulations (x-axis: Control, free liposome, free TXT, TXT-loaded in bulk and MF liposome, respectively) at a certain drug concentration of 1.0 μg mL−1
Although different values of IC50 have been reported for Taxotere®, these microfluidic-formed liposomes showed better cellular uptake and lower IC50 value against MCF-7 cells, as compared to some other TXT formulations (Table S1, ESM). Based on the IC50 value of encapsulated Taxotere for 24 h, we have chosen 0.5 μg mL−1 as experimental concentration with regard to the amount of 1-octanol presented in liposomes (50 µM). According to Wilkemeyer et al. (2000) studies, 1-octanol was categorized as inactive alcohol at a concentration of 0.05 mM without any significant effect on cell–cell adhesion, which is in agreement with our results.
3.6 Cellular uptake study
Cellular uptake studies provide essential information about the mechanisms of interaction between liposomes and cells, the localization of its encapsulated material in the cell, and the quantity of liposome internalized. Liposomes can deliver their content to the cells via four mechanisms: (1) adsorption on a cell membrane, followed by diffusion of the active component into the cell; (2) endocytosis mechanism; (3) fusion of liposomal membrane to the cell membrane; and (4) lipid exchange by transfer of lipophilic compounds from the liposomal bilayer to the cell membrane (Ducat et al. 2011; Malam et al. 2009). These mechanisms of liposome–cell interaction are variables and depend on the phospholipid type, its concentration in the liposomal membrane, the function of the cells, and the probe types (Prasad et al. 2005). One of the strategies for studying the penetration of liposome formulations in cells or membrane fusogenicity is an encapsulation of hydrophilic fluorescent dye such as CF in the inner cavity of liposomes. We used CF loaded-liposomes (size ~ 7 µm) to consider cellular uptake for different time periods (i.e. 30 min and 2 h). When liposomes are taken up by cells and lysed, their contents are diluted in the cytoplasm, allowing the cellular fluorescence to increase significantly. Indeed, the liposomes lost their internal fluorescence within a few minutes after incubation with the cell, due to uptake and interactions with the cell membrane. Chen et al. suggested a membrane-mediated pathway for cellular uptake of hydrophobic molecules preloaded in micelles. The plasma membrane provides a temporal residence for hydrophobic molecules before their delivery to target intracellular destinations (Chen et al. 2008). However, no accumulation of hydrophilic molecules into the cell membrane was observed before internalization. Therefore, our results are illustrated in Fig. 5A(a, b) and supplementary Movies 2 and 4; the total cell luminosity is sharply increased indicating that liposome binds to the cell and dye internalization take place rather than accumulation in cell membrane after 2 h incubation.
A Internalization imaging of CF loaded-liposomes in MCF-7 cells in a trapping chip after different time periods: a 30 min and b 2 h incubation. (left) Bright-field microscopy image, (middle) Green fluorescence of CF (λex: 450 nm), liposomes showed with the red sign, (right) Merged images, scale 100 µm. (also see graphical abstract), B the quantitative cellular uptake of CF-loaded liposomes in a time-dependent manner at the cell regions (color figure online)
The results of qualitative uptake were further corroborated by quantitative determination of the liposome interaction with the cell membranes. As a more rigorous way to show “the total luminosity is increased”, the fluorescent intensity profile at various time points is plotted in Fig. 5B. Interestingly, further enhancement in the fluorescent intensity was noticed at 2 h in comparison with 30 min, which showed time-dependent increase in cell uptake of CF-loaded liposomes by MCF-7 cells. In addition, increase in the incubation time does not cause remarkable changes in the CF fluorescence intensity.
This pathway of dye cellular uptake in our study was confirmed by Kushwah et al. with MCF-7 cells in which the presence of cellular uptake inhibitors suggested endocytic cellular uptake as a primary mechanism (Kushwah et al. 2018). A similar effect was observed by cellular trafficking of liposomes, demonstrating uptake through the endo-lysosomes track (Chaudhari et al. 2012). Furthermore, Papahadjopoulos et al. reported that fusion could not occur between the cellular and the liposomal membrane when liposomes are reconstituted by pure phosphatidylcholine (Papahadjopoulos et al. 1973), which in our work is pure POPC as well. The last mechanism between the liposome and cellular membranes is lipid exchange, which can occur in the absence of liposome uptake (Ducat et al. 2011). These experiments clearly show that 1-octanol-based liposomes not only can efficiently encapsulate drugs but also present a proper function of the lipid bilayer to interact with the membrane of plasma.
3.7 Apoptotic assay
An accurate interpretation of apoptotic assay such as apoptotic index (AI) has been evaluated as prognostic biomarkers in cancers and may affect therapeutic decisions and influence clinical research but their exact clinical value remains unclear. There are major side effects in the development of anti-cancer drugs include unexpected toxicity, insufficient penetration of the drug into cancer tissue, and the lack of a favorite effect on the target. Therefore, the observed cell cytotoxicity results and intracellular uptake of Taxotere-liposomes were further confirmed by the apoptosis assay. The site direct drug delivery has been developed to deliver carriers within tumor cells via receptor-mediated endocytosis. The damage of plasma membrane integrity and increase permeability in the latest stages of cell death was detected by the staining of end-point markers, resulting from necrotic or apoptotic processes. Therefore, early apoptotic cells detected by the vital dye in conjunction with PE Annexin V. 7-AAD excluded from intact membranes of live cells, whereas the dead and damaged membranes of cells are permeable to it. Quantitative estimation of apoptosis was further evaluated via calculating the apoptosis index, the ratio of the red (7-AAD, a measure of late apoptotic cells) to yellow (Annexin V-PE conjugate, a measure of early apoptotic cells) fluorescence intensity. As compared to free TXT (apoptotic index 0.60, Fig. 6a), the larger portions of cells were found to be apoptotic when treated with liposomes (apoptotic index 1.01, Fig. 6b), showing higher drug uptake and cell cytotoxicity. Additionally, the apoptotic index for cells exposed to TXT-loaded microfluidic liposomes in 96-well plates is comparable within the chip and untreated cells and TXT-loaded bulk liposomes used as control groups (Fig. S8).
Apoptosis assay of A free drug versus B microfluidic liposomal drug against MCF-7 cells; whereas first window a depicts the bright field image of representative cells, b yellow channel depicts fluorescence from PE Annexin V (early apoptosis marker dye), and c red channel shows the fluorescence from 7-AAD (late apoptosis marker dye). 7-AAD has a high DNA binding constant and is efficiently excluded by intact cells. When excited in 488 nm (green filter), the fluorescence is detected in the far-red range of the spectrum (650 nm). The apoptosis index was measured as the ratio of fluorescence intensity from the red channel to the yellow channel. Scale bar 100 µm (color figure online)
3.8 Kinetic of cytotoxicity assays in single-cell level
Cell culture on-a-chip system which supports continuous perfusion of cells over extended periods of time can be used to conduct high-throughput screening (Fig. 7). Therefore, a qualitative on-chip analysis of cancer cell death was performed using time-lapse imaging, every 1 h over a period of 6 h.
Time-lapse imaging of apoptosis at a single-cell level. MCF-7 cells were cultured on-a-chip and exposed to Taxotere-loaded liposomes at a concentration less than IC50 equivalent to Taxotere (0.5 μg mL−1) for up to 6 h in the continuous presence of a the plasma membrane permeability marker 7-AAD, b composite with bright field and red squares denote c the magnified cell (5 times) undergoing apoptosis (color figure online)
The first detectable apoptosis morphologic event was found in about 1 h, including loss of cell–cell contacts. The next stages of apoptosis are to continue cell dehydration, shrinkage, rounding up and loss of attachment with the substrate (2–4 h); finally, the permeability of the plasma membrane is increasing to 7-AAD (6 h) as a marker of end-point assays for live against dead cells. The latter events indicate the ultimate instability of plasma membrane structure during apoptosis. After entering the cells, it bound to the nucleic acids by intercalation, and weak nuclear fluorescence is probably from binding to RNA and cytoplasmic staining that might be due to the uptake of 7-AAD by mitochondria, lysosomes or endosomes. Kinetic imaging from the end-point of a single-cell on-a-chip provides a better dynamic observation of apoptotic cell death. We suggest that this simple bioassay allows for easy tracking of the differences in stochastic cell death sensitivity, which often remains undetectable with conventional end-point analysis. The clinical validation and development of combinatorial regimens for struggle malignancy can result from such a dynamic analysis of cancer cell death. An advantage of flow cytometry (FCM) is a quick analysis that can reach tens of thousands of cells per second (several times faster than a microfluidic process). However, the common problems of conventional FCM are its complexity, cost, limited portability, and the requirement for trained personnel. Moreover, each flow cytometric analysis requires a considerable number of cells (typically > 104 cells. mL−1), and the processing of samples prior to analysis is time-consuming, which often involves several centrifugation steps that connect cells to additional stress. Whereas, a considerably reduced number of cells needed in microfluidic systems (even 20-fold), providing a significant advantage for the studies of primary cancer cells (Wlodkowic et al. 2011). In addition, the ability to monitor single cancer cells in a rare population provides the development of personalized treatments in which the dosages of the drug can be patient-defined and determined for the therapy of individual disease (Sims and Allbritton 2007). Most importantly, reducing the processing steps of the sample obtained with such a kinetic protocol is essential for the storage of apoptotic cells (Wlodkowic et al. 2009; Xia et al. 2019). The complexity of conventional cell culture protocols can reduce by a simple microfluidic device and can also provide time-lapse studies on the apoptosis of adherent cells. The latter feature is a special advantage to determine the kinetics of drug potency and enable for sequential pharmacological stimulations and real-time analysis of cellular physiology while providing a keen decrease in the use of reagents and samples. Finally, we adopt a set of standard fluorescence assays with dynamic, timing bioassays that are able to perform traditional tasks in a more practical, faster, and cheaper way.
4 Conclusions
In summary, this microfluidic approach can be useful for developing liposome-based immunoassays, producing giant liposome with 6.7 ± 0.5 µm in size. The drug was encapsulated at molar ratio (drug: lipid) up to 1:25 with efficiency 65.49 ± 3.08% in a single process step and maximum residence time was about 3 min in the chip. Furthermore, it has the ability to optimize the formulation of liposomal drugs with high drug concentrations, minimal waste reagent at the point of care, and great stability more than 80% after storage for 2 weeks. Importantly, our system is a good model to be used for drug screening at single-cell level and the transfer of traditional bio-analytical methods such to a micro-fabricated format increases the resolution of bio-analysis and the sampling throughput as well as the possibility of designing automated systems with innovative capabilities.
References
Akhtartavan S, Karimi M, Karimian K, Azarpira N, Khatami M, Heli H (2019) Evaluation of a self-nanoemulsifying docetaxel delivery system. Biomed Pharmacother 109:2427–2433
Becucci L et al (2015) Interaction study of phospholipid membranes with an N-glucosylated β-turn peptide structure detecting autoantibodies biomarkers of multiple sclerosis. Membranes 5:576–596
Bobone S et al (2016) Activity and selectivity of host defense peptides: a complex interplay of multiple equilibria. In: Journal of Peptide Science, 2016. Wiley 111 River ST, Hoboken 07030-5774, NJ USA, pp S36–S36
Bozyczko-Coyne D, McKenna B, Connors T, Neff N (1993) A rapid fluorometric assay to measure neuronal survival in vitro. J Neurosci Methods 50:205–216
Bruus H (2008) Theoretical microfluidics, vol 18. Oxford University Press, Oxford
Chaudhari KR et al (2012) Bone metastasis targeting: a novel approach to reach bone using Zoledronate anchored PLGA nanoparticle as carrier system loaded with Docetaxel. J Controll Rel 158:470–478
Chen H, Kim S, Li L, Wang S, Park K, Cheng J-X (2008) Release of hydrophobic molecules from polymer micelles into cell membranes revealed by Förster resonance energy transfer imaging. Proc Natl Acad Sci 105:6596–6601
Cortes JE, Pazdur R (1995) Docetaxel. J Clin Oncol 13:2643–2655
Cummings BS, Wills LP, Schnellmann RG (2012) Measurement of cell death in Mammalian cells. Curr Protoc Pharmacol 56:12.18. 11–12.18. 24
Deng N-N, Mou C-L, Wang W, Ju X-J, Xie R, Chu L-Y (2014) Multiple emulsion formation from controllable drop pairs in microfluidics. Microfluid Nanofluid 17:967–972
Deng N-N, Yelleswarapu M, Huck WT (2016) Monodisperse uni-and multicompartment liposomes. J Am Chem Soc 138:7584–7591
Deshpande S, Dekker C (2018) On-chip microfluidic production of cell-sized liposomes. Nat Protoc 13:856
Deshpande S, Caspi Y, Meijering AE, Dekker C (2016) Octanol-assisted liposome assembly on chip. Nat Commun 7:10447. https://doi.org/10.1038/ncomms10447
Deshpande S, Birnie A, Dekker C (2017) On-chip density-based purification of liposomes. Biomicrofluidics 11:034106
Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D (1999) Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol Rev 51:691–744
Ducat E, Evrard B, Peulen O, Piel G (2011) Cellular uptake of liposomes monitored by confocal microscopy and flow cytometry. J Drug Deliv Sci Technol 21:469–477
Edwards KA, Bolduc OR, Baeumner AJ (2012) Miniaturized bioanalytical systems: enhanced performance through liposomes. Curr Opin Chem Biol 16:444–452
He T, Liang Q, Zhang K, Mu X, Luo T, Wang Y, Luo G (2011) A modified microfluidic chip for fabrication of paclitaxel-loaded poly (l-lactic acid) microspheres. Microfluid Nanofluid 10:1289–1298
Hood RR, DeVoe DL (2015) High-throughput continuous flow production of nanoscale liposomes by microfluidic vertical flow focusing. Small 11:5790–5799
Hood R, Vreeland W, DeVoe DL (2014) Microfluidic remote loading for rapid single-step liposomal drug preparation. Lab Chip 14:3359–3367
Immordino ML, Brusa P, Arpicco S, Stella B, Dosio F, Cattel L (2003) Preparation, characterization, cytotoxicity and pharmacokinetics of liposomes containing docetaxel. J Controll Rel 91:417–429
Kastner E, Verma V, Lowry D, Perrie Y (2015) Microfluidic-controlled manufacture of liposomes for the solubilisation of a poorly water soluble drug. Int J Pharm 485:122–130
Kazayama Y, Teshima T, Osaki T, Takeuchi S, Toyota T (2015) Integrated microfluidic system for size-based selection and trapping of giant vesicles. Anal Chem 88:1111–1116
Kong F, Zhang X, Hai M (2014) Microfluidics fabrication of monodisperse biocompatible phospholipid vesicles for encapsulation and delivery of hydrophilic drug or active compound. Langmuir 30:3905–3912
Kuramoto H et al (2008) On-chip fabrication of mutifunctional envelope-type nanodevices for gene delivery. Anal Bioanal Chem 391:2729–2733
Kushwah V, Jain DK, Agrawal AK, Jain S (2018) Improved antitumor efficacy and reduced toxicity of docetaxel using anacardic acid functionalized stealth liposomes. Colloids Surf B 172:213–223
Loos WJ, Baker SD, Verweij J, Boonstra JG, Sparreboom A (2003) Clinical pharmacokinetics of unbound docetaxel: role of polysorbate 80 and serum proteins. Clin Pharmacol Ther 74:364–371
Malam Y, Loizidou M, Seifalian AM (2009) Liposomes and nanoparticles: nanosized vehicles for drug delivery in cancer. Trends Pharmacol Sci 30:592–599
Matosevic S, Paegel BM (2011) Stepwise synthesis of giant unilamellar vesicles on a microfluidic assembly line. J Am Chem Soc 133:2798–2800
Mijajlovic M, Wright D, Zivkovic V, Bi J, Biggs MJ (2013) Microfluidic hydrodynamic focusing based synthesis of POPC liposomes for model biological systems. Colloids Surf B 104:276–281
Mohammadi SS, Vaezi Z, Shojaedin-Givi B, Naderi-Manesh H (2019) Chemiluminescent liposomes as a theranostic carrier for detection of tumor cells under oxidative stress. Anal Chim Acta 1059:113–123
Muthu MS, Kulkarni SA, Xiong J, Feng S-S (2011) Vitamin E TPGS coated liposomes enhanced cellular uptake and cytotoxicity of docetaxel in brain cancer cells. Int J Pharm 421:332–340
Otsu N (1979) A threshold selection method from gray-level histograms. IEEE Trans Syst Man Cybernet 9:62–66
Papahadjopoulos D, Poste G, Schaeffer B (1973) Fusion of mammalian cells by unilamellar lipid vesicles: influence of lipid surface charge, fluidity and cholesterol. BBA Biomembr 323:23–42
Prasad TK, Rangaraj N, Rao NM (2005) Quantitative aspects of endocytic activity in lipid-mediated transfections. FEBS Lett 579:2635–2642
Rezaei N, Mehrnejad F, Vaezi Z, Sedghi M, Asghari SM, Naderi-Manesh H (2020) Encapsulation of an endostatin peptide in liposomes: stability, release, and cytotoxicity study. Colloids Surf B Biointerfaces 185:110552
Sajeesh P, Sen AK (2014) Particle separation and sorting in microfluidic devices: a review. Microfluid Nanofluid 17:1–52
Shum HC, Lee D, Yoon I, Kodger T, Weitz DA (2008) Double emulsion templated monodisperse phospholipid vesicles. Langmuir 24:7651–7653
Sims CE, Allbritton NL (2007) Analysis of single mammalian cells on-chip. Lab Chip 7:423–440
Smallwood I (1996) Handbook of organic solvent properties (Arnold, London) Smallwood handbook of organic solvent properties 1996
Sohail MF et al (2018) Advancements in the oral delivery of Docetaxel: challenges, current state-of-the-art and future trends. Int J Nanomed 13:3145
Stella L et al (2016) The role of thermodynamics in the activity and selectivity of antimicrobial peptides. Biophys J 110:75a–76a
Teh S-Y, Khnouf R, Fan H, Lee AP (2011) Stable, biocompatible lipid vesicle generation by solvent extraction-based droplet microfluidics. Biomicrofluidics 5:044113
Vaezi Z et al (2019) Aggregation determines the selectivity of membrane-active anticancer and antimicrobial peptides: the case of killerFLIP. BBA Biomembr 1862(2):183107
Van Swaay D (2013) Microfluidic methods for forming liposomes. Lab Chip 13:752–767
Walde P, Cosentino K, Engel H, Stano P (2010) Giant vesicles: preparations and applications. ChemBioChem 11:848–865
Wilkemeyer MF, Sebastian AB, Smith SA, Charness ME (2000) Antagonists of alcohol inhibition of cell adhesion. Proc Natl Acad Sci 97:3690–3695
Wlodkowic D, Cooper JM (2010) Tumors on chips: oncology meets microfluidics. Curr Opin Chem Biol 14:556–567
Wlodkowic D, Skommer J, McGuinness D, Faley S, Kolch W, Darzynkiewicz Z, Cooper JM (2009) Chip-based dynamic real-time quantification of drug-induced cytotoxicity in human tumor cells. Anal Chem 81:6952–6959
Wlodkowic D, Khoshmanesh K, Sharpe JC, Darzynkiewicz Z, Cooper JM (2011) Apoptosis goes on a chip: advances in the microfluidic analysis of programmed cell death. Anal Chem 83:6439–6446
Wolbers F, Andersson H, Van den Berg A, Vermes I (2004) Apoptosis induced kinetic changes in autofluorescence of cultured HL60 cells-possible application for single cell analysis on chip. Apoptosis 9:749–755
Xia A et al (2019) A fixed cytometer chip for identification of cell populations and real-time monitoring of single-cell apoptosis under gradient UV radiation. Microfluid Nanofluid 23:78
Zhang T et al (2017) Polysialic acid and Pluronic F127 mixed polymeric micelles of docetaxel as new approach for enhanced antitumor efficacy. Drug Dev Ind Pharm 43:1827–1835
Zhao L, Cao J-T, Wu Z-Q, Li J-X, Zhu J-J (2013) Lab-on-a-Chip for anticancer drug screening using quantum dots probe based apoptosis assay. J Biomed Nanotechnol 9:348–356
Acknowledgements
We are very thankful to Prof. Alireza Ghassempour and Prof. Peyman Salehi (medicinal plants and drugs research institute, Shahid Beheshti University) for kindly providing us with the purified Taxotere. The authors would like to kindly acknowledge all the supports and funding from the University of Tarbiat Modares (Grant IG-39708).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There are no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 2 (AVI 88720 kb)
Supplementary material 3 (AVI 7535 kb)
Supplementary material 4 (MP4 10816 kb)
Supplementary material 5 (MP4 2354 kb)
Rights and permissions
About this article
Cite this article
Vaezi, Z., Sedghi, M., Ghorbani, M. et al. Investigation of the programmed cell death by encapsulated cytoskeleton drug liposomes using a microfluidic platform. Microfluid Nanofluid 24, 48 (2020). https://doi.org/10.1007/s10404-020-02353-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-020-02353-3