Abstract
Environment determines the distribution and prevalence of vector-borne pathogens due to its direct and indirect effects on the hosts, vectors, and pathogens. To investigate the relationship between Usutu virus occurrence and host biodiversity and to characterize the nidus of infection, we used field-based measures of host diversity and density (all birds and only passerines), vector abundance, landscape and Usutu virus prevalence (mosquito infection rate), an emergent disease with a similar cycle to West Nile virus. We collected 908,237 female mosquitoes in an area of 54,984 ha in the Doñana National Park, southern Spain. We identified the mosquitoes and screened them for viruses, censused birds, characterized landscape and climatic variables, and then modeled the presence and infection rate of the virus in relation to host, vector, climatic, and landscape variables. Monthly Usutu presence, detected in Culex perexiguus, was positively related to Passeriformes richness and secondarily to NDVI in the previous month. Our results suggest that Usutu prevalence may be higher when and where host (passerine) richness was high, and thus challenging the conventional idea that host biodiversity reduces flavivirus amplification.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction and Purpose
Emerging infectious diseases are transmissible diseases whose geographic range, host range, or prevalence have increased in recent years. Most of them have a zoonotic origin, i.e., they are caused by pathogens shared with wild or domestic animals or originate from wildlife (Jones et al. 2008; Taylor et al. 2001). The spatial distribution of vector-borne pathogens arises from underlying variation in the biological and physical conditions that support the pathogen and its vectors and reservoirs. The nidus of infection must contain the competent vectors, the competent reservoirs, the susceptible recipient hosts, the pathogen, and the influence of environmental factors that contribute to transmission (Reisen 2010). This concept led to the emergence of landscape epidemiology, aimed at identifying the ecological settings where vector, host, and pathogen intersect within a permissive climate favoring pathogen circulation (Reisen 2010). In multi-vector, multi-host pathogens, where vector(s)-host(s) interactions are highly heterogeneous (Woolhouse et al. 1997), analyzing these complex vector–host–pathogen–environment systems and their interactions require an interdisciplinary approach and modern analytical tools (Lambin et al. 2010). Different studies have modeled pathogen distribution based on climatic variables (Hay et al. 2002; Caminade et al. 2014; Marcantonio et al. 2015), but only a few have incorporated host information (Kilpatrick et al. 2006). Our aim, therefore, was to integrate data on climate, landscape, vectors, and hosts in order to model transmission risk for a vector-borne pathogen. We have previously shown how landscape and climatic variables determine the mosquito populations in our study area, the Doñana National Park, and its surroundings (Roiz et al. 2014, 2015) and have also studied the vector–host interactions (Muñoz et al. 2012), which, together with vector competence, greatly determine disease amplification (Kilpatrick et al. 2005; Hamer et al. 2009).
In this work, we screened for West Nile virus (WNV) and Usutu virus (USUV) presence in mosquitoes, but as we detected USUV but not WNV, we focus on the circulation of USUV. Usutu virus is an emerging mosquito-borne flavivirus of African origin, first isolated in Culex univitattus in South Africa in 1959, and later detected in birds, mosquitoes, and humans (Williams et al. 1964). In Europe, USUV is widely distributed in Italy, Austria, Hungary, Spain, Switzerland, the Czech Republic, France, the UK, Germany, and Poland (Weissenböck et al. 2007) and is considered as endemic in some areas (Calzolari et al. 2013). Usutu has caused some mortalities in wild birds, and the number of neuroinvasive cases in humans has increased in recent years (Vázquez et al. 2011; Santini et al. 2015). The USUV cycle is similar to West Nile virus (WNV) cycle, replicating successfully in many avian hosts and several mosquito vectors (Nikolay 2015). Like WNV, it is mostly transmitted by Culex mosquitoes, and in Spain, USUV has been detected in Culex pipiens and Culex perexiguus (Busquets et al. 2008; Vázquez et al. 2011).
In this paper, we analyze the distribution and prevalence of Usutu virus in the most important wetland area in SW Spain in relation to vectors, hosts, landscape, and climate variables.
Methods
Study Area
Doñana is the most important wetland in Western Europe. The protected area extends over more than 1060 km2 of marshes, sand dunes, scrubland, and Mediterranean forests. Doñana is a major site for migrating birds and has been declared as a Biosphere Reserve, a Ramsar Site, and a UNESCO World Heritage Site. Considered one of the most important reserves and most outstanding protected areas in Europe, it receives around 350,000 visitors per year. The Park has a Mediterranean subhumid climate with rainy winters and dry summers. Usutu was detected in Cx. perexiguus in 2009 (Vázquez et al. 2011), and the fieldwork was carried out between March and November 2010.
Sampling Design, Remote Sensing, and Mosquito Identification
We used a stratified sampling design, considered an adequate method (Reisen and Lothrop 1999) to distribute the mosquito traps over six ecological units in the study area: marshland, scrubland, sand dunes, fish ponds, rice fields, and crops (total surface area 54,984 ha). Each of the six ecological units was divided into 1 × 1 km squares and 20% of the quadrates of each unit were randomly selected. As some of these randomly selected quadrates (less than 1%) could not be easily reached by walking or by car, we exchanged them by others, with easier access, in the same ecological unit chosen with the same randomization procedure. Each quadrate was located in the field using a PDA device with GPS and ArcPad software. Each sampling station was located in each of these quadrats and consisted of a BG-Sentinel trap with BG-lure and a container of dry ice to generate a continuous CO2 flow, a trapping method that has been previously demonstrated as efficient as CDC traps for Culex species (Roiz et al. 2012). Trap number and distribution were adapted to the inundation regimes to omit areas inaccessible due to extensive flooding. A greater number of quadrates were sampled through the spring as the marsh dried until September when the maximum numbers of quadrates were sampled. During April, May, and June, traps were transported by horse and placed on two transects across the flooded marshes. Due to the environmental homogeneity of the ricefields and the difficulty of maintaining the traps in these areas because of theft, fewer traps were located there. A total of 112 traps were distributed according to the surface of the main habitats: 47 in marshland, 31 in scrubland, 15 in sand dunes, 10 in fish ponds, 6 in crops, and 3 in ricefields. Sampling stations were located in open areas near vegetation and operated one night per month (from March to November 2010), for a total of 716 trap nights. Details of the methodology of mosquito identification are described in detail by Roiz et al. (2015). A total of 908,237 female mosquitoes were captured, but only a proportion of them were tested for the presence of flaviviruses (Table 1). The most common species were Cx. theileri (47.2%) and Oc. caspius (40.2%), followed by Cx. modestus (10.7%), An. atroparvus (1.02%), Cx. perexiguus (0.67%), Cx. pipiens (0.2%), and Oc. detritus (0.06%).
Remote Sensing and Climatic Variables
Remote sensing variables were extracted from landsat images obtained from the USGS (U.S. Geological Survey) Geographic Information Centre (http://glovis.usgs.gov/). Twelve images were selected according to availability and cloud cover, spanning the full hydrologic year (30 August 2009, 17 October 2009, 2 November 2009, 4 December 2009, 6 February 2010, 3 April 2010, 5 May 2010, 6 June 2010, 30 June 2010, 1 August 2010, 10 September 2010, and 5 November 2010). Hydroperiod, a variable that quantifies the number of days that each pixel (900 m2) remained flooded, was estimated from all the available images for the period of 30 August 2009–September 2010 (hydrologic season). Inundation area was calculated as the number of pixels that were flooded inside each buffer. Areas unsuitable for mosquito larval development, such as salt ponds, the sea, and the river, were removed from the analysis. Normalized difference vegetation index (NDVI) was calculated from the images as a normalized ratio of the red and near-infrared bands. NDVI, which is commonly used to determine vegetation covertures, is a measure of the relationship between these two wavelength bands, from which photosynthetically active radiation can be calculated. We estimated several landscape variables from satellite images (see modeling strategy) over a buffer of 2000 m away from the mosquito traps (as 2000 m explained the highest amount of variance in mosquito abundance when compared to buffers of 1000, 500, 250, and 100 m, see, Roiz et al. 2015). We calculated the following variables: (1) monthly inundation surface and (2) monthly NDVI the same month and the month before. For these variables, the mean and standard deviation at the 2000 m buffer are 5.88 ± 0.98 for monthly inundation surface and 0.07 ± 0.06 for monthly NDVI. Using ARCGIS 10.1 software, we extracted the data for the various buffers using the spatial analyst tool Zonal Statistics as a table for raster files and the geoprocessing intersect tool for vector files.
Climatic data (temperature and rainfall) were extracted from Palacio de Doñana meteorological station (http://icts.ebd.csic.es/en/datos-meteorologicos). Data were pooled by month, and the mean monthly temperature values and the total monthly rainfall were calculated, as in the previous works (Roiz et al. 2014).
Host Density
Abundance of avian hosts was quantified by making five six-minute fixed point counts at each mosquito trapping place (one at the trap itself and the other four 150–200 m in each cardinal direction). Counts were made from early in the morning until 4 h after sunrise within 7 days of mosquito trapping. Both visual and auditory contacts in the front of the observer (180°) were counted, and the distance to the bird was recorded with a telemeter. Density for each avian species was calculated using the DISTANCE program (Thomas et al. 2010). Count data were available for 29 trapping stations, and we related the averaged counts of the stations less than 2000 m away from the mosquito traps (as 2000 m is a representative buffer, see, Roiz et al. 2015). Therefore, data from 63 of the traps inside these buffer areas were related to host census data. Avian richness and density and Passeriformes richness and density were computed from the count data. From census data, we estimated species richness as the number of different species detected, density as the number of individuals counted, and diversity estimated as the Shannon diversity index (Logiudice et al. 2003). Shannon diversity index ranges between 0 (when all the individuals observed belong to a single species) and is maximized when individuals are evenly distributed across all the species present in the area. Species richness, density, and diversity were calculated based on data for all avian species and only for the order Passeriformes. Although information on host competence for Usutu virus is scarce, we hypothesized based on infection prevalence data that Passeriformes are an important group of competent hosts for Usutu virus (as for West Nile virus) and that mammals are not competent hosts (Nikolay 2015; Ezenwa et al. 2006).
Virus Detection
Viral RNA was extracted from pools of up to 50 unfed female mosquitoes with an RNeasy miniKit (Qiagen extraction Kit (Qiagen). Flavivirus presence was determined by RT-PCR following Sánchez-Seco et al. (2005) protocol modified by adding HotMaster Polymerase enzyme (Promega) to the nested PCR to avoid nonspecific amplicons. Positives results were confirmed using a generic RT-Nested-PCR (Vázquez et al. 2012), and the flavivirus amplified was identified by sequencing of both the generated fragments. Minimum infection rate (MIR) was calculated using the mosqSurv Software (http://www.cdc.gov/westnile/resourcepages/mosqSurvSoft.html). Due to economic limitations and based on the results of the previous studies (Muñoz et al. 2012), we analyzed all the pools of Culex modestus, Cx. perexiguus, and Cx. pipiens, only a fraction of the pools of Oc. caspius, and excluded Cx. theileri pools from molecular analysis (Table 1). Our previous work in the area indicates that Oc. caspius and Cx. theileri, due to its high tendency to feed on mammals, are not relevant vectors of West Nile virus and probably for the other avian flavivirus as Usutu (Muñoz et al. 2012; Martínez-de la Puente et al. 2018).
Modeling Strategy, Data Exploration, and Statistical Analysis
We analyzed Usutu monthly presence in relation to host, vector, climate, and landscape characteristics. We examined the relationship between mosquito, host, and environmental monthly data from the current and the previous months (Inundation surface, NDVI, and rainfall of the previous month). Results of the data exploration together with a principal component analysis (PCA) are presented in Table S1 and Figures S1, S2, S3, S4, and S5. We modeled the monthly presence of Usutu with a generalized linear mixed model with binomial distribution, trap location as random variable, and mosquito pool size as an offset variable in relation to the following explanatory variables: (1) Cx. perexiguus abundance, (2) flooded surface, (3) flooded surface in the previous month, (4) NDVI, (5) NDVI in the previous month, (6) Passeriform richness, (7) Passeriform abundance, (8) Passeriform diversity (Shannon index), (9) avian richness, (10) avian abundance, (11) avian diversity (Shannon index), (12) relative Passeriform abundance in relation to total avian abundance, (13) mean temperature, (14) rainfall, (15) rainfall in the previous month, and (16) a null model including only the intercept.
Model Selection
We fitted univariate models to avoid collinearity and model overfitting. We based model selection on the AICc criterion (corrected Akaike information criteria) and reported the AIC weights as an estimate of the relative support for each model. We calculated the explained deviance as (null model deviance − residual deviance)/null model deviance. Statistical analyses were carried out and figures generated in R version 2.14.2 (Venables and Smith 2009) using several packages (mass, ade4, lattice, pscl, ncf, rms, odds ratio, akima, MuMin).
Results
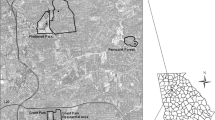
Having analyzed all the selected samples (2802 pools; 81,339 female mosquitoes), we detected Usutu virus in nine pools of Cx. perexiguus (seven in July, two in August) captured in seven different locations (six in scrubland and one in sand dunes). All pools analyzed for the other mosquito species were negative for USUV (Table 1, Fig. 1).
Map of the study area and the results of Usutu virus detection inside the pools of the vector Cx. perexiguus. Red circles indicate the locations of the traps with positive detections of Usutu virus in Cx. perexiguus. Green circles indicate the locations of the traps with uninfected pools of Cx. perexiguus. White circles indicate the locations of the traps without detection of the vector Cx. perexiguus. Sampling is described in methods. Urban areas correspond to the villages situated at the border of the study area (Color figure online).
We analyzed the relationship between monthly Usutu presence and all the climatic (temperature and rainfall), landscape (NDVI, inundation surface), vector (vector abundance) and host (avian and Passeriformes density, richness, and diversity) variables, and the relationship among variables (Table S1, Figs. S1–S5). Bird host variables, landscape, and vector abundance variables resulted as relatively relevant to determine Usutu presence, based on PCA results. Most of the variables are correlated. The first axis (horizontal) explained 35% on which they belong the host variables (avian and passerine richness, and density positively related), the landscape variables (NDVI and NDVI before positively related and inundation area and inundation area before negatively correlated). The second axis, that explained 16% of the variance, is dominated by climatic variables (positively to mean temperature and negatively to rainfall). Secondarily, vector abundance is controlled by both the axis 1 and the axis 2.
Based on the results of GLM analysis, Usutu presence was positively related to Passeriform richness, with an AICc weight (relative importance) of 54% (Table 2, Fig. 2), followed by NDVI in the previous month (10% of the total). The effect size for Passeriforme richness on the probability of Usutu detection increases after a value of 20 to 40. The results and effect sizes of other variables are presented in Table 2 and Fig. S5.
Discussion
The nidus of infection for a vector-borne disease is determined by the pathogen(s), the vector(s) and host(s) populations, landscape and climate, and the interactions between these components (Reisen 2010). For our study, we used a field-based measure of disease risk (virus presence in vector populations) and measures of wildlife biodiversity (species richness and Shannon diversity index) together with vector, landscape, and climatic variables.
Monthly Usutu presence was positively related to Passeriformes richness. From all the landscape and climatic variables considered, only NDVI of the month before was positively related to the presence of Usutu virus in Cx. perexiguus. This relationship probably reflects the positive relationship between NDVI and Cx. perexiguus presence and abundance reported in the same study area by Roiz et al. (2015). Unfortunately, while climatic and landscape variables can be easily measured over large surfaces, vector and host communities are harder to study. Having several potential vector species, this study highlights that Cx. perexiguus (a highly ornithophilic and opportunistic mosquito species (Muñoz et al. 2012)) may play an important role in the circulation of Usutu virus in southern Europe, something that needs to be addressed in further studies.
We found a positive relationship between Passeriformes richness and Usutu virus monthly prevalence, explaining more variance than any other climatic, landscape, vector, and host variables. Our results demonstrate that Passeriformes may be an important group for the amplification of Usutu virus, and have, in fact, been indicated as a key group of competent hosts for West Nile, a phylogenetically related virus with a similar ecology (Nikolay 2015). However, the results may be treated with some caution, due to the limited prevalence of the virus and the absence of Usutu host competence data.
The “dilution effect” hypothesis suggested that biodiversity and disease transmission may be negatively related to increasing diversity reducing the number of contacts resulting in successful pathogen transmission (Keesing et al. 2010). Consequently, it has been suggested that loss of biodiversity may increase the transmission rate of pathogens (Keesing et al. 2010; Begon 2008). Studies have been published supporting this idea for Lyme disease (LoGiudice et al. 2003), West Nile virus (Ezenwa et al. 2006), and helminths (Johnson and Thieltges 2010). However, others suggest that the diversity–disease relationship is idiosyncratic and depends on local factors, and cannot, therefore, be considered as a general phenomenon (Salkeld et al. 2013; Wood and Lafferty 2013; Randolph and Dobson 2012). Although a recent meta-analysis supported the generality of the dilution effect (Civitello et al. 2015), this idea is controversial (Salkeld et al. 2013). In fact, it is theoretically possible for high host diversity to either dilute or amplify disease prevalence, depending on host competence and composition (Keesing et al. 2006), but they are few field studies analyzing these relationships, e.g., Johnson and Thieltges (2010) and Salkeld et al. (2013). We report an amplification effect of host diversity on a multi-host, multi-vector, mosquito-borne disease. Usutu prevalence was higher when and where passerine richness was higher. A simple explanation for this result is that pathogen amplification increases with the addition of competent hosts, in this case, an increase in the richness and density of Passerines (Ezenwa et al. 2006; Randolph and Dobson 2012; Keesing et al. 2006). The higher abundance of passerines may increase the number of encounters between infected mosquitoes and competent hosts (Allan et al. 2009). We failed to find any relationship with avian abundance, richness, and diversity contrary to the predictions of the dilution effect for these results.
Therefore, rather than biodiversity per se, it is more likely that the specific composition of the community and the role of individual host species and their interactions with other hosts, vectors, and the pathogen are more influential in determining local disease risk leading to negative (dilution), positive (amplification), or nonsignificant relationships with biodiversity (Kilpatrick et al. 2005; Hamer et al. 2009; LoGiudice et al. 2003; Salkeld and Lane 2010). Straightforward studies of how the local infection rates are determined by the host used by vectors and community force of infection (the heterogeneity of vector host feeding together with the variation in vertebrate reservoir competence of the host community) rather than simply host diversity or richness are needed to clarify the underlying mechanisms as demonstrated for West Nile (Hamer et al. 2011). In addition, the contribution of relevant hosts, as Passeriformes, and particular groups, as Turdidae, may be investigated (Vázquez et al. 2011; Tolsá et al. 2018). Pathogens, such as Usutu, with multiple hosts and vectors are systems where the role of diversity and host–vector community interactions on pathogen amplification becomes more complicated and therefore interesting to characterize. Our data support an amplification effect modulated by the nidus of infection. Detailed data on host competence and blood-feeding preferences of mosquitoes together with host community composition and reservoir competence are needed to confirm this biodiversity–disease relationship in a multi-vector, multi-host, mosquito-borne disease.
References
Allan BF, Langerhans RB, Ryberg WA, Landesman WJ, Griffin NW, Katz RS, et al. (2009) Ecological correlates of risk and incidence of West Nile virus in the United States. Oecologia 158: 699–708; https://doi.org/10.1007/s00442-008-1169-9
Begon, M. (2008) Effects of host diversity on disease dynamics. Infectious Disease Ecology: Effects of Ecosystems on Disease and of Disease on Ecosystems. https://doi.org/10.1515/9781400837885.
Busquets N, Alba A, Allepuz A, Aranda C, Nuñez JI. (2008) Usutu virus sequences in Culex pipiens (Diptera: Culicidae), Spain. Emerging Infectious Diseases, 14(5): 861. https://doi.org/10.3201/eid1405.071577
Calzolari M, Bonilauri P, Bellini R, Albieri A, Defilippo F, Tamba M, et al. (2013) Usutu virus persistence and West Nile virus inactivity in the Emilia-Romagna region (Italy) in 2011. PLoS One, 8: e63978; https://doi.org/10.1371/journal.pone.0063978.
Caminade C, Kovats S, Rocklov J, Tompkins AM, Morse AP, Colón-González FJ, et al. (2014) Impact of climate change on global malaria distribution. Proceedings of the National Academy of Sciences 111: 3286–3291; https://doi.org/10.1073/pnas.1302089111.
Civitello DJ, Cohen J, Fatima H, Halstead NT, Liriano J, McMahon TA, et al. (2015) Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proceedings of the National Academy of Sciences 112: 8667–8671; https://doi.org/10.1073/pnas.1506279112.
Ezenwa VO, Godsey MS, King RJ, Guptill SC. (2006) Avian diversity and West Nile virus: testing associations between biodiversity and infectious disease risk. Proceeding of the Royal Society of London B 273: 109–117 https://doi.org/10.1098/rspb.2005.3284.
Hamer GL, Kitron UD, Goldberg TL, Brawn JD, Loss SR, Ruiz MO, et al. (2009) Host selection by Culex pipiens mosquitoes and West Nile virus amplification. American Journal of Tropical Medicine and Hygiene 80: 268–278; https://doi.org/10.4269/ajtmh.2009.80.268.
Hamer GL, Chaves LF, Anderson TK, Kitron UD, Brawn JD, Ruiz MO, et al (2011) Fine-scale variation in vector host use and force of infection drive localized patterns of West Nile virus transmission. PLoS One (8): 23767. https://doi.org/10.1371/journal.pone.0023767.
Hay SI, Cox J, Rogers DJ, Randolph SE, Stern DI, Shanks GD, et al. (2002) Climate change and the resurgence of malaria in the East African highlands. Nature 415: 905–909; https://doi.org/10.1038/415905a.
Johnson PTJ, Thieltges DW. (2010) Diversity, decoys and the dilution effect: how ecological communities affect disease risk. Journal of Experimental Biology 213: 961–970; https://doi.org/10.1242/jeb.037721.
Jones KE, Patel NG, Levy MA, Storeygard A, Balk D, Gittleman JL, Daszak P. (2008) Global trends in emerging infectious diseases. Nature 451: 990–993; https://doi.org/10.1038/nature06536.
Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, et al. (2010) Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468: 647–652; https://doi.org/10.1038/nature09575.
Keesing F, Holt RD, Ostfeld RS. (2006) Effects of species diversity on disease risk. Ecology Letters 9: 485–498; https://doi.org/10.1111/j.1461-0248.2006.00885.x.
Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. (2006) Host heterogeneity dominates West Nile virus transmission. Proceedings of the Royal Society of London B, 273: 2327–2333; https://doi.org/10.1098/rspb.2006.3575.
Kilpatrick AM, Kramer LD, Campbell SR, Alleyne EO, Dobson AP, Daszak P. (2005) West Nile virus risk assessment and the bridge vector paradigm. Emerging Infectious Diseases 11: 425–429; https://doi.org/10.3201/eid1103.040364.
Lambin EF, Tran A, Vanwambeke SO, Linard C, Soti V. (2010) Pathogenic landscapes: interactions between land, people, disease vectors, and their animal hosts. International Journal of Health Geographic 9: 1; https://doi.org/10.1186/1476-072x-9-54.
LoGiudice K, Ostfeld RS, Schmidt KA, Keesing F. (2003) The ecology of infectious disease: effects of host diversity and community composition on Lyme disease risk. Proceedings of the National Academy of Sciences. 100: 567–571.
Marcantonio M, Rizzoli A, Metz M, Rosà R, Marini G, Chadwick E, Neteler M. (2015). Identifying the environmental conditions favouring West Nile Virus outbreaks in Europe. PLoS One, 10(3): e0121158. https://doi.org/10.1371/journal.pone.0121158
Martínez-de la Puente J, Ferraguti M, Ruiz S, Roiz D, Llorente F, Pérez-Ramírez E, et al. (2018) Mosquito community influences West Nile virus seroprevalence in wild birds: implications for the risk of spillover into human populations. Scientific Reports, 8 (1): 2599.
Muñoz J, Ruiz S, Soriguer R, Alcaide M, Viana DS, Roiz D, et al. (2012) Feeding patterns of potential West Nile virus vectors in south-west Spain. PloS One, 7: e39549; https://doi.org/10.1371/journal.pone.0039549.
Nikolay B (2015) A review of West Nile and Usutu virus co-circulation in Europe: how much do transmission cycles overlap? Transactions of the Royal Society of Tropical Medicine and Hygiene. 109: 609-618; https://doi.org/10.1093/trstmh/trv066.
Randolph SE, Dobson ADM. (2012) Pangloss revisited: a critique of the dilution effect and the biodiversity-buffers-disease paradigm. Parasitology. 139: 847–863; https://doi.org/10.1017/s0031182012000200.
Reisen WK, Lothrop HD (1999) Effects of sampling design on the estimation of adult mosquito abundance. Journal of the American Mosquito Control Association 15: 105–114.
Reisen WK. (2010) Landscape epidemiology of vector-borne diseases. Annual Review of Entomology. 55: 461–483; https://doi.org/10.1146/annurev-ento-112408-085419.
Roiz D, Roussel M, Muñoz J, Ruiz S, Soriguer R, Figuerola, J. (2012) Efficacy of mosquito traps for collecting potential West Nile mosquito vectors in a natural Mediterranean wetland. American Journal of Tropical Medicine and Hygiene. 86: 642–648; https://doi.org/10.4269/ajtmh.2012.11-0326.
Roiz D, Ruiz S, Soriguer R, Figuerola J. (2014) Climatic effects on mosquito abundance in Mediterranean wetlands. Parasites and Vectors 7: 333; https://doi.org/10.1186/1756-3305-7-333.
Roiz D, Ruiz S, Soriguer R, Figuerola J. (2015) Landscape effects on the presence, abundance and diversity of mosquitoes in mediterranean wetlands. PloS One, 10: e0128112; https://doi.org/10.1371/journal.pone.0128112.
Salkeld DJ, Lane RS. (2010) Community ecology and disease risk: lizards, squirrels, and the Lyme disease spirochete in California, USA. Ecology, 91: 293–298; https://doi.org/10.1890/08-2106.1.
Salkeld DJ, Padgett KA, Jones JH. (2013) A meta‐analysis suggesting that the relationship between biodiversity and risk of zoonotic pathogen transmission is idiosyncratic. Ecology Letters 16: 679–686; https://doi.org/10.1111/ele.12101.
Sánchez-Seco MP, Rosario D, Domingo C, Hernández L, Valdes K, Guzmán MG, Tenorio A. (2005) Generic RT-nested-PCR for detection of flaviviruses using degenerated primers and internal control followed by sequencing for specific identification. Journal of Virological Methods. 126: 101–109; https://doi.org/10.1016/j.jviromet.2005.01.025.
Santini M, Vilibic-Cavlek T, Barsic B, Barbic L, Savic V, Stevanovic V, et al. (2015) First cases of human Usutu virus neuroinvasive infection in Croatia, August–September 2013: clinical and laboratory features. Journal of Neurovirology. 21: 92–97; https://doi.org/10.1007/s13365-014-0300-4.
Taylor LH, Latham SM, Mark EJ. (2001) Risk factors for human disease emergence. Philosophical Transactions of the Royal Society B. 356: 983-9; https://doi.org/10.1098/rstb.2001.0888.
Thomas L, Buckland ST, Rexstad EA, Laake JL, Strindberg S, Hedley SL, et al. (2010) Distance software: design and analysis of distance sampling surveys for estimating population size. Journal of Applied Ecology. 47: 5-14; https://doi.org/10.1111/j.1365-2664.2009.01737.x.
Tolsá MJ, García-Peña GE, Rico-Chávez O, Roche B. Suzán G. (2018) Macroecology of birds potentially susceptible to West Nile virus. Proceedings of the Royal Society B. 285: 1893: 20182178. https://doi.org/10.1098/rspb.2018.2178.
Vázquez A, Jiménez-Clavero MA, Franco L, Mantke OD, Sambri V, Niedrig M, et al. (2011) Usutu virus–potential risk of human disease in Europe. EuroSurveillance. 16: 1-5.
Vázquez A, Sánchez-Seco MP, Palacios G, Molero F, Reyes N, Ruiz S, et al. (2012) Novel flaviviruses detected in different species of mosquitoes in Spain. Vector Borne and Zoonotic Diseases. 12: 223-229; https://doi.org/10.1089/vbz.2011.0687.
Venables WN, Smith DM. (2009) The R development core team. An Introduction to R. Notes on R: A Programming Environment for Data Analysis and Graphics Version 2.10.1.
Weissenböck H, Chvala-Mannsberger S, Bakonyi T, Nowotny N. (2007) Emergence of Usutu virus in Central Europe: Diagnosis, Surveillance and Epizootiology. Emerging Pests and Vector-Borne Diseases in Europe. Wageningen: Wageningen Academic, pp 153–168; https://doi.org/10.3920/978-90-8686-626-7.
Williams MC, Simpson DI, Haddow AJ, Knight EM. (1964) The Isolation of West Nile. Virus from Man and of Usutu Virus from the Bird Biting Mosquito Mansonia aurites (Theobald) in the Entebbe Area of Uganda. Annals of the Tropical Medicine and Parasitology. 58 (3): 367-74; https://doi.org/10.1080/00034983.1964.11686258.
Wood CL, Lafferty KD. (2013) Biodiversity and disease: a synthesis of ecological perspectives on Lyme disease transmission. Trends in Ecology and Evolution. 28: 239-247; https://doi.org/10.1016/j.tree.2012.10.011.
Woolhouse ME, Dye C, Etard JF, Smith T, Charlwood JD, Garnett GP, et al. (1997) Heterogeneities in the transmission of infectious agents: implications for the design of control programs. Proceedings of the National Academy of Sciences. 94: 338-42; https://doi.org/10.1073/pnas.94.1.338.
Acknowledgements
Oscar González carried out all the vertebrate censuses, while Juani Moreno and Antonio Magallanes from the Mosquito Control Service (Huelva, Spain) and Esmeralda Perez Morueta, Mayca Lozano, Carlos Moreno, and Cristina Pérez from the Estación Biológica de Doñana (Sevilla, Spain) helped with mosquito capture and identification. Discussions with Jean-François Guégan, Benjamin Roche, Gabriel Garcia-Peña, and Carlo Costantini improved this paper.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Roiz, D., Vázquez, A., Ruiz, S. et al. Evidence that Passerine Birds Act as Amplifying Hosts for Usutu Virus Circulation. EcoHealth 16, 734–742 (2019). https://doi.org/10.1007/s10393-019-01441-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10393-019-01441-3