Abstract
The dynamics of DNA molecules in highly confined nanoslits under varying electric fields are studied using dissipative particle dynamics method, and our results show that manipulation of the electrical field can strongly influence DNA mobility. The mobility of DNA μ scales with electric field E as \( \mu = \mu^{\text{H}} - k_{1} e^{{ - E/E_{c} }} . \) And the data points for different DNA lengths finally approach each other in strong fields, which suggest that the sensitivity to chain length is almost lost. To explain the unusual field-dependent phenomena, we analyze the time evolution of DNA configurations under different fields. For strong driving potentials when the system is dominated by the electric driving force, the DNA chains are more likely to hold coiled configurations. For weak driving potential when the random diffusion forces dominate, we see frequent dynamic transitions between stretched and coiled configuration, which may increase the drag resistance, therefore reduce the mobility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Nanofluidic systems have been developed as potential tools for the analysis, detection, and separation of chemical and biological agents with broad applications in biology and medicine (Eijkel and van den Berg 2004; Abgrall and Nguyen 2008). Gel has been conventionally used as a separating medium in DNA electrophoresis (Lumpkin et al. 1985; Chu and Wang 1991; Brahmassandra et al. 2001). But it has been proposed that gel-less separation of DNA using nanochannel electrophoresis may significantly improve both the speed and separation of DNA (Baldessari and Santiago 2006; Balducci et al. 2006). Single well-designed and controlled nanochannels are ideal physical modeling systems to study fluidics in a precise manner. For instance, Jo et al. (2007) showed that DNA stretch is possible in relatively large (100 nm) nanoslits by using low-ionic-strength buffers. And Cross et al. (2007) performed DNA separation using 19-nm deep nanochannels, and he proposed a simple theoretical model to describe the length-dependent mobility for 2–10 kbp DNA.
For the electrophoresis conducted in small constriction size channel or under strong driving electric field, theoretical and experimental studies beyond the equilibrium theories are rather important for studying the relevant processes (Stein et al. 2006; Salieb-Beugelaar et al. 2008). For instance, for pressure-driven individual DNA molecules transporting through 175 nm–3.8 μm high silica channels (Stein et al. 2006); DNA mobility increased with molecular length in channels larger than the molecular radius of gyration, whereas it was practically independent of molecular length in thin channels. And in a recent experiment (Salieb-Beugelaar et al. 2008), when the applied field is as high as 30–200 kV/m, DNA molecules in 20 nm nanoslits showed highly reduced mobility because some DNA were delayed/trapped inside the nanoslits, and there were even repeated hooking with the wall surface and elongation during the translocation process. Meller et al. (2001) investigated the voltage-driven DNA translocation through nanopores, in which he found a quadratic dependence of mobility on electric field \( \mu = k_{0} + k_{1} \left( {E - E_{0} } \right)^{2} \), where \( E_{0} \) is a threshold potential and k 0 is a small additive constant. Streek et al. (2004) used Brownian dynamics simulation to study DNA migration in an electrophoretic microchannel device designed by Han et al. (1999) and Han and Craighead (2000) for the separation of DNA molecules. He reproduced the experimental observation that the mobility increases with the length of the DNA. And he also reported that longer chains are more likely to diffuse out of their main path into the corners of the box and remain trapped. Pan et al. (2010) conducted dissipative particle dynamics (DPD) simulations and verified the observation in Han et al.’s (1999) and Han and Craighead’s experiments (2000), but he also claimed that the corner trapping is not a contributor to the DNA separation process as reported by Streek et al. (2004).
Experiments on DNA electrophoresis in nanofilters at high fields showed some band-inverted behavior (Fu et al. 2006, 2007), in which the relative mobility increases with the electric field. However, our understanding of the high field electrophoresis is still incomplete. Since, we still do not have a satisfactory model for the electrophoretic transport of DNA in strong confinement under strong field, further investigation is necessary. In this article, we investigate the field-dependent mobility of DNA in nanoslits. Our data show the influence of the electrical field on the configuration and movement of DNA molecules with different lengths. A new model is proposed to predict the field dependence of the DNA mobility in nanoslits under strong electric field.
2 Simulation model
2.1 DPD model
DPD is a mesoscopic method that bridges the gap between the atomistic scale that is accessible by molecular dynamics (MD) simulations and the macroscopic scale described by continuum methods. It was introduced by Hoogerbrugge and Koelman (1992) as a novel method to simulate complex fluids, and it has gained significant theoretical support and refinement thereafter (Groot and Warren 1997). Like the MD method, the DPD method describes the materials by ensembles of particles, and every particle is defined by its position, velocity, and mass. Based on Newton’s equation of motion, if the mass of particles is normalized to 1, the time evolution of the positions and velocities of DPD particles are calculated by
where \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {r}_{i} \) and \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {v}_{i} \) are the position and velocity vectors of particle i, \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {F}_{i}^{\text{ext}} \) is the external force on the particle i, and \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {F}_{i}^{\text{int}} \) is the total interparticle force acting on particle i, consisting of three parts:
The conservative force, dissipative force, and random force on particle i due to the presence of other particles are given by
The conservative force is a soft repulsion with the maximum repulsion \( a_{ij} \) between particles i and j,
where \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {r}_{ij} = \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {r}_{i} - \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {r}_{j} ,\,r_{ij} \) is the distance between particles i and j, \( \hat{r}_{ij} \) is the unit vector directed from particle j to i, and r c is the cut-off radius.
The dissipative force and random force, on the other hand, are characterized by the weight functions \( \omega^{\text{D}} \) and \( \omega^{\text{R}} \) which vanish if \( r \ge r_{\text{c}} , \)
where \( \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {v}_{ij} = \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {v}_{i} - \overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\rightharpoonup}$}} {v}_{j} \) and \( \theta_{ij} \) is a white noise. \( \gamma \) and \( \sigma \) are the coefficients coupled by \( \gamma = \frac{{\sigma^{2} }}{{2k_{\text{B}} T}} \) where k B T is the Boltzmann temperature, and the weight functions are coupled by \( \omega^{\text{D}} (r) = [\omega^{\text{R}} (r)]^{2} . \) According to the fluctuation–dissipation theorem, the above relation is necessary for thermodynamic equilibrium at the specific temperature. A widely adopted weight function is given by,
The DPD parameters of conservative force are given by \( a^{\text{ss}} = 375,\,a^{\text{bb}} = 10,\,a^{\text{bs}} = 1,\,a^{\text{sw}} = a^{\text{bw}} = 12.75. \) For dissipative force and the random force, \( \sigma^{\text{ss}} = \sigma^{\text{bb}} = 3,\,\sigma^{\text{bs}} = 0.5. \) Here s represents solvent particle, b the bead, and w the wall. Kasiteropoulou et al. (2011) investigated the DPD parameters affecting planar nanochannel flow, and he found that the fluid-wall conservative force \( a^{\text{sw}} \) affect both fluid velocity and pressure distribution.
DNA chain can be represented as a series of beads linked through springs after coarse-graining. And a variety of numerical models have been developed to represent the restoring force of the spring, including Hookean spring, the inverse Langevin function spring, the finitely extensible non-linear elastic (FENE) spring and the wormlike chain (WLC). In particular, the WLC model is well suited for describing stiff polymers and is effectively used to model DNA molecules. Perkins et al. (1995) found the mechanical properties of DNA molecule in an aqueous solution can be realistically modeled using the WLC model, where the attraction spring force between successive beads i and j is expressed as
Here, l is the maximum length of one chain segment and P is the effective persistence length, which are both chosen to be 50 nm. Symeonidis et al. (2005) also studied the different bead-spring representations for the excluded volume effects, among which he found that the parameters for the WLC were consistent with the rest of the models. Here, we choose the maximum segment length to be 2.778\( [\sigma ], \) leading to a correspondence of 20, 35, 50, 69, and 103 bead chains for DNAs of 2.8, 5, 7.2, 10, and 15 kbp, respectively.
Then density of solvent is chosen to be 0.1, the cut-off radius r c is set to 2.0. Our parameter choices give an energy unit \( [\varepsilon ] = k_{\text{B}} T = 4.14 \times 10^{ - 21} \,{\text{J}}, \) for T = 300 K, a length scale \( [\sigma ] = 18\,{\text{nm}}, \) and a mass unit \( [m] = 2 \times 10^{ - 14} \,{\text{kg}}. \) The time scale is calculated as \( [t] = \sqrt {m\sigma^{2} /\varepsilon } = 3.95 \times 10^{ - 15} \,{\text{S}}, \) and each time step is \( \Updelta t = 0.01[t] \) (Kumar et al. 2009).
2.2 Implementation of electroosmotic flow (EOF)
When a large DNA molecule enters a tight nanoslit, the molecule has to overcome an entropic barrier. Once inside the channel, it will experience friction with the wall and the fluid inside the nanoslit. Furthermore, the DNA transport and conformations will be influenced by the charged surface and the resulting potential in the electrical double layers (EDL). Experiments (Pennathur and Santiago 2005) proved that the transport of analyte ions is a function of ion valence, EDL thickness and surface charge density in the case of order 10 nm channels. The characteristic thickness of EDL is the Debye length, which is typically formulated as
where \( k_{\text{B}} \) is the Boltzmann constant, \( \varepsilon_{0} ,\,\varepsilon_{b} \) are the permittivity of vacuum and the dielectric constant of fluid, respectively, C represents the ionic strength, e is the electron charge, and z is the valence number.
When the Debye length is small compared to the characteristic length scale of the channel, the electric field E and flow velocity U EOF are proportional, with the proportionality coefficient being constant throughout the domain.
where \( \eta \) is the fluid viscosity, \( \xi \) is the zeta potential (Hunter 1981), and E is the local electric field. If the surface is negatively charged, the net excess of positive ions in the EDL will draw the liquid along because of viscous interactions, which results in flow toward the cathode. Because of the fluid motion around the molecule, the molecule is pushed backward in the opposite direction.
To simulate the flow near the wall, an effective bounce-back boundary condition is employed in our DPD simulation for particles that penetrate the wall (Duong-Hong et al. 2008a). The new position and velocity of a particle crossing the wall are given by
Here d r is the normal distance from the particle to the boundary and n w is the normal vector on the wall directing into the simulation domain, U wall is the velocity of the wall. The velocity U wall is locally assigned to the wall particles, and the “moving wall” therefore drags the fluid particles by viscosity.
We employed a simple method to effectively simulate the local EOF generated by the counterions around the DNA molecules (Duong-Hong et al. 2008b). The electrostatic interactions and the resulting fluid shearing mainly occur within the Debye layer in DNA electrophoresis. When the Debye length \( \lambda_{\text{D}} \) is smaller than the radius of gyration R g of molecules, the interactions between the counterions and the DNA molecules needs to be considered. When an electric field is applied, the DNA will be attracted to the cathode and the counterions will be attracted toward the anode. Accurate modeling of the process requires explicit modeling of counterion charges, which is too expensive to carry out. Since the electrophoresis is the converse of electroosmosis (Fig. 1), we assumed that if the distance between a solvent particle and the DNA is within the Debye length \( \lambda_{\text{D}} \), the former acquires the same amount of charge that one DNA-bead possesses, but it is positive rather than negative. Therefore, the solvent particle will be subject to an electric force that is equal to the one exerted on the DNA particle, but in the opposite direction. This model was proven to work well for DNA electrophoresis without creating any fluid motion in the surrounding area (Duong-Hong et al. 2008b). They included the approximation of the complex solvent–DNA interactions in the simulation and obtained unperturbed velocity profile of local EOF. And it was shown that the free-draining flow can be effectively modeled without compromising the computational efficiency of the method.
3 Results and discussion
We performed 3D DPD simulation in slits with DNA molecules at varying applied electric field. Periodic boundary conditions are employed in both x and y directions, so there is no lateral confinement of DNA molecules. From the displacement of DNA during electrophoresis, the mobility of the center of mass of the DNA was calculated, as shown in Fig. 2a. The mean value and standard deviation of the mobility for each DNA length, as shown in Fig. 2b, suggests that the dependence of the mobility of DNA chain on nanoslit height is almost negligible.
To obtain the field dependence of DNA mobility, we conducted simulations of DNA electrophoresis across 54 nm nanoslits under different electric fields. We found that the movement of molecules differed as a function of the applied field. As we can see from Fig. 3, when the applied field is low (E* < 5), DNA mobility increases rapidly with a slight increase of the field. But the mobility eventually levels off with a further increase of the field, and the data points for different DNA lengths begin to approach each other at around E* = 16, which suggest that the sensitivity to chain length is almost lost at high fields. The high field mobility \( \mu^{\text{H}} \) of all three datasets is the same, which is around \( 6.7 \times 10^{ - 9} \,{\text{m}}^{2} /{\text{V}}\,{\text{s}}. \) The mobility data for DNA in 54 nm nanoslits are fitted using \( \mu = \mu^{\text{H}} - k_{1} e^{{ - E/E_{c} }} \) (Table 1). The theoretical curves are very close to the data, and the fit for shorter chains (2.8 kbp) shows better agreement with the data than longer chains (15 kbp).
The mobility data are plotted as function of DNA length in Fig. 4. The data suggest that lower field leads to a higher mobility difference, or a higher selectivity. With the increase of applied field, DNA mobility become less length dependent, and DNA chains ranging from 2.8 to 15 kbp can no longer be distinguished when E* > 12.
The field dependence of DNA mobility may come from the different configuration of DNA chain at different applied fields, which leads to different drag resistance with buffer and with wall surface. Figures 5 and 6 (shown in the length unit of \( [\sigma ] = 18\,{\text{nm}} \)) suggest that DNA chains are more likely to hold coiled configurations in strong applied fields than in weak fields. For applied field as low as E* = 0.5, frequent dynamic transitions between stretched and coiled configuration are observed for all DNA chains, probably because that the random diffusion forces dominate the system when the driving field is not strong enough. Since, more charges are exposed on the surface when DNA is stretched than when coiled, those coil-stretching dynamics for weak field could be the reason for the high drag resistance in weak field.
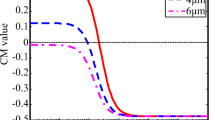
The frequent coil-stretching dynamics of DNA chain in weak field can also be reflected in the more quantitative analysis of the directional radius of gyration of DNA chain, which is plotted as a function of the center of mass position in x direction in Fig. 7.
4 Conclusions
DNA molecules in highly confined nanoslits were investigated via DPD simulations. The nanochannels have vertical dimensions of 54 nm, and DNA chains with lengths varying between 2.8 and 15 kbp were studied. Our results show that manipulation of the electrical field can strongly influence DNA mobility, and the mobility \( \mu \) scales with electric field E as \( \mu = \mu^{\text{H}} - k_{1} e^{{ - E/E_{c} }} . \) The data points for different DNA lengths approach each other with increase of fields, which suggest that the sensitivity to chain length is almost lost when the field is very strong. To explain the unusual field-dependent phenomena, we analyzed the DNA configurations during the electrophoresis. The results show that the DNA chains are more likely to hold coiled configurations for strong driving potentials when the system is dominated by the electric driving force, while for weak driving potential when the random diffusion forces dominate, there are frequent dynamic transitions between stretched and coiled configuration. The stretched-coiled configuration transitions in weak field may increase the drag resistance of DNA with buffer and wall surface, therefore reduce the mobility. We also found that lower field leads to higher selectivity.
References
Abgrall P, Nguyen NT (2008) Nanofluidic devices and their applications. Anal Chem 80:2326–2341
Baldessari F, Santiago JG (2006) Electrophoresis in nanochannels: brief review and speculation. J Nanobiotechnol 4:12
Balducci A, Mao P, Han J, Doyle PS (2006) Double-stranded DNA diffusion in slitlike nanochannels. Macromolecules 39:6273–6281
Brahmassandra SN, Durke DT, Mastrangelo CH, Burns MA (2001) Mobility, diffusion and dispersion of single-stranded DNA in sequencing gels. Electrophoresis 22:1046–1062
Chu B, Wang Z (1991) DNA electrophoretic mobility and deformation in agarose gels. J Non Cryst Solids 131:685–692
Cross JD, Strychalski EA, Craighead HG (2007) Size-dependent DNA mobility in nanochannels. J Appl Phys 102:024701
Duong-Hong D, Wang J, Liu GR et al (2008a) Dissipative particle dynamics simulations of electroosmotic flow in nano-fluidic devices. Microfluid Nanofluid 4:219–225
Duong-Hong D, Han J, Wang J et al (2008b) Realistic simulation of combined DNA electrophoretic flow and EOF in nano-fluidic devices. Electrophoresis 29:4880–4886
Eijkel JCT, van den Berg A (2004) Nanofluidics: what is it and what can we expect from it? Microfluid Nanofluid 1:249–267
Fu J, Yoo J, Han J (2006) Molecular sieving in periodic free-energy landscapes created by patterned nanofilter arrays. Phys Rev Lett 97:018103.1
Fu J, Schoch RB, Stevens AL et al (2007) A patterned anisotropic nanofluidic sieving structure for continuous-flow separation of DNA and proteins. Nat Nanotechnol 2:121–128
Groot RD, Warren PB (1997) Dissipative particle dynamics: bridging the gap between atomistic and mesoscopic simulation. J Chem Phys 107:4423–4435
Han J, Craighead HG (2000) Separation of long DNA molecules in a microfabricated entropic trap array. Science 288:1026–1029
Han J, Turner SW, Craighead HG (1999) Entropic trapping and escape of long DNA molecules at submicron size constriction. Phys Rev Lett 83:1688–1691
Hoogerbrugge PJ, Koelman JMVA (1992) Simulating microscopic hydrodynamic phenomena with dissipative particle dynamics. Europhys Lett 19:155
Hunter RJ (1981) Zeta potential in colloid science: principles and applications. Academic, New York
Jo K, Dhingra DM, Odijk T, Pablo J, Graham MD et al (2007) A single-molecule barcoding system using nanoslits for DNA analysis. PNAS 104:2673–2678
Kasiteropoulou D, Karakasidis TE, Liakopoulos A (2011) Dissipative particle dynamics investigation of parameters affecting planar nanochannel flows. Mater Sci Eng B. doi:10.1016/j.mseb.2011.01.023
Kumar A, Asako Y, Abu-Nada E, Krafczyk M, Faghri M (2009) From dissipative particle dynamics scales to physical scales: a coarse-graining study for water flow in microchannel. Microfluid Nanofluid 7:467–477
Lumpkin J, Dejardin P, Zimm BH (1985) Theory of gel electrophoresis of DNA. Biopolymers 24:1573–1593
Meller A, Vivon L, Branton D (2001) Voltage-driven DNA translocations through a nanopore. Phys Rev Lett 86:3435–3438
Pan H, Ng TY, Li H, Moeendarbary E (2010) Dissipative particle dynamics simulation of entropic trapping for DNA separation. Sens Actuators A 157:328–335
Pennathur S, Santiago J (2005) Electrokinetic transport in nanochannels: 2. Experiments. Anal Chem 77:6782–6789
Perkins TT, Smith DE, Larson RG, Chu S (1995) Stretching of a single tethered polymer in a uniform flow. Science 268:83–87
Salieb-Beugelaar GB, Teapal J et al (2008) Field-dependent DNA mobility in 20 nm high nanoslits. Nano Lett 8:1785–1790
Stein D, van der Heyden FHJ et al (2006) Pressure-driven transport of confined DNA polymers in fluidic channels. PNAS 103:15853–15858
Streek M, Schmid F, Duong TT, Ros A (2004) Mechanisms of DNA separation in entropic trap arrays: a Brownian dynamics simulation. J Biotechnol 112:79–89
Symeonidis V, Karniadakis GE, Caswell B (2005) Dissipative particle dynamics simulations of polymer chains: scaling laws and shearing response compared to DNA experiments. Phys Rev Lett 95:076001
Acknowledgment
This study is funded by Singapore-MIT Alliance (Computational Engineering Program).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yan, K., Chen, YZ., Han, J. et al. Dissipative particle dynamics simulation of field-dependent DNA mobility in nanoslits. Microfluid Nanofluid 12, 157–163 (2012). https://doi.org/10.1007/s10404-011-0859-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10404-011-0859-5