Abstract
Background
We previously demonstrated the relationship of human microbiome Fusobacterium nucleatum with unfavorable clinical outcomes and inferior chemotherapeutic responses in esophageal cancer. Global DNA methylation is associated with the occurrence and development of various cancers. In our previous study, LINE-1 hypomethylation (i.e., global DNA hypomethylation) was associated with a poor prognosis in esophageal cancer. As the gut microbiota may play crucial roles in the DNA methylation of host cells, we hypothesized that F. nucleatum might influence LINE-1 methylation levels in esophageal cancer.
Methods
We qualified the F. nucleatum DNA using a quantitative PCR assay and LINE-1 methylation via a pyrosequencing assay using formalin-fixed paraffin-embedded specimens from 306 esophageal cancer patients.
Results
Intratumoral F. nucleatum DNA was detected in 65 cases (21.2%). The LINE-1 methylation scores ranged from 26.9 to 91.8 (median = 64.8) in tumors. F. nucleatum DNA was related to the LINE-1 hypomethylation of tumor lesions in esophageal cancer (P < 0.0001). The receiver operating characteristic curve analysis showed that the area under the curve was 0.71 for F. nucleatum positivity. Finally, we found that the impact of F. nucleatum on clinical outcomes was not modified by LINE-1 hypomethylation (P for interaction = 0.34).
Conclusions
F. nucleatum alters genome-wide methylation levels in cancer cells, which may be one of the mechanisms by which F. nucleatum affects the malignant behavior of esophageal cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal cancer is one of the leading causes of cancer-related death worldwide. Despite the development of multidisciplinary treatments, including surgery, immunotherapy, chemotherapy, radiotherapy, and chemoradiotherapy, the prognosis of patients with esophageal cancer remains unfavorable [1]. The limited improvement in treatment outcomes with these conventional therapies has prompted the search for innovative therapeutic strategies [2, 3]. Importantly, epigenetic changes and alterations in the gut microbiome are reversible and can be targets for cancer therapy or chemoprevention [4,5,6].
Gut microbiota is a highly advanced research field that has attracted much attention in recent years because of its reported association with various diseases, such as obesity, inflammatory bowel disease and cancers. Fusobacterium nucleatum (F. nucleatum) is a gram-negative non-spore-specific anaerobic bacterium and a constituent of the oral microbiomes [7]. It can adhere to the oral cavity as well as migrate to and colonize the intestinal tract, which is closely related to the occurrence and development of various types of cancers, including esophageal cancer [8, 9]. F. nucleatum can activate the host cell cancer-related signal pathway and promote the proliferation and metastasis of cancer cells [10,11,12]. We have previously reported that the high amount of F. nucleatum DNA is related to a poor prognosis in patients with esophageal cancer [13]. In addition, F. nucleatum confers chemoresistance to esophageal cancer cells via the modulation of autophagy. However, the mechanism by which F. nucleatum contributes to esophageal cancer malignancy is not yet completely clear.
DNA methylation changes associated with human tumors are site-specific CpG island promoter hypermethylation and global DNA hypomethylation. Promoter hypermethylation can silence tumor suppressor genes, DNA mismatch repair genes (e.g., MLH1), or DNA repair genes (e.g., MGMT), thereby contributing to esophageal carcinogenesis [14]. Global DNA hypomethylation appears to play an important role in genomic instability, leading to cancer development [5]. As long interspersed element-1 (LINE-1 or L1; a repetitive DNA retrotransposon) constitutes approximately 17% of the human genome, the level of LINE-1 methylation is regarded as a surrogate marker of global DNA methylation [5]. LINE-1 methylation is highly variable, and the strong relationships between LINE-1 hypomethylation and unfavorable prognosis have been shown in many types of human cancers, such as esophageal cancer [15,16,17,18].
The gut microbiome can alter DNA methylation in host cells through a variety of mechanisms. Therefore, we hypothesized that the level of LINE-1 methylation in esophageal cancer tissues might be influenced by F. nucleatum. To our knowledge, this is the first study to focus on the relationship between F. nucleatum and LINE-1 methylation in esophageal cancer. This study provides new insights into the correlation between the amount of F. nucleatum DNA and the methylation level of LINE-1. Therefore, we inferred that the enrichment of F. nucleatum is associated with host tumor epigenetic modification in esophageal cancer.
Materials and methods
Study cohort
We analyzed 306 formalin-fixed paraffin-embedded (FFPE) specimens of esophageal cancer tissues from patients who underwent resection. The TNM stage was determined according to the American Joint Committee on Cancer Staging Manual (7th edition) [19]. We interviewed patients at their first visit for information about their smoking and drinking history and assessed whether they had ever smoked or drank alcohol (Yes or No). Table 1 shows the clinical features of the study cohort. Written informed consent was obtained from each patient, and the procedures were approved by the Institutional Review Board of Kumamoto University (#1272).
Quantitative real-time polymerase chain reaction (qPCR) for F. nucleatum
H&E-stained slides of the tumors were reviewed by one pathologist, who marked the areas of the tumor and normal mucosa. H&E-stained tissue sections from each case were scraped off slides for DNA extraction. The DNA was extracted using a QIAamp DNA FFPE tissue kit (Qiagen, Valencia, CA, USA). Whole genome amplification of genomic DNA was performed by PCR using random 15-mer primers for subsequent genetic analyses. We determined the amount of F. nucleatum DNA by qPCR assay. The nus G gene of F. nucleatum and the reference human gene SLCO2A1 were amplified using custom-made TaqMan primer/probe sets (Applied Biosystems, Waltham, Massachusetts, USA). Assays were performed in a 384-well optical PCR plate. DNA was amplified and detected using a LightCycler® 480 Instrument II (Roche, Basel, Switzerland). The amount of F. nucleatum DNA in each tissue was normalized relative to SLCO2A1 [13]. In this study, 0 (i.e., cases with no F. nucleatum DNA detected at all) are classified as negative and > 0 (i.e., cases with even a little F. nucleatum DNA detected) are classified as positive.
Pyrosequencing for LINE-1 methylation
Bisulfite treatment of genomic DNA was carried out using an EpiTect Bisulfite kit. PCR and subsequent LINE-1 pyrosequencing were performed using a PyroMark Kit (Qiagen) [20]. This assay amplifies a region of LINE-1 element (position 305–331 in accession No. X58075), which includes four CpG cites. The PCR condition was 45 cycles of 95 °C for 20 s, 50 °C for 20 s and 72 °C for 20 s, followed by 72 °C for 5 min. The biotinylated PCR product was purified and made single-stranded to act as a template in a pyrosequencing reaction, using the Pyrosequencing Vacuum Prep Tool (Qiagen). Pyrosequencing reactions were performed in the PyroMark Q24 System (Qiagen). The nucleotide dispensation order was: ACT CAG TGT GTC AGT CAG TTA GTC TG. The amount of C relative to the sum of the amounts of C and T at each CpG site was calculated as percentage (i.e., 0–100). The average of the relative amounts of C in the four CpG sites was used as overall LINE-1 methylation level in a given tumor (Fig. 1).
Pyrosequencing assay used to measure the LINE-1 methylation level. Upper panel shows LINE-1 hypomethylated tumor (methylation level, 32.5). Lower panel shows LINE-1 hypermethylated tumor (methylation level, 76.0). The % (in blue) are the proportion of C at each CpG site after bisulfite conversion, and the methylation level of each CpG site was estimated by the proportion of C (%). The overall LINE-1 methylation level was calculated as the average of the proportions of C (%) at the 4 CpG sites
Pyrosequencing to measure promoter methylation of MGMT and MLH1
Pyrosequencing for MGMT and MLH1 was performed using the PyroMark kit (Qiagen) [21]. For each sample, the average of the four and five CpG islands were calculated, respectively. 182 cases could be analyzed for MGMT and MLH1 methylation, because sufficient amounts of biotinylated PCR product were available (supplemental table). For MGMT, a clear pyrogram was not obtained in one case.
Statistical analysis
All statistical analyses were performed using JMP, version 10 (SAS Institute, Cary, NC, USA). All P values were two sided. Fisher’s exact test and Student’s t test were utilized to compare mean values for all variables. The area under the receiver operating characteristic (ROC) curve was calculated using the variables for F. nucleatum DNA and LINE-1 methylation. In this study, patients were followed up as outpatients every 1–3 months after discharge until death or December 2021. Cancer-specific survival was defined as the period from the date of surgery to the date of death by esophageal cancer. The Kaplan–Meier method was used to describe the distribution of esophageal cancer-specific survival time, and the log-rank test was performed. Until the time of censoring, censored subjects are considered “at risk”, and thus continue to contribute towards the calculation of percent survival. survival curve was accompanied by a table giving the actual numbers of patients involved. Interactions were evaluated using the Wald test for confounding the respiratory morbidity variable with another variable of interest.
Results
We utilized 306 cases of patients who underwent resection of esophageal cancer at Kumamoto University Hospital and qualified the relative amounts of F. nucleatum DNA in the esophageal cancer tissues using qPCR. We divided the patients into an F. nucleatum-negative group (n = 241, 78.8%) and an F. nucleatum-positive group (n = 65, 21.2%), according to their F. nucleatum DNA status. There was no significant difference in the clinicopathological features of the patients in terms of age, sex, preoperative performance status, alcohol, smoking, and preoperative therapy (all P > 0.05); however, the advanced stage was significantly associated with F. nucleatum positivity (P = 0.0023) (Table 1). We have already reported in a previous paper that F. nucleatum affects the prognosis of esophageal cancer [13]. Similarly, in this cohort, we found that F. nucleatum-positive esophageal cancer cases had a significantly poorer prognosis than negative cases [log-rank P = 0.0046; univariate hazard ratio (HR) = 2.07, 95% confidence interval (CI) 1.22–3.41, P = 0.0082] (Supplemental figure).
We also measured the LINE-1 methylation level in the tumor lesions using pyrosequencing technology (Fig. 1). The distribution of the LINE-1 methylation level in the 306 cases was as follows: mean, 62.2; median, 62.0; SD, 12.7; range, 26.9–91.8; interquartile range, 53.3–71.4 (all 0–100 scale). LINE-1 methylation levels were distributed approximately normally. There was no significant correlation between LINE-1 methylation and any of the clinical features (age, sex, preoperative performance status, alcohol, smoking, or preoperative therapy). In this study, cases with methylation levels higher than median value 62.0 were classified as hypermethylated cases and those with lower levels as hypomethylated cases. In this cohort, LINE-1 hypomethylated cases had a predominantly poorer prognosis than hypermethylated cases (log-rank P = 0.027; univariate HR = 1.73, 95% CI 1.06–2.86, P = 0.027) (Supplemental figure).
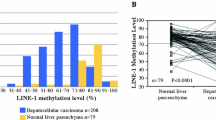
Based on our hypothesis that the level of LINE-1 methylation in esophageal cancer tissues might be influenced by F. nucleatum, we next examined the relationship between F. nucleatum and LINE-1 methylation in esophageal cancer. We found that the positivity of F. nucleatum DNA in a tumor was significantly associated with LINE-1 hypomethylation (P < 0.0001 by paired t test) (Fig. 2A). Similar results were obtained when squamous cell carcinoma and adenocarcinoma were analyzed separately [P < 0.001 for squamous cell carcinoma (Fig. 2B); P = 0.013 for adenocarcinoma and others (Fig. 2C)]. In addition, the level of F. nucleatum DNA (as a continuous variable) was related to the LINE-1 methylation level (P = 0.011) (Fig. 2D). The ROC curve analysis showed that the area under the curve was 0.71, P < 0.0001 for detecting F. nucleatum positivity in a tumor (Fig. 3). These findings certainly support the clear relationship between F. nucleatum and LINE-1 hypomethylation in esophageal cancers.
A Assessment of LINE-1 methylation scores in the Fusobacterium nucleatum-negative and -positive groups in the tumor tissue of 306 patients with esophageal cancer. P values were determined using a t test. B Correlation between the tumor LINE-1 methylation level and the amount of F. nucleatum DNA in esophageal squamous cell carcinoma. C Correlation between the tumor LINE-1 methylation level and the amount of F. nucleatum DNA in esophageal adenocarcinoma and other cancers. D Correlation between the tumor LINE-1 methylation level and the amount of F. nucleatum DNA (as a continuous variable) in esophageal cancer
We determined whether the influence of F. nucleatum on cancer-specific survival was modified by LINE-1 hypomethylation. We found that the effect was not significantly modified by LINE-1 hypomethylation (P for interaction = 0.34). In the LINE-1 hypermethylated cases, the F. nucleatum-positive cases experienced significantly shorter cancer-specific survival than the F. nucleatum-negative cases (log-rank P = 0.0049; univariate HR = 3.09, 95% CI 1.27–6.84, P = 0.015) (Fig. 4A). In the LINE-1 hypomethylated cases, the F. nucleatum-positive cases experienced shorter cancer-specific survival than the F. nucleatum-negative cases, though this was not statistically significant (log-rank P = 0.32; univariate HR = 1.39, 95% CI 0.70–2.64, P = 0.33] (Fig. 4B).
To test whether F. nucleatum specifically alters LINE-1 methylation in cancer cells, we evaluated the relationship between F. nucleatum and the methylation of MGMT and MLH1 promoter region. We obtained valid results for MGMT methylation in 181 cases and for MLH1 methylation in 182 cases using pyrosequencing technology and found that there was no significant relationship with F. nucleatum positivity (P = 0.12 for MGMT methylation level and P = 0.66 for MLH1 methylation level) (Fig. 5). This indicates that F. nucleatum has a specific effect on LINE-1 methylation level (i.e., global DNA hypomethylation).
Discussion
We conducted this study to examine the relationship between F. nucleatum (a bacterial species in the gut microbiome) and LINE-1 hypomethylation (i.e., global DNA hypomethylation) among 306 patients with resected esophageal cancers. We could demonstrate that F. nucleatum DNA was significantly related to the LINE-1 hypomethylation of tumor lesions in esophageal cancer, but not to the methylation of tumor suppressor genes (i.e., MGMT and MLH1) promoter region. Increasing evidence suggests that both the gut microbiome and epigenetic changes play important roles in esophageal cancer development and could be therapeutic targets. Importantly, the gut microbiome can alter DNA methylation in host cells through a variety of mechanisms. To the best of our knowledge, this is the first study showing the relationship between F. nucleatum and LINE-1 hypomethylation in human cancers.
F. nucleatum is a gram-negative anaerobic bacterium found in the human oral and gastrointestinal tract [7]. F. nucleatum is an opportunistic pathogen, not only involved in inflammatory processes, such as periodontitis, inflammatory bowel disease, pancreatic abscess, premature birth, and liver abscess, but also involved in the progression of cancer [8]. The ability of F. nucleatum to adhere to epithelial cells might be one of the possible reasons that it promotes tumor development [10,11,12]. The abundance of F. nucleatum correlates with poor prognosis in patients with gastrointestinal cancer, further supporting its role in imparting aggressive tumor phenotype [8]. We previously reported that F. nucleatum is associated with shorter survival and an inferior chemotherapeutic response in esophageal cancer, suggesting its potential role as a prognostic or predictive biomarker [13, 22]. In addition, utilizing in vitro and in vivo models, we have demonstrated that F. nucleatum invaded esophageal cancer cells and induced the NF-κB pathway through the NOD1/RIPK2 pathway, leading to tumor progression [23]. NOD1 is a member of the NOD-like receptor family of proteins, which functions to detect peptidoglycan and stimulate host responses to limit bacterial infection. We are of course aware that F. nucleatum may affect esophageal cancer malignancy through various complex mechanisms other than this pathway. In particular, the recent finding that the gut microbiota influences DNA methylation in host cells led us to design this study.
In cancer cells, DNA methylation can be changed in two ways: global DNA hypomethylation and site-specific CpG island promoter hypermethylation. LINE-1 constitutes approximately 17% of the human genome, and its methylation level is well-correlated with the global DNA methylation status [15]. LINE-1 methylation is highly variable, and the strong relationships between LINE-1 hypomethylation and unfavorable prognosis have been shown in many types of human cancers, such as esophageal cancer [15,16,17,18]. We have previously demonstrated the prognostic impact of LINE-1 hypomethylation in esophageal cancer, supporting its potential role as a prognostic marker [18]. We also found that LINE-1-hypomethylated tumors showed highly frequent genomic gains at various loci containing candidate oncogenes, such as CDK6 [24]. These findings certainly support the importance of global DNA hypomethylation (i.e., LINE-1 hypomethylation) in esophageal cancer development. Importantly, in this study, we demonstrated that F. nucleatum affects genome-wide methylation levels, but not promoter region methylation levels of tumor suppressor genes (MGMT and MLH1). Mismatch repair is one of the main DNA repair systems that relates to the homologous MutLS bacterial system (human MutS and MutL proteins) [25]. MLH1 (mutL homolog 1) is a human gene that plays a key role in the DNA duplication error reparation process, and likewise, it also plays a pivotal role in preserving genomic stability. Methylguanine–DNA methyltransferase (MGMT) is a specific DNA damage repair protein which plays a key role in maintaining normal cell physiology and genomic stability. Methylation of this promoter is a key predictor of whether alkylating agents can effectively control tumor cell progression [26]. It has been reported that promoter hypermethylation can silence these tumor suppressor genes, thereby contributing to the development of esophageal cancer. Of course, we understand that further validation is needed to determine whether F. nucleatum does not affect methylation of promoter regions of other cancer-related genes. In addition, further validation is needed to determine whether the relationship with LINE-1 methylation is F. nucleatum-specific or can occur in other oral bacteria.
Experimental studies have demonstrated that the gut microbiota plays an important role in the DNA methylation of host cells. Sobhani revealed that colorectal cancer-associated microbiota could induce the methylation of host genes, which contribute to epigenetic modification [27]. Kim et al. also found that the composition of the gut microbiome could play a role in persistent epigenetic modification of the liver [28]. Hattori et al. used an antibiotic to prohibit the tumorigenesis of colorectal cancer through aberrant DNA methylation induced by inflammation [29]. Although studies that focus on the link between F. nucleatum and epigenetic changes in human malignancies are rare, a population-based study by Koi et al. reported that F. nucleatum was associated with genomic hypermutation independent of the CpG island methylator phenotype (CIMP) and BRAF mutations [30]. Another study showed the relationship of Fusobacterium with wild-type TP53, hMLH1 methylation, genomic hypermutation, CHD7/8 mutation and the CIMP phenotype [31]. These studies strongly suggest the contribution of Fusobacterium to epigenetic alterations. This is the first study, to our knowledge, to reveal the relationship between the gut microbiome and epigenetic alterations in esophageal cancers. In this study, we demonstrated that the amount of intratumoral F. nucleatum DNA was related to the LINE-1 methylation of tumor lesions in esophageal cancer. This result may suggest that F. nucleatum may promote the development of esophageal cancer by regulating genome-wide DNA methylation. Nonetheless, we acknowledge that it is not clear how LINE-1 hypomethylation (i.e., genome-wide hypomethylation) is mediated in the process by which F. nucleatum affects the prognosis of esophageal cancer. We have demonstrated that F. nucleatum contributes to esophageal cancer malignancy via the NF-κB pathway, but we also believe that it influences cancer progression by other different mechanisms. We acknowledge that further validation is needed in this regard. In addition, the relationship between the localization of F. nucleatum in tumor tissue and the level of LINE-1 methylation should also be examined in the future.
These results highlight the epigenetic modification induced via gut microbiota modulation as a potential mechanism for exploring the pathogenesis of esophageal cancer. As the diagnostic and therapeutic applications of host cell epigenetic modification regulated by the gut microbiota have become increasingly popular, our study may provide information for pursuing targeted approaches for the etiology of esophageal cancer. In addition, considering that both gut microbiota and DNA methylation are reversible and have attracted attention as targets for disease therapy, we believe that this study has clinical significance.
Data availability
All presented data are available from the corresponding author upon reasonable request.
References
Baba Y, Yoshida N, Shigaki H, et al. Prognostic impact of postoperative complications in 502 patients with surgically resected esophageal squamous cell carcinoma: a retrospective single-institution study. Ann Surg. 2016;264(2):305–11.
Kato K, Cho BC, Takahashi M, et al. Nivolumab versus chemotherapy in patients with advanced oesophageal squamous cell carcinoma refractory or intolerant to previous chemotherapy (ATTRACTION-3): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20(11):1506–17.
Baba Y, Nomoto D, Okadome K, et al. Tumor immune microenvironment and immune checkpoint inhibitors in esophageal squamous cell carcinoma. Cancer Sci. 2020;111(9):3132–41.
Baba Y, Iwatsuki M, Yoshida N, et al. Review of the gut microbiome and esophageal cancer: pathogenesis and potential clinical implications. Ann Gastroenterol Surg. 2017;1(2):99–104.
Baba Y, Watanabe M, Baba H. Review of the alterations in DNA methylation in esophageal squamous cell carcinoma. Surg Today. 2013;43(12):1355–64.
Roy S, Trinchieri G. Microbiota: a key orchestrator of cancer therapy. Nat Rev Cancer. 2017;17(5):271–85.
Bashir A, Miskeen AY, Hazari YM, et al. Fusobacterium nucleatum, inflammation, and immunity: the fire within human gut. Tumour Biol J Int Soc Oncodev Biol Med. 2016;37(3):2805–10.
Liu Y, Baba Y, Ishimoto T, et al. Progress in characterizing the linkage between Fusobacterium nucleatum and gastrointestinal cancer. J Gastroenterol. 2019;54(1):33–41.
Castellarin M, Warren RL, Freeman JD, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22(2):299–306.
Abed J, Emgard JE, Zamir G, et al. Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe. 2016;20(2):215–25.
Yang Y, Weng W, Peng J, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating toll-like receptor 4 signaling to nuclear factor-kappaB, and up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152(4):851-866 e24.
Rubinstein MR, Wang X, Liu W, et al. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/beta-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14(2):195–206.
Yamamura K, Baba Y, Nakagawa S, et al. Human microbiome Fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin Cancer Res. 2016;22(22):5574–81.
Zhao R, Casson AG. Epigenetic aberrations and targeted epigenetic therapy of esophageal cancer. Curr Cancer Drug Targets. 2008;8(6):509–21.
Baba Y, Yagi T, Sawayama H, et al. long interspersed element-1 methylation level as a prognostic biomarker in gastrointestinal cancers. Digestion. 2018;97(1):26–30.
Harada K, Baba Y, Ishimoto T, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol. 2015;22(4):1280–7.
Shigaki H, Baba Y, Watanabe M, et al. LINE-1 hypomethylation in gastric cancer, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastric Cancer. 2013;16(4):480–7.
Iwagami S, Baba Y, Watanabe M, et al. LINE-1 hypomethylation is associated with a poor prognosis among patients with curatively resected esophageal squamous cell carcinoma. Ann Surg. 2013;257(3):449–55.
Rice TW, Blackstone EH, Rusch VW. 7th edition of the AJCC cancer staging manual: esophagus and esophagogastric junction. Ann Surg Oncol. 2010;17(7):1721–4.
Iwagami S, Baba Y, Watanabe M, et al. Pyrosequencing assay to measure LINE-1 methylation level in esophageal squamous cell carcinoma. Ann Surg Oncol. 2012;19(8):2726–32.
Felsberg J, Thon N, Eigenbrod S, et al. Promoter methylation and expression of MGMT and the DNA mismatch repair genes MLH1, MSH2, MSH6 and PMS2 in paired primary and recurrent glioblastomas. Int J Cancer. 2011;129(3):659–70.
Yamamura K, Izumi D, Kandimalla R, et al. Intratumoral Fusobacterium nucleatum levels predict therapeutic response to neoadjuvant chemotherapy in esophageal squamous cell carcinoma. Clin Cancer Res. 2019;25(20):6170–9.
Nomoto D, Baba Y, Liu Y, et al. Fusobacterium nucleatum promotes esophageal squamous cell carcinoma progression via the NOD1/RIPK2/NF-kappaB pathway. Cancer Lett. 2022;530:59–67.
Baba Y, Watanabe M, Murata A, et al. LINE-1 hypomethylation, DNA copy number alterations, and CDK6 amplification in esophageal squamous cell carcinoma. Clin Cancer Res. 2014;20(5):1114–24.
Kadyrov FA, Dzantiev L, Constantin N, et al. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126(2):297–308.
Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5(3):223–31.
Sobhani I, Bergsten E, Couffin S, et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc Natl Acad Sci U S A. 2019;116:24285–95.
Kim H, Worsley O, Yang E, et al. Persistent changes in liver methylation and microbiome composition following reversal of diet-induced non-alcoholic-fatty liver disease. Cell Mol Life Sci. 2019;76(21):4341–54.
Hattori N, Niwa T, Ishida T, et al. Antibiotics suppress colon tumorigenesis through inhibition of aberrant DNA methylation in an azoxymethane and dextran sulfate sodium colitis model. Cancer Sci. 2019;110(1):147–56.
Koi M, Okita Y, Carethers JM. Fusobacterium nucleatum infection in colorectal cancer: linking inflammation, DNA mismatch repair and genetic and epigenetic alterations. J Anus Rectum Colon. 2018;2(2):37–46.
Tahara T, Yamamoto E, Suzuki H, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74(5):1311–8.
Acknowledgements
This work was supported in part by JSPS KAKENHI, grant numbers 20H03755 and 17KK0195 (to Y.B.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Ethical approval
This study was approved by the institutional review board of Kumamoto University (#1272). Written informed consent was obtained from all patients. Our study was performed as per the principles of the Declaration of Helsinki.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baba, Y., Hara, Y., Toihata, T. et al. Relationship between gut microbiome Fusobacterium nucleatum and LINE-1 methylation level in esophageal cancer. Esophagus 20, 704–712 (2023). https://doi.org/10.1007/s10388-023-01009-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10388-023-01009-9