Abstract
Microfluidic systems are increasingly popular for rapid and cheap determinations of enzyme assays and other biochemical analysis. In this study reduced order models (ROM) were developed for the optimization of enzymatic assays performed in a microchip. The model enzyme assay used was β-galactosidase (β-Gal) that catalyzes the conversion of Resorufin β-d-galactopyranoside (RBG) to a fluorescent product as previously reported by Hadd et al. (Anal Chem 69(17): 3407–3412, 1997). The assay was implemented in a microfluidic device as a continuous flow system controlled electrokinetically and with a fluorescence detection device. The results from ROM agreed well with both computational fluid dynamic (CFD) simulations and experimental values. While the CFD model allowed for assessment of local transport phenomena, the CPU time was significantly reduced by the ROM approach. The operational parameters of the assay were optimized using the validated ROM to significantly reduce the amount of reagents consumed and the total biochip assay time. After optimization the analysis time would be reduced from 20 to 5.25 min which would also resulted in 50% reduction in reagent consumption.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Enzymatic assays are used both for determination of enzymatic activity and for quantification of compounds, which can be a substrate or an inhibitor of an enzymatic reaction. Miniaturization of the existing commercial enzyme assays in microtiter plates reduces not only the cost of analysis by over 90% but also may increase the number of sample analyzed per unit time in comparison to analysis in 3 mL cuvettes (Vermeir et al. 2007a). A further step forward towards integration of unit operations is microfluidics, which allows integrating sample injection, mixing, reaction and detection of the above analysis on a single device with limited human intervention (Li 2004). As a result, microfluidic biochips are increasingly popular for rapid and cheap determinations of enzyme kinetics and biochemical analysis (Hadd et al. 1997; Schilling et al. 2002; Bilitewski et al. 2003).
Microfluidics-based biochips have a high degree of complexity due to the need to simultaneously handle different streams on the chip (Bilitewski et al. 2003; Xu and Ewing 2005; Su et al. 2006). Also because of the downscaling, the surface and interfacial phenomena become increasingly important (Polson and Hayes 2001; Squires and Quake 2005) and fluidic and material properties may rapidly and regionally change (Seiler et al. 1994; Bayraktar and Pidugu 2006; Chein et al. 2006). Thus, precise manipulation of the liquid flows in such microfluidic networks is vital to the functionality and performance of these devices and will require correspondingly complex fluidic designs and flow control strategies (Karniadakis et al. 2005; Squires and Quake 2005; Su et al. 2006). Computer aided simulation tools have proven their significance in the design and optimization of fluidic structures (Ermakov et al. 1998; Barak-Shinar et al. 2004; Vermeir et al. 2005, 2007b). This is because mathematical models increase the level of understanding of the system and ultimately decrease the cost and time of the design stages significantly (Chatterjee and Aluru 2005; Erickson 2005). Numerical models for fluid flow in microchips have been widely reported in literature (Ermakov et al. 1998; Chen and Santiago 2002; Li 2004; Bayraktar and Pidugu 2006; Krishnamoorthy et al. 2006) and computational fluid dynamics (CFD) models were also presented to analyze fluid flow and enzyme kinetics in continuous flow biosensors (Barak-Shinar et al. 2004; Lammertyn et al. 2006). For a full insight into biochip systems one should combine the relevant phenomena such as microfluidics, species transport and heat transfer with the relevant chemical and biological reaction kinetics.
Despite the fact that a full numerical simulation approach such as CFD provides detail and accurate understanding of the device behavior, its use to evaluate the performance of a microfluidic system comprising many fluidic channels and /or complex biochemical process needs a prohibitive amount of computational resources, which makes this approach unsuitable for total system evaluation (Qiao and Aluru 2002; Chatterjee and Aluru 2005). Compact or reduced order modeling (ROM) approaches are much faster than the CFD modeling approach and have been shown accurate enough to capture the fundamental physical characteristics and system behavior and used for preliminary optimization (Wang et al. 2005; Su et al. 2006).
Based on electric current analogy (Chatterjee and Aluru 2005) reduced order models have been developed to evaluate fluid flow in the network of microchannels of a biochip and to quickly study diffusive mixing in electrokinetically driven passive mixers and steady state enzymatic analysis (Wang et al. 2005, 2007). Chein et al. (2006) proposed a reduced model to estimate temperature buildup associated with joule heating in an electrokinetically driven microfluidic system. In this article, we will revise those models for enzyme based microfluidic assays in electrokinetically controlled transient system. We will couple the fluid flow, energy transport and species transport equations with an enzyme kinetic model for quick optimization of a system having channel dimensions in the ten micrometer range. The developed models will be validated against CFD models and enzyme assay experiments previously reported in literature (Hadd et al. 1997). We will demonstrate that enzyme assays in such a device can be well studied using reduced order models that accurately describe the reactions and transport processes. For the device considered here, the operational parameters (flow rates and injection time) of the biochip were iteratively optimized in terms of the amount of reagents, experimental time and accuracy of the estimated kinetic parameters. Beyond a significant reduction in design time and cost spent for prototyping and refining fluidic layouts, such models aid to better understand phenomena in the whole microfluidic systems and facilitate the development of novel devices.
2 Theoretical analysis and numerical methods
2.1 Enzyme assay and microfluidic chip layout
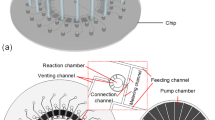
Our model system was based on experiments presented by Hadd et al. (1997) which is summarized here. The experiment was designed for the measurement of enzyme kinetics using a continuous flow microfluidic system. The enzyme used was β-Gal, a hydrolase enzyme of industrial relevance that catalyzes the hydrolysis of β-galactosides into monosaccharides. In this specific experiment, β-Gal (Escherichia coli, 780 units/mg and 540 kDa) was used to catalyze the conversion of β-d-galactopyranoside (RBG) to resorufin, which can be detected optically. The basis lay-out of the biochip is depicted in Fig. 1a. Reagents were filled in the respective reservoirs which were interconnected by the network of microchannels. Electrical contact between the solution and multi terminal high-voltage power supply was achieved using platinum wires placed in the reservoirs and the flow directions and flow rates were controlled by monitoring the potential applied at these electrodes. The concentration of the substrate in the reaction channel was determined by the on chip T-type sample dilution unit, integrated in to the cross channel. The substrate, RBG, supplied from reservoir 1 was diluted by the buffer from reservoir 2 and was mixed with enzyme β-galactosidase (from reservoir 3) and buffer (from reservoir 4) at the second channel intersection; resorufin was subsequently produced in the reaction channel. The total flow rate in the reaction channel was 14 nL min−1 (associated with an applied electric field of 220 V cm−1), where 40% was contributed from the mixing channel and 30% from each of the side channels (buffer and enzyme reservoirs). The potentials applied at reservoir 1 and 2 were manipulated in real time to produce a dynamic change of the substrate concentration (i.e., step increments of final substrate concentration in the reaction channel) keeping the flow rates in the mixing and reaction channel the same. The assay was performed at an ambient temperature of 294 K. The reaction product, resorufin, was optically measured (absorbance at 571 nm wavelength) using a charged coupled device (CCD) mounted on an optical microscope positioned at 20 mm downstream of the reaction channel. This was done to demonstrate the use of microchips for continuous and efficient kinetic analysis and also showed the possibility of biochips for online analysis applications. Further details can be found in Hadd et al. (1997).
2.2 CFD model formulation
2.2.1 Fluid flow
Reagents are transported electrokinetically. When an external electric potential field is applied along the axial direction of the channel, the liquid starts to move as a result of the interaction between the net charge density in the electric double layer (EDL) of the channel and the applied electric field. The fluid flow in the microfluidic system is governed by the Navier–Stokes equations with appropriate body force terms. However, the driving force of electro-osmotic flow is present only within the electric double layer EDL (1–100 nm thickness), while the characteristic size of microfluidic channels is in the range of 10–100 μm (Li 2004; Karniadakis et al. 2005). Therefore, the flow field within the EDL can be neglected and the electro-osmotic flow, u eo , can be considered to be induced by a moving wall with slip velocity given by the Smoluchowski equation (Krishnamoorthy et al. 2006):
with ϕ the electrical potential which can be found from the Laplace equation
The meaning of all symbols is explained in nomenclature and the values of the variables used in the simulation are presented in Table 1. Using the slip velocity approach, the steady state profile of fully developed electrokinetic flow is governed by the Navier–Stokes equations shown below.
When the depth of the channel is much smaller than the length and width, gradients for the dependent variables that exist along the channel depth will not be significant (Ermakov et al. 1998; Chen and Santiago 2002; Li 2004). In this case, two-dimensional (2D) models were sufficient to study the given system.
2.2.2 Species transport
The species involved in this assay were RBG, β-galactosidase and resorufin. There exist three basic modes of mass transfer relevant to the present application: diffusion, convection and electrokinetic migration. Therefore, the mass conservation equation for the species was given by (Barak-shinar et al. 2004):
The diffusion coefficient of β-gal, RBG and resorufin at 298 K are 2.7 × 10−11, 4.3 × 10−10 and 4.8 × 10−10 m2 s−1 respectively (Hadd et al. 1997; Schilling et al. 2002).
For the relevant enzymatic reactions, the reaction rate, r resorufin was modeled by the Michaelis–Menten equation (Marangoni 2003; Lammertyn et al. 2006):
Unless otherwise stated the kinetic values used in the simulation were 320 μM and 54 s−1 for K M and k cat respectively (Hadd et al. 1997).
2.2.3 Joule heating
Electroosmotic flows always involve volumetric Joule heating when an electric field is applied across the conducting media (Erickson et al. 2003; Chein et al. 2006; Tang et al. 2004). The fluid temperature is mainly affected by the rate of heat generated (in this case joule heating), external cooling mechanism, microchannel geometry, velocity of the fluid and thermophysical properties of fluid and channel wall. Within the liquid, the 3D energy equation in the presence of electroosmotic flow effect is given by (Tang et al. 2004; Hu et al. 2006):
The thermo-physical properties including the solution electrical conductivity, viscosity, dielectric constant and thermal conductivity and zeta potential, sample species mass diffusivity and electrophoretic mobility were considered to be temperature dependent, shown in Table 1 (Erickson et al. 2003; Tang et al. 2004, 2006; Venditti et al. 2006).
2.3 Reduced order model
2.3.1 Fluid flow
Microfluidic devices can be depicted as integrated electric circuits using the analogy of fluid and current flows (Qiao and Aluru 2002; Kohlheyer et al. 2005). The overall microchip system was represented by a network of components, the channels, connected by “nodes” of zero resistance. The channels in the microchip then act as electric resistors, which results in the equivalent resistor network (Fig. 1b). The electric resistance of a solution filled in a simple straight channel, R is given by:
Using Ohm’s law, the electric current I, the applied potential and the electrical resistance are related as (Seiler et al. 1994; Hadd et al. 1997):
The potential at any point in a channel was calculated by applying Kirchoff’s rule (current balance) in the system. In a straight channel, the electroosmotic velocity can be analytically predicted using the Helmholtz-Smoluchowski equation given by Eq. 1 without solving the Navier–Stokes equations (Qiao and Aluru 2002; Kohlheyer et al. 2005; Krishnamoorthy et al. 2006). Therefore, the electrokinetic fluid flow in the microfluidic system was evaluated using Eqs. 1 and 8 that is by making mass balance and current balance (Kirchhoff’s law) at the intersection of the channels.
2.3.2 Species and energy transport
In the microfluidic devices mixing mainly depend on the molecular diffusion due to the fact flow is restricted to the laminar region where turbulence or chaotic advection was absent. This might required a very long mixing channel depending the width,w, of the microchannel, analyte diffusion coefficient, D, and velocity,u (i.e. mixing length, L = w 2 u/2D). For assay performed in microchannel of less than 50 μm width, diffusion mixing is adequate to initiate enzyme reaction (He et al. 2001). They claimed that by using a 200 μm length static mixing system, complete mixing can be achieved within second in microdevice up to 100 μm width. Johnson et al. (2002) also reported that significant mixing was achieved within 443 μm length at a flow rate as high as 8.1 mm s−1using series of slanted wells at the junction of a microchannel. For numerical analysis of different types of micromixers the readers refered to the book by Li (2004). Taking in to account the aforementioned facts, enzyme assay in the microdevice of width 35 μm and 20 mm long reactor, can be modeled by reduceing the species transport equation, Eq. 4, to a one-dimensional (1D) form assuming the diffusion effect is minor:
where u i = u eo + u ep,i is the net flow of the species which is a combination of electroosmotic flow and electrophoretic flow (u ep,i = μ ep,i∇ϕ) respectively. The efficiency of the one dimensional convection-diffusion-reaction model was previously evaluated (He and Hauan 2006) for steady-state processes and here we extended it by including the Michaelis–Menten enzyme kinetic model and used for transient system analysis. More is given in Sect. 3.3.
Previous studies have shown that the time for the temperature field to reach steady state is very short, less than one second (Tang et al. 2006). The energy transport equation presented in Eq. 6 was then simplified by Chein et al. (2006) to predict the fluid temperature distribution of the electrokinetic flow in microchannel:
where the second term of the right hand side represents heat losses at the edges of the channel to the glass chip substrate and its environment. Based on boundary conditions, analytical solution can be found for Eq. 10.
In optimization of the enzyme assay aforementioned ROMs were used. The flow rate for a specific electric field in the reaction channel was calculated using the Smoluchowski equation Eq. 1 and one dimensional energy transport equation Eq. 10. Hence, the velocity and temperature profile along the channel was used in the one dimensional species transport equation Eq. 9. Using the conservation law (Kirchhoff’s rule) and the given flow ratio of each streams the potential at each reservoir can be calculated.
2.4 Boundary and initial conditions
The models were applied to the microfluidic chip illustrated in Fig. 1a. Since the microfluidic chip was made of glass, insulation boundary conditions were applied at the channel walls in the Laplace equation. The electrode at reservoir 5 was grounded. The potentials applied at reservoir 1 and 2 were manipulated in real time to produce a dynamic change of the substrate concentration (i.e., step increments of final substrate concentration in the reaction channel) keeping the flow rates in the mixing and reaction channel the same. All the channels have constant cross-sectional area and electrical conductivity of the solution is assumed to be constant, as the concentration of substrate is much less than the running buffer. Simultaneous application of conservation equations for mass flow Eq. 1, substrate and current flow Eq. 8 at the two junctions of the microchannel networks (as represented in Fig. 1b) and using the flow information, flow rates contributed from each streams expressed by a i, the potentials applied at the respective reservoirs were determined (Qiao and Aluru 2002; Chatterjee and Aluru 2005):
where a 1, a 2 and a 3 are flow ratios with respect to the total flow rate in the reaction channel (0.4, 0.3 and 0.3 for mixing stream, stream 3 and 4 respectively); L i is the length of the ith channel, L M and L R are the length of the mixing and reaction channel respectively; E R is the electric field strength in the reaction channel; C RBG,R and C RBG,1 are the concentration of RBG in the reaction channel (ranging from 14 to 122 μM) and in reservoir 1, respectively. For a given flow rate determined by E R, the potentials at all reservoirs were calculated using Eq. 11. The potential at reservoir 1 and 2 increased step by step to change the concentration of substrate in the reaction channel and such boundary condition can be easily implemented in COMSOL.
An initial condition of zero velocity, zero potential and zero concentrations for all species were set. In electrokinetically driven open system, the absence of hydrostatic pressure gradient was model by setting atmospheric pressure at all the inlets and outlet. At the channel walls, slip velocity given by Eq. 1 was implemented. For the species transport equation, all the walls were set to a zero mass flux (i.e. impermeable walls) and the concentration of each species was specified at the respective inlets (340 μM of RBG, 2.28 nM of β-Gal). Since the concentrations of species were not known at the outlet, the gradient of the concentration of these species at this boundary was set to zero. For energy, the initial condition and the temperature at all inlet reservoirs were set to reference temperature value (294 K). For a sufficiently long channel both flow and temperature are assumed to reach their fully developed state at the outlet of the channel such that the temperature gradient was set to zero. In the 3-D model that includes both the channel and the chip material, the heat dissipated from the system to the surrounding was modeled by a heat flux boundary condition (heat flux, q = U 0(T −T ∞ )), which was specified at the outer walls of the chip. U 0 covers the heat resistance for both external air, convective heat transfer, as well as the channel wall with Δx thick, conduction heat transfer.
2.5 Numerical solution and optimization procedure
The sets of models used in this study were summarized in Table 2. 3D heat transfer analysis was performed on part of the device (Model I in Table 2) and result compared with the reduced order model to estimate the temperature rise associative with joule heating. As the depth of the channel was much smaller than the length and the width, 2D models were sufficient to study the given system (Ermakov et al. 1998; Chen and Santiago 2002; Li 2004). Hence, a CFD model was developed considering the whole biochip system using the 2D geometry shown in Fig. 1a. This geometry was not appropriate to investigate energy transport in the system. However, it is possible to study the effects of joule heating in the flow direction by implementing one-dimensional heat transfer model. This was modeled by including the heat dissipation as source term in the 2D heat transfer model equation while all walls were considered thermally insulated (Model II in Table 2). Taking into account all temperature dependent thermo-physical properties of both fluid and channel walls, these equations were solved in the finite element code, COMSOL Multiphysics 3.3 (COMSOL AB, Stockholm, Sweden) on an AMD Opteron Cluster node with 3.8GB of RAM on 2D computational domina. Time for the temperature and flow field to reach steady state is very short. Hence, steady state energy and flow equations were solved first and then coupled with the transient species transport equations for a relative tolerance equal to 10−8 and with an appropriate time step chosen by the solver.
In the ROM approach the Smoluchowski equation Eq. 1,Ohm’s law Eq. 8, the reduced energy transport equation Eq. 10 and 1D convective-diffusive-reaction equation Eq. 9 with Michaelis–Menten enzymatic reaction model equation Eq. 5 were coupled in 1D geometry of length equal to the size of the reaction channel (Model III in Table 2). Once the ROM was validated against the CFD simulation results and experimental values, we demonstrated its applicability for fast microfluidic biochip optimization. The ROM was used to optimize the operational parameters of the biochip in terms of amount of reagents, experimental time while the accuracy of the device remain high (described by kinetic parameters estimated from each simulation). The optimization was implemented in Matlab 7.0.1. An interface script between Matlab and COMSOL multi-physics was written so that in every iterations of the optimization algorism the ROM was solved. The optimization procedures are summarized using the flow diagram given in Fig. 2. For every possible combination of injection time and flow rate, the ROM was computed and the concentration profile for product at the detector was extracted; then kinetic parameters were calculated using a Lineweaver-Burk plot. The kinetic parameters (\( k^{{{\text{est}}}}_{{{\text{cat}}}} \)and \( K^{{{\text{est}}}}_{{\text{M}}} \)) estimated from the in silico assay for the given injection time and flow rate values were compared with the experiment values and the simulation was repeated until the percentage error calculated by Eq. 12 is less than 0.5%.
Flow diagram for the optimization procedure. The optimization routine finds injection time and flow rate that minimize the percentage error given by Eq. 12. t inj and V are injection time and flow rate respectively
3 Results and discussion
3.1 Model validation
Application of electric field results in Joule heating. The temperature rise associated with Joule heating may affect the assay performance by changing physicochemical properties of the fluid, which alter the electroosmotic velocity, the mass diffusivity and electrophoresis mobility of species. The 3D CFD simulation was done for heat transfer in the reaction channel, where the electric field is highest (refer to Model I in Table 2), and the result is presented together with the reduced order model in Fig. 3. According to our simulation analysis shown in Fig. 3a, the Joule heating effect in the current assembly will not be significant for this specific enzyme kinetics to change drastically. This is in agreement to previous works (Tang et al. 2004; Krishnamoorthy et al. 2006) where the effect of Joule heating was not significant in small microchannels with size range between 30 and 50 μm. This is due to the larger surface area to volume ratio resulting in good heat exchange with the environment to remove the heat generated. Thus, the electroosmotic velocity did not deviate from its normal plug-like profile throughout the working region considered in this study (flow rate from 0.945 to 37.8 nL min−1). Looking at Fig. 3a, the reduced order model over predicted the Joule heating effect with respect to the detailed 3D model. This is due to oversimplification of boundary conditions to represent cross-stream heat loss in the reduced order model. Chein et al. (2006) argued that even though the proposed reduced model Eq. 10 over predicted the temperature profile compared with the 3D CFD simulation, the results agreed well with the experimental results. The temperature in the biochip is highest at the center line of the chip and decreases towards the external surface of the chip, where heat dissipates to the environment (Fig. 3b).
If the reduced energy transport equation shows a small temperature change for the maximum possible electric field applied to the system, it would not be important to couple the energy transport equation to the model set. However, the current trend of biochip construction is mainly towards polymer material, in which thermal dissipation is low. Therefore, for such microchips with larger sized microchannels Joule heating does play a significant role (Erickson et al. 2003; Tang et al. 2004; Hu et al. 2006). Then, coupling of the reduced heat transfer equation with fluid flow and species transport equations is desirable to include its effects on the performance of the device and techniques that improve heat dissipation capability of the chip could be considered (Zhang et al. 2004; Tang et al. 2004).
In this on-chip biocatalytic assay study, the production rate was evaluated as the change in resorufin produced over the incubation time (this is the time it takes for the product of the reaction to travel from where the substrate and enzyme start to mix to the detector). In Fig. 4 the normalized signal of resorufin from 2D CFD simulation (result with diamond marker) was presented with the experimental profile reported by Hadd et al. (1997). In this case the concentration of RBG in the reaction channel varied in the range from 14 to 122 μM while 1.37 nM of β-Gal was used. A good agreement between the simulation and the experimental results observed over the entire range considered except at the beginning of the experiment which cannot be explained.
Normalized resorufin concentration profile detected at 20 mm downstream of the reaction channel for equal increments in concentration of input substrate for three different β-Gal concentrations (0.352, 0.685 and 1.37 nM). The solid lines are ROM simulation results and experimental values are marked with different markers. Result with diamond marker is from the 2D CFD simulation. ROM compares well to the experimental values (injection time: 175 s, flow rate: 14 nL min−1)
3.2 Mixing performance of the biochip
In the laminar microfluidic system, mixing of the different compounds was based on diffusion only. Figure 5a displays the concentration contours of the RBG in the mixing and reaction channel and resorufin formed at the beginning of the reaction channel and concentration profile of β-Gal across the channel at different location downstream the reaction channel (Fig. 5b). The CFD simulation confirmed that RBG was completely mixed in the mixing channel before it reached the second crossing. β-Gal takes a relatively long time (6 s, or an equivalent downstream distance of 4.3 mm) to diffuse half of the width of the channel, as a result of its relatively small diffusion coefficient. Nevertheless, the reaction starts close to the crossing (Fig. 5a) because the diffusion time of RBG is not larger than 1 s at a flow rate of 14 nL min−1. Therefore, the limiting effect of mixing is minor.
Concentration contour of the RBG and Resorufin (a) in the microfluidic channel. Substrate mix very well before entering the reaction channel while enzyme takes relatively long to cross the channel width. Concentration profile of enzyme across the channel at different location along the reaction channel (b)
Because the ROM approach assumes instantaneous mixing, comparison of ROM predictions to those obtained with CFD and experiments will also help to judge the effect of mixing. Figure 4 also shows the normalized signal of resorufin for three enzyme concentrations (0.352, 0.685 and 1.37 nM) while the concentration of RBG equally increased from 14 to 122 μM. The solid lines are simulation results from the ROM whereas doted lines with different markers represent the experimental results. The ROM simulation results compared well with the respective experimental results. The difference between ROM and CFD was negligible, but the ROM was many times (64 times) faster in CPU time than the 2D CFD simulation.
Increasing flow rate will shorten the reaction time and extend the mixing length of species in the microchannel; as a consequence the amount of products produced will be lower. To investigate this, both ROM and 2D simulation was done at higher flow rate (33.1 nL min−1 for injection time of 35 s) and results are presented in Fig. 6. The concentration contours of β-Gal, RBG and resorufin are presented (from top (i) to bottom (iii) respectively) in a part of the system. As shown, RBG mix with buffer and β-Gal as soon as it gets into the reaction channel so that resorufin was formed in the diffusion region of two reagents very close to the crossing. The signal obtained from this simulation had also clear steps (Fig. 6b). At this flow rate the mixing length of RBG increased from 1 to 2.23 mm, nevertheless, comparison of the 2D CFD result with ROM proved that the effect of transverse diffusion on the assay performance is insignificant for this particular process within 20 mm long reaction channel. In addition, during on-chip kinetic analysis complete conversion of the substrate would not be important as far as the detector is sensitive enough and signals are clearly changed (see Fig. 6b) with respect to the change in the substrate concentration at the inlet of the reactor. To improve the sensitivity of the device at higher flow rate condition, the concentration of enzyme could be increased so that the amount of product produced would be within the detection limit of the device. Mixing in this microchip, thus, is sufficient not to limit the performance of the assay. On the other hand, numerical analysis revealed that the ROM over predicted the CFD simulation results at higher flow rate for larger channel width and short reaction channel (result not shown). For such cases, the system needs a mixer integrated with the reaction channel (Hadd et al. 1997; He et al. 2001).
Concentration contour for β-Gal, RBG and Resorufin (from top to bottom) after the second crossing in the biochip for flow rate of 33.1 nL min−1 and injection time of 35 s (a). Concentration profile of resorufin obtained from simulation using ROM and 2D model (b). The concentration of RBG range from 14 to 122 μM for 0.685 nM of β-Gal. The kinetic parameters calculated out of it were 320.4 μM and 53.76 s−1 for K M and k cat respectively
3.3 Biochip optimization
Using the conditions specified in the experiment by Hadd et al. (1997), 120 pg of β-Gal and 7.5 ng of RBG were required with a total analysis time of 20 min. Here we attempted to reduce both the assay time and reagent consumed per assay without affecting the quality of the signal obtained from the experiment. A good quality signal often implies a good accuracy of the kinetic parameters estimated from it, which is associated with the performance of the device. The process parameters that were optimized in this manuscript were the flow rate and the injection time. For this purpose, we have performed in silico experiments, using the known kinetic parameters of the given assay.
The total assay time, which depends on the injection time for each substrate concentration steps, has to be as short as possible but the injection time for each stepped-increment of substrate should also be sufficient to fill the reaction channel with the new concentration of substrate. This is to avoid dilution of the resorufin produced from consecutive injections. When the injection time is too short, the signal will not have clear steps (shown in Fig. 7) and computation of the kinetic values from the result will not be reliable. This is clearly demonstrated using the numerical simulation presented in Fig. 7, where different values of injection time were taken keeping the flow rate constant. As shown from these results there would not be a clear signal for kinetic studies when the injection time is below the threshold of 50 s.
A Lineweaver–Burk plot (Fig. 8) was reproduced from the simulation result (presented in Figs. 7 and 9) for an injection time of 75 s and flow rate of 14 nL min−1. The Lineweaver–Burk plot is a linear form of the enzyme kinetic model of Eq. 5 and is a plot of 1/r resorufin versus 1/C RBG. The rate associated with every substrate concentration, r resorufin, was determined by dividing the concentration of resorufin obtained at the respective plateau per assay time. Then the K M and V max value were calculated from the y-intercept (i.e. 1/V max) and the slope of the line which is equal to K M/V max. For injection times larger than 75 s the kinetic constants recalculated from the simulation were equal to the original values (K M = 320 μM and k cat = 54 s−1). At 75 s injection time, the value of K M only deviated by 2.7% from the true value. This is used as an indicative threshold for the accuracy of the system for the operating conditions specified.
Lineweaver–Burk plot from ROM simulations (injection time was 75 s; flow rate was 14 nL min−1 and concentration of β-Gal was 0.685 nM). The K M and k cat values calculated from this plot were 328.5 μM and 54 s−1 respectively which are close to the true values (K M = 320 μM and k cat = 54 s−1). r = r resorufin [μM s−1] is the rate of resorufin produce
Simulations were done for different flow rates above and below the previous value, 14 nL min−1, keeping the injection time of substrate constant (75 s in this case). Result is presented in Fig. 9. The sensor response increased as the flow rate decreased due to the increase in reaction time. However, the kinetic values obtained at a reduced flow rate (10.5 nL min−1) are 296 μM and 50 s−1 which deviate by about 7.5%. This is because at lower flow rates, the steady-state plateaus corresponding to the different levels of substrate concentration are too short and eventually disappeared (i.e. the signal became a straight line), shown in Fig. 9. On the other hand, by increasing the flow rate the amount of product will be smaller. For instance, the K M and k cat calculated from simulation with 75 s injection time and a 25% increase in flow rate (equal to 17.5 nL min−1) were 318.7 μM and 54 s−1, respectively, which is still close to the original value. As shown from simulation result, signal becoming smaller at increasing flow rate and at some point the amount of product will be below the detection limit of the device. It is also important to consider the effects of Joule heating associated with the rise of applied potential in order to increase the flow rate. The maximum rise in system temperature computed from analytical model presented should be checked not to be much larger than the optimum temperature range of the enzyme used. Moreover, increasing flow rate means increasing the amount of reagents used per experiment. Therefore, the significance of increasing the flow rate for the performance of the device might not be so essential unless the injection time would be further reduced.
The aforementioned results lead us to develop a general optimization algorithm that considers broad ranges of injection time (from 5 to 200 s) and flow rates (0.945 to 37.8 nL min−1). In every iteration the kinetic parameters were estimated from the Lineweaver–Burk plot and the % error given by Eq. 12 was calculated (See flow diagram in Fig. 2.). In Fig. 10 percentage errors were plotted for a range of injection times and flow rates. Looking at Fig. 10, there exist a large workable region where the signal has a clear shoulder, which results in a very low percentage error. The percentage error at the lower boundary in the workable region was around 0.5%. Now what we need was the combination of flow rate and injection time in the workable region that consumed less reagents and performed in short time.
In Table 3 the amount of reagent required and total time to finish one enzyme assay experiment is presented. The injection time and the flow rate were taken from the lower boundary of the workable region where the error was below 0.5%.The first and last value were taken to check the extreme combinations and had percentage error about 2.5 and 4% respectively. As can be seen from this table the amount of reagent consumed seems lower in the region close to A (largest injection times and lowest flow rates) and B (lowest injection times and highest flow rates). For instance, a flow rate of 33.1 nL min−1 and an injection time of 35 s. This resulted in the RBG and β-Gal consumption to drop by half while, more importantly, the total assay time reduced from 20 to 5.25 min. For this combination the calculated kinetic values were 320.4 μM and 53.76 s−1 for K M and k cat respectively. In addition, the electric field for this flow rate is 520 V/cm and the change in temperature as a result of Joule heating (shown in Fig. 3) is not large enough to affect the enzyme assay. Thus, the combination of the highest possible flow rate and a short injection time (region B) can reduce the analysis time considerably.
ROMs, hence, are sufficiently accurate to illustrate the fundamental physical characteristics of the system and used for preliminary optimization. Further, once the lab-on-chip is optimized, CFD simulations can always be carried out for a more detailed investigation. Therefore, the time and money spent in prototyping to optimize the sample delivery, reaction kinetics and thermal and separation performance can be significantly reduced. Simulation based design also aids to understand coupling between those phenomena and retrieve detailed information, which could be impossible to access by experiment. For instance, in the study of Hadd et al. (1997), the possible effects of Joule heating on the assay performance was not addressed. The flow ratio of the reagents can be further optimized and spatial information about the concentration of unconverted substrates, intermediate and final products, and mixing efficiency can be comprehensively investigated using the mathematical models.
4 Conclusions
While miniaturization as such improves performance and analysis time with respect to the existing wet lab methods, we have demonstrated that biochips themselves can be considerably improved based on computer simulations with mathematical models that accurately describe the reactions and transport processes in the fluidic system. In this article we have shown how enzymatic assays in microfluidic biochip can be optimized in silico using the reduced order model (ROM). The results from ROM agreed well with both 2D CFD simulations and experimental values while the CPU time reduced significantly. The operational parameters of the assay were optimized using the validated ROM to considerably reduce the amount of reagents consumed and the total biochip assay time. After optimization the analysis time would be reduced from 20 to 5.25 min which would also cut the consumption of reagents by 50%. Hence, such modeling approaches are important to transform the existing and many other bioassays in to high performance multiplexed biochips format aimed at multi-component analysis systems that have a wide range of diagnostic applications. However, the reduced order model for species transport analysis is limited to systems having microchannel size in ten micrometer rang where mixing time is short. This is due to the effect of diffusion mixing on the performance of the assay.
Abbreviations
- A c :
-
cross-sectional area of the channel (m2)
- C p :
-
specific heat capacity of fluid (J kg−1 K−1)
- C i :
-
concentration of species i(mol m−3)
- C β-Gal :
-
concentrations of the enzyme (mol m−3)
- C RBG :
-
concentrations of substrate (mol m−3)
- D i :
-
diffusion coefficient of the ith species (m2 s−1)
- E :
-
externally applied electric field (V m−1)
- E x :
-
externally applied electric field in the x direction (V m−1)
- F :
-
Faraday constant (C mol−1)
- h :
-
convective heat transfer coefficient of the air (W m−2 K−1)
- I :
-
electric current (A)
- k :
-
thermal conductivity of the fluid (W m−1 K−1)
- k w :
-
thermal conductivity of the glass wall (W m−1 K−1)
- k cat :
-
rate constant (s−1)
- K M :
-
Michaelis–Menten constant (mol m−3)
- L :
-
length (m)
- a i :
-
The flow ratio of each streams with respect to the total flow in the reaction channel
- p :
-
pressure (Pa)
- P :
-
perimeter of the micro channel (m)
- r resorufin :
-
reaction rate of the resorufin (mol m−3 s−1)
- R :
-
electric resistance of solution in the channel (Ω)
- R g :
-
universal gas constant (J mol−1 K−1)
- t :
-
time (s)
- T :
-
temperature (K)
- T ∞ :
-
surrounding temperature (K)
- T 0 :
-
reference temperature (298 K)
- U 0 :
-
overall heat transfer coefficient (W m−2 K−1)
- Δx :
-
thickness of the microchannel (m)
- u :
-
the velocity vector (m s−1)
- u eo :
-
electroosmotic slip velocity (m s−1)
- V max :
-
maximum reaction rate (mol m−3 s−1)
- z i :
-
valence number, −1 for β-Gal and Resorufin and zero for RBG
- ϕ :
-
applied potential (V)
- ε 0 :
-
permittivity of vacuum (C2 N−1 m−2)
- μ eo :
-
electroosmotic mobility (m2 V s−1)
- ρ :
-
density of the fluid (kg m−3)
- η :
-
dynamic viscosity of fluid (kg m−1 s−1)
- σ :
-
electrical conductivity of fluid (S m−1)
- ζ :
-
zeta potential (V)
- ε r :
-
dielectic constant of the medium
- μ ep,i :
-
electrophoretic mobility of the ith species (m2 V−1 s−1)
- EDL:
-
electric double layer
- EOF:
-
electroosmotic flow
- ROM:
-
Reduce order model
- CFD:
-
Computational fluid dynamics
- 2D:
-
Two-dimensional
- 1D:
-
One-dimensional
- β-Gal:
-
β-galactosidase
- RBG:
-
Resorufin β-d-galactopyranoside
References
Barak-Shinar D, Rosenfeld M, Rishpon J, Neufeld T, Abboud S (2004) Computational fluid dynamic model of diffusion and convection processes in electrochemical sensor. IEEE Sens J 4(1):65–71
Bayraktar T, Pidugu SB (2006) Characterization of liquid flows in microfluidic systems. Int J Heat Mass Transf 49:815–824
Bilitewski U, Genrich M, Kadow S, Mersal G (2003) Biochemical analysis with microfluidic systems. Anal Bioanal Chem 377:556–569
Chatterjee AN, Aluru NR (2005) Combined circuit/device modeling and simulation of integrated microfluidic systems. J Microelectromech Syst 14(1):81–95
Chein R, Yang YC, Lin Y (2006) Estimation of joule heating effect on temperature and pressure distribution in electrokinetic-driven microchannel flows. Electrophoresis 27:640–649
Chen C, Santiago JG (2002) A planar electroosmotic micro pump. J Microelectromech Syst 11(6):672–683
Erickson D (2005) Towards numerical prototyping of labs-on-chip: modeling for integrated microfluidic devices. Microfluid Nanofluid 1:301–318
Erickson D, Sinton D, Li D (2003) Joule heating and heat transfer in poly(dimethylsiloxane)microfluidic systems. Lab chip 3(3):141–149
Ermakov SV, Jacobson SC, Ramsey JM (1998) Computer simulations of electrokinetic transport in microfabricated channel structures. Anal Chem 70(21):4494–4504
Hadd AG, Raymond DE, Halliwell JW, Jacobson SC, Ramsey JM (1997) Microchip device for performing enzyme assays. Anal Chem 69(17):3407–3412
He X, Hauan S (2006) Microfluidic modeling and design for continuous flow in electrokinetic mixing-reaction channels. AIChE J 52(11):3842–3851
He B, Burke BJ, Zhang X, Zhang R, Regnier FE (2001) A picoliter-volume mixer for microfluidci analysis systems. Anal chem 73:1942–1947
Hu G, Xiang Q, Fu R, Xu B, Venditti R, Li D (2006) Electrokinetically controlled real-time polymerase chain reaction in microchannel using joule heating effect. Analytica Chimica Acta 557:146–151
Johnson TJ, Ross D, Locascio LE (2002) Rapid microfluidic mixing. Anal Chem 74(1):45–51
Karniadakis G, Beskok A, Aluru N (2005) Microflows and nanoflows: fundamentals and simulation. Springer, USA
Krishnamoorthy S, Feng J, Henry AC, Locascio LE, Hickman JJ, Sundaram S (2006) Simulation and experimental characterization of electroosmotic flow in surface modified channels. Microfluid Nanofluid 2:345–355
Kohlheyer D, Besselink GAJ, Lammertink RGH, Schlautmann S, Unnikrishnan S, Schasfoort RBM (2005) Electro-osmotically controllable multi-flow microreactor. Microfluid Nanofluid 1:242–248
Lammertyn J, Verboven P, Veraverbeke EA, Vermeir S, Irudayaraj J, Nicolaï BM (2006) Analysis of fluid flow and reaction kinetics in a flow injection analysis biosensor. Sens Actuators B Chem 114:728–736
Li D (2004) Electrokinetics in microfluidics. Elsevier, Amsterdam
Marangoni AG (2003) Enzyme kinetics: a modern approach. Wiley, Hoboken
Polson NA, Hayes MA (2001) Microfluidics controlling fluids in small places. Anal Chem 73(11):312–319
Qiao R, Aluru NR (2002) A compact model for electroosmotic flows in microfluidic devices. J Micromech Microeng 12:625–635
Schilling EA, Kamholz AE, Yager P (2002) Cell lysis and protein extraction in a microfluidic device with detection by fluorogenic enzyme assay. Anal Chem 74:1798–1804
Seiler K, Fan ZH, Fluri K, Harrison DJ (1994) Electroosmotic pumping and valveless control of fluid flow with in a manifold of capillaries on a glass chip. Anal Chem 66:3485–3491
Squires TM, Quake SR (2005) Microfluidics: fluid physics at the nanoliter scale. Rev Mod Phys 77:977–1026
Su F, Chakrabarty K, Fair RB (2006) Microfluidics-based biochips: technology issues, implementation platforms, and design-automation challenges. IEEE Trans Comput Aided Des Integr Circ Syst 25(2):211–223
Tang GY, Yang C, Chai CK, Gong HQ (2004) Numerical analysis of the thermal effect on electroosmotic flow and electrokinetic mass transport in microchannels. Anal Chim Acta 507:27–37
Tang G, Yan D, Yang C, Gong H, Chai JC, Lam YC (2006) Assessment of Joule heating and its effects on electroosmotic flow and electrophoretic transport of solutes in microfluidic channels. Electrophoresis 27:628–639
Venditti R, Xuan X, Li D (2006) Experimental characterization of the temperature dependence of zeta potential and its effect on electroosmotic flow velocity in microchannels. Microfluid Nanofluid 2:493–499
Vermeir S, Verboven P, Atalay YT, Nicolaï BM, Irudayaraj I, Lammertyn J (2005) Computational fluid dynamics model for optimal flow injection analysis biosensor design. In: Proceedings of the 4th IEEE conference on sensors, Irvine, USA, pp 365–368
Vermeir S, Nicolaï BM, Jans K, Maes G, Lammertyn J (2007a) High-throughput microplate enzymatic assays for fast sugar and acid quantification in apple and tomato. J Agric Food Chem 55:3240–3248
Vermeir S, Nicolaï BM, Verboven P, Van Gerwen P, Baeten B, Hoflack L, Vulsteke V, Lammertyn J (2007b) Microplate differential calorimetric biosensor for ascorbic acid analysis in food and pharmaceuticals. Anal chem 79:6119–6127
Wang Y, Lin Q, Mukherjee T (2005) A model for laminar diffusion-based complex electrokinetic passive micromixers. Lab chip 5:877–887
Wang Y, Bedekar AS, Krishnamoorthy S, Siddhaye SS, Sundaram S (2007) System-level modeling and simulation of biochemical assays in lab-on-a-chip devices. Microfluid Nanofluid 3(3):307–322
Xu H, Ewing AG (2005) High-throughput enzyme assay on a multichannel microchip using optically gated sample introduction. Electrophoresis 26(24):4711–4717
Zhang Y, Bao N, Yu X-D, Xu J-J, Chen H-Y (2004) Improvement of heat dissipation for polydimethylsiloxane microchip electrophoresis. J Chromatogr A 1057:247–251
Acknowledgments
This research was funded by a Ph.D. Grant of the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT-Vlaanderen), by the Fund for Scientific Research Flanders (FWO G.0298.06 and FWO 1.5.136.05), and by the KU Leuven through a bilateral scientific collaboration between Flanders and Russia (BIL/05/47).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Atalay, Y.T., Verboven, P., Vermeir, S. et al. Design optimization of an enzymatic assay in an electrokinetically-driven microfluidic device. Microfluid Nanofluid 5, 837–849 (2008). https://doi.org/10.1007/s10404-008-0291-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10404-008-0291-7