Abstract
Strong phosphine resistance in Tribolium castaneum is due to point mutations in DNA that code for amino acid changes of P45S and/or G131S in the enzyme dihydrolipoamide dehydrogenase (DLD). One allele coding for P45S is the most common in all phosphine-resistant US populations and in one strain from Brazil (TCBR), whereas another allele, G131S, occurs only in Australian beetles. Dose-mortality studies found that the TCBR strain was more resistant to phosphine than US populations. To investigate strong resistance mutations in TCBR, we sequenced cDNA for DLD in TCBR and compared results with a US population from Kansas. The common P45S mutation was detected in both populations, but two additional mutations G131D and V167A were identified only from TCBR. We used a CAPS marker (Cleaved Amplified Polymorphic Sequence) for P45S, herein designated M1, to survey this resistance allele in TCBR. We also developed a marker for the G131D mutation, designated M2. Only two genotypes, R1R1S2S2 (homozygous for resistance at M1, but homozygous susceptible at the M2 site) and R1S1R2S2 (heterozygous for resistance at M1 and M2) existed in TCBR. However, phosphine resistance levels were similar between individuals with the two genotypes. Beetles with strong resistance in TCBR may be homozygous for either the presence of the common P45S allele, or heterozygous for two resistance alleles at a second locus, G131D. The strong resistance phenotype in TCBR may be expressed differently based on previously unknown mutations at the DLD locus, but this will require further research to resolve.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Key message
-
Tribolium castaneum is a stored product pest with resistance to the fumigant phosphine. This research studied genes associated with the strong form of resistance.
-
A Brazilian population of T. castaneum has the strongest resistance to phosphine ever documented, at least 250-times more difficult to kill than beetles from a fully susceptible population.
-
We report another nonsynonymous mutation in the gene that codes for strong resistance in the Brazilian population. Genotypes for the new mutation together with the common mutation may generate the very strong resistance phenotype.
-
PCR markers for the two strong resistance genes provide tools to study the genes in pest populations to assist in phosphine resistance management.
Introduction
Phosphine gas, hydrogen phosphide (PH3), has been one of the most effective and commonly used mitigation agents for stored grain insects and other pests worldwide for many decades. Resistance to phosphine evolved in several species of stored grain insects and records of its occurrence and geographic range are increasing (Champ and Dyte 1976; Zettler and Cuperus 1990; Rajendran 1999; Collins et al. 2001; Cao et al. 2003; Benhalima et al. 2004; Pimentel et al. 2010; Lorini et al. 2007; Opit et al. 2012; Ahmad et al. 2013; Nayak et al. 2013; Jittanun and Chongrattanameteekul 2014; Koçak et al. 2015; Sağlam et al. 2015; Cato et al. 2017; Konemann et al. 2017; Afful et al. 2018). Tribolium castaneum (Coleoptera: Tenebrionidae) is one of the most common pests of stored grains and value-added grain products worldwide, and phosphine resistance has been reported for this species in many countries (Champ and Dyte 1976; Tyler et al. 1983; Zettler and Cuperus 1990; Rajendran 1999; Ren et al. 1994; Benhalima et al. 2004; Hagstrum et al. 2012; Opit et al. 2012; Schlipalius et al. 2012; Chen et al. 2015; Kaur et al. 2015). Documentation of widespread phosphine resistance in T. castaneum and other grain pest species has stimulated research on simple laboratory assays, so-called quick tests for resistance (Nayak et al. 2013), and also on molecular markers to detect the presence of resistance genes (Chen et al. 2015).
Recent research on phosphine resistance genes determined that there are generally two resistance phenotypes referred to as “weak resistance” and “strong resistance” in T. castaneum that are coded for by partially recessive alleles at each of the two unlinked loci (Jagadeesan et al. 2013, 2016; Nguyen et al. 2016). Weak resistance is expressed in insects homozygous for a resistance allele at the rph1 locus, while the strong resistance phenotype relies on a synergistic interaction in individuals that are homozygous for resistance at rph1 and also homozygous for resistance at the rph2 locus. Weak resistance can be detected through LC50 estimates for PH3 toxicity in the range of 5–50-fold higher than LC50 for susceptible insects, while the strong phenotype may exhibit resistance levels more than 100-fold higher than susceptible insects. Most recently, Schlipalius et al. (2018) identified the gene coding for rph1 weak resistance as a cytochrome b5 fatty acid desaturase (Cyt-b5-r) using the combination of next-generation sequencing, variant analysis and linkage analysis in T. castaneum and three other species of stored product insect pests. The genetic basis for strong phosphine resistance in T. castaneum and also in Rhyzopertha dominica (Coleoptera: Bostrichidae) was found to be due to one or more point mutations for a single amino acid change in a core metabolic enzyme called dihydrolipoamide dehydrogenase (DLD) (Schlipalius et al. 2012). Using these data, we also identified a single nucleotide mutation in the DLD gene, which led to a single amino acid mutation in the enzyme in T. castaneum and R. dominica, respectively, from US-resistant populations (Oppert et al. 2015; Chen et al. 2015). Transcriptome sequencing found the P45S polymorphism in the FAD binding region of the DLD gene of a Brazilian T. castaneum strain (TCBR) that was reported to be 475-fold resistant to phosphine, and it was speculated that this mutation could be associated with the strong resistance in this strain (Oppert et al. 2015). Chen et al. (2015) identified several strongly phosphine-resistant populations of T. castaneum using the modified FAO bioassay (Food and Agriculture Organization 1975) and also had high frequencies of the P45S mutation for strong resistance.
The high frequency of phosphine-resistant individuals and initial dose-mortality analysis in TCBR suggested that many beetles had a strong resistance phenotype. The transcriptome sequencing study found that the P45S mutation was at a 65–69% frequency in non-exposed or phosphine exposed TCBR beetles (Oppert et al. 2015). The first objective of the current study was to further characterize the phosphine resistance phenotype of the TCBR strain. The second objective was to determine which mutations in the DLD gene were associated with strong resistance in TCBR, and to identify any additional alleles associated with this phenotype in the TCBR strain. Lastly, from sequence information on potential new resistance alleles, we assessed the potential of a cleaved amplified polymorphic sequence (CAPS; Yeam et al. 2005) marker for the detection of phosphine-resistant alleles in this strain.
Materials and methods
Insects
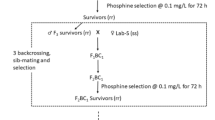
Laboratory colonies originated from three different populations of T. castaneum were used in this research. The first population was a laboratory colony obtained in May 2005 from grain samples at Bom Despacho County, State of Minas Gerais, Brazil (Pimentel et al. 2007) and referred to here as TCBR. The remaining populations were from the USA and collected at Minneapolis, Kansas, designated TCKS; and a phosphine-susceptible laboratory strain maintained at the USDA ARS Center for Grain and Animal Health Research and originally from Savannah, Georgia, referred to here as Lab-S (Chen et al. 2015). Adult insects were maintained in the laboratory on a diet of 95% all-purpose wheat flour and 5% Brewer’s yeast (wt:wt), at 27 °C, 60% RH and a light:dark cycle of 14:10. Adults were frozen at − 80 °C prior to analysis.
Dose-mortality experiments to characterize strong phosphine resistance
The strength of phosphine resistance in adults from the TCBR strain was determined and compared to that of adults from the TCKS and Lab-S strains in a series of dose-mortality experiments at concentrations of phosphine ranging from 2 ppm to 1000 ppm, with methods similar to those conducted by Opit et al. (2012). For each of the three strains, we exposed groups of 50 adults that were 2–6 wk old and held in ventilated 5-ml glass vials that had 20 g of laboratory diet. We exposed replicate vials of 50 beetles each (n = 3 for a total of 150 beetles) at each of the phosphine concentrations tested using the discriminating dose bioassay methods described by the Food and Agriculture Organization (1975). Fumigation chambers were 3.8-L gas-tight glass jars held at 25 °C and fumigated for 20 h, then ventilated and held for 14 d in clean air under standard rearing conditions to allow for delayed mortality or recovery of beetles following the fumigation. Surviving beetles were designated as resistant to the gas concentration tested compared to those killed in the same treatment. Application of phosphine to fumigation jars at target concentrations for bioassays, followed by quantitative analyses of the resulting concentrations, was conducted according to the methods of Opit et al. (2012) and Chen et al. (2015). Phosphine dose-mortality data, for which the dependent variable was % mortality and the independent variable was phosphine concentration, were subjected to probit regression using PoloPlus (LeOra Software, Petaluma, CA) (LeOra Software 2005) to determine the LC50 and LC99 values and their 95% confidence intervals (CIs). Resistance ratio values indicating the relative level of resistance, whether weak or strong, in a given population were calculated by dividing the LC50 of the test population, TCBR and TCKS, by the LC50 for the Lab-S population.
Amplification and sequencing of the DLD gene
Total RNA and poly(A) RNA isolations followed the methods described in Chen et al. (2015). Briefly, total RNA was isolated from a group of 25 individuals from each population of T. castaneum using Trizol reagent (Life Technologies, Gaithersburg, MD, USA). Poly(A) RNA was purified from total RNA using the RevertAid First Strand cDNA Synthesis Kit according to the manufacturer’s instruction (Thermo Scientific, Waltham, MA, USA). The DLD gene from T. castaneum was then amplified using cDNA as a template in 25 µl reaction volume as described in Chen et al. (2015). Briefly, the mixture included 12.5 µl Master Mix, 1 µl each corresponding forward and reverse primers at concentrations of 10.0 µmol, 2 µl cDNA template (at approximately 50 ng/µl) and 8.5 µl ddH2O using Thermo Scientific™ PCR MasterMix polymerase kit (Thermo Fisher Scientific). The PCR temperature program was the following: denaturation at 95 °C for 5 min; 36 cycles at 95 °C for 15 s, 58 °C for 30 s and 72 °C for 2 min for denaturation, annealing and extension, respectively; and a final extension at 72 °C for 10 min. PCR products were purified from the gels using QIAEX II® Gel Extraction Kit according to the manufacturer’s instruction (QIAGEN Science, MD, USA). The purified PCR products were sequenced directly using ABI 3700 DNA Sequencer at the Kansas State University DNA Sequencing Facility (Manhattan, KS USA). The cDNA of DLD from T. castaneum was assembled from the sequence reads using DNAStar (Madison, WI, USA), and the deduced amino acid sequences were obtained using the www.expasy.com website. Nucleotide sequences and deduced amino acid sequences from T. castaneum were aligned against that of a population from Garfield County in Oklahoma (Chen et al. 2015; Opit et al. 2012) and an Australian population, QTC931 (Schlipalius et al. 2012) using Clustal W2 software (Katoh et al. 2002).
Development of a resistance marker
Cleaved amplified polymorphic sequence (CAPS) marker assays were designed to detect two SNPs (single nucleotide polymorphisms) in phosphine-resistant T. castaneum, responsible for amino acid changes of P45S, referred to as M1, and G131D, referred to below as M2, found in the DLD gene sequence of T. castaneum from Brazil. A fragment from each of the relevant gene sequences was amplified in a 25 µl reaction volume (12.5 µl Master Mix, 1 µl each forward and reverse primer, 2 µl gDNA template and 8.5 µl ddH2O) using the Thermo Scientific PCR MasterMix polymerase kit as described earlier. Forward and reverse primers to amplify the regions containing M1 and M2 of T. castaneum (Table 1) were used in the PCRs for the critical SNPs responsible for the P45S and G131D mutations, respectively. The PCR temperature program for the CAPS assays was as follows: denaturation at 95 °C for 5 min; 30 cycles at 95 °C for 15 s, 54.2 °C for 30 s and 72 °C for 45 s for denaturation, annealing and extension, respectively; and a final extension at 72 °C for 10 min. The amplified 392 bp PCR product containing the SNP for the P45S mutation and the 347 bp PCR product containing the SNP for the G131D mutation were subjected to separate digestions using the restriction enzymes MboI and BsmAI (New England Biolabs, MA USA), respectively, in 10 µl reactions containing 8 µl of PCR product, 1 µl reaction buffer and 1 U restriction enzyme. The reactions were then incubated at 37 °C for MboI and 60 °C for BsmAI for 15 min according to manufacturer’s instruction.
Sequencing the DLD containing the SNP marker (M2) for G131D in the TCBR strain
In order to confirm whether the SNPs coding for guanidine to adenine at nucleotides 390 and 392 in the DLD gene sequence also exist in the TCBR strain, PCR products were sequenced following Chen et al. (2015).
Cloning and sequencing
In order to confirm that the guanine-to-adenine mutations at nucleotide positions 390 and 392 of DLD were found in heterozygous individuals, a pair of primers (Table 1) flanking the mutation region were designed to amplify the target fragment. High fidelity DNA polymerase (PrimeSTAR® from TaKaRa BIO Inc., Japan) was used to amplify the DLD fragment in DNA extracted from different beetle populations. The PCR products were sub-cloned into pGEMT Easy vector (Promega, Madison, WI, USA). Twenty-three positive clones were randomly selected for plasmid extraction (Miniprep kit from Zymo Research, Irvine, CA, USA). The purified plasmids were sequenced by the Sanger method, and the sequences were analyzed by Sequencher 4.4 (Gene Codes, Ann Arbor, MI, USA) to detect the point mutations.
Genotyping the dead individuals from the dose response bioassay
To investigate whether the phosphine resistance levels in individual beetle from the TCBR strain with one mutation that are homozygous at the P45S locus, and those with two mutations that are heterozygous at both P45S and G131D were similar, dead individuals from each phosphine concentration treatment in the dose-morality experiment were collected following the assay and subjected to the molecular marker analysis as described above (a total of 60 dead beetles across 5 doses).
Results
Relative toxicity of Phosphine for T. castaneum populations
Results of dose-mortality bioassays and probit analyses are presented in Fig. 1 and Table 2, respectively. Resistance ratios calculated using LC50 estimates indicate that the TCBR strain was 8.3-fold more resistant than the TCKS strain, which up to that time (Chen et al. 2015; Cato et al. 2017) was the most resistant T. castaneum population characterized in the USA. When compared against the Lab-S strain, the TCKS strain exhibited a 31.1-fold resistant whereas the TCBR strain exhibited a 258.5-fold resistance.
Dose-mortality curves for PH3 fumigation in the red flour beetle, T. castaneum from Brazil (TCBR), Minneapolis, Kansas (TCKS) and a laboratory-susceptible population (Lab-S). The LC50 and LC99 values for each strain were estimated with probit analyses (Table 2) using data shown here. Each symbol represents the mean of three replicates
Identification of SNPs for DLD Mutations
Only the previously identified P45S mutation was found in the TCKS strain. However, two additional mutations, G131D and V167A, also were identified in the TCBR strain in addition to P45S (Fig. 2). We could not find commercially available restriction enzymes matching the V167A mutation. However, mutations from guanine to adenine at nucleotides 390 and 392 were the result of a gagac sequence, which is the recognition site for the restriction enzyme BsmAI and therefore a potential diagnostic marker.
Partial sequences of the DLD gene from the red flour beetle, TCBR strain, with numbering below corresponding to the DLD gene sequence from the full length gene. a Determination of mutations in the genomic DNA and amino acid sequences of the DLD gene from TCBR strain compared to the laboratory-susceptible strain (Lab-S). Three mutations, P45S (tct versus cct), G131D (gac versus ggc) and V167A (gct versus gtt), were identified from the TCBR strain that differed from the susceptible strain and b partial DNA sequence corresponding to the second mutation in heterozygous individuals, c partial DNA sequence corresponding to the second mutation in homozygous at P45S individuals; and d partial DNA sequence of a clone corresponding to the second mutation in heterozygous individuals. A second CAPS marker was developed based on gagac sequence from nucleotides 389–393
A molecular marker for the G131D mutation in the TCBR strain
There was g to a substitution at nucleotide 392 of the DLD gene sequence in heterozygous individuals of the TCBR strain (Fig. 2b), whereas no such substitution was detected for the same sequence in the homozygous R1R1 individual (i.e., homozygous for the allele for resistance at the first marker location, M1) of the TCBR strain (Fig. 2c). We further confirmed the co-existence of g to a substitution at nucleotides 390 and 392 by sequencing the clones from heterozygous individuals (Fig. 2d). In addition, the synonymous mutation at nucleotide 390 was validated using Clustal W multiple sequence alignment (S1 Table). Taken together, BsmAI can be used as the restriction enzyme for the molecular marker determination of the G131D mutation (M2) in TCBR individuals (Fig. 3).
Amplification of P45S and G131D regions of the DLD coding sequence in two individuals from the TCBR strain using specific pairs of primers, shown in gels (a, b), respectively. The digests at locations for the point mutations responsible for key amino acid changes were made with the restriction enzymes MboI and BsmAI in (c, d), respectively. Genotypes in lane 1 are homozygous and those in lane 2 are heterozygotes. M is the quantitative ladder for base pair numbers in each gel
PH3-resistant level in TCBR homozygous and heterozygous individuals
The proportions of TCBR adults with a homozygous-resistant genotype for just the M1 mutation compared to those with heterozygous genotypes for both the M1 and M2 mutations from the concentration-mortality experiment were similar (Fig. 4), which suggested that the resistance phenotypes of homozygous R1R1S2S2 individuals and heterozygous R1S1R2S2 individuals were similar. Although we observed both the homozygous and heterozygous genotypes for M1 resistance in several individuals, none of the 60 beetles analyzed were homozygous for the presumed resistance allele at the M2 marker.
Proportions of homozygous and heterozygous individuals in the TCBR strain (number analyzed in parentheses) killed at each of the concentrations in the concentration-mortality bioassays (Fig. 1) and recognized as either homozygous R1R1S2S2 (for M1 only) or heterozygous R1S1R2S2 (for M1 and M2) using the CAPS analyses
Discussion
Our results further confirm that strong phosphine resistance of T. castaneum in TCBR is mainly associated with high frequencies of the point mutation responsible for the amino acid change of P45S in the DLD gene, as previously described (Oppert et al. 2015; Chen et al. 2015). The P45S mutation in DLD is widely distributed in Australia (Schlipalius et al. 2012), China (Lu et al. 2018), India (Kaur et al. 2015) and the USA (Chen et al. 2015). However, the G131S mutation is not widespread and has been characterized only from Australian populations of T. castaneum. For the TCBR strain, we have identified that the P45S mutation and a nearby mutation in the same sequence, G131D, coexist in this strongly resistant strain. The mutation in the DLD gene at location 390–392 results in a nonsynonymous mutation of G131D and occurs in the same position as the mutation for G131S in resistant Australian beetles, which suggests that the loss of glycine at this position in the DLD protein is important for strong phosphine resistance.
The occurrence of strong phosphine resistance in populations of T. castaneum can be predicted by genotyping sampled individuals using previously designed CAPS markers (Chen et al. 2015). Strong phosphine-resistant phenotypes are associated with high R-allele frequencies at the M1 site of P45S. In addition to the CAPS marker analysis based on P45S described by Oppert et al. and Chen et al. (2015), we now report another CAPS marker for the G131D mutation detected in the TCBR strain. In terms of the CAPS marker analysis of the TCBR strain studied here, there were just two genotypes observed, homozygous R1R1S2S2 and heterozygous R1S1R2S2 and there were no G131D homozygotes (S1S1R2R2) observed. We observed the ratio of the genotypes to be about 3:7 in the population, and this distribution of the genotypes in the population does not fit the Hardy–Weinberg equilibrium. This phenomenon is also confirmed by the genotypes of the offspring from the crossings between heterozygous R1S1R2S2 parents (Hubhachen et al., unpublished data). Experiments in our laboratory are underway to investigate why there are no homozygous S1S1S2S2 individuals in the population when about 70% of individuals are heterozygous R1S1R2S2.
The LC50 value reported here for the TCBR strain was 258.5-fold higher than that of the susceptible laboratory population, and 31.1-fold higher than TCKS, which had been the strongest phosphine-resistant population collected in North America at the time of Chen et al. (2015), and it is twice the 127-fold resistance reported for another USA population studied more recently (the “Red Level AL” population of Cato et al. 2017). The higher level of resistance in the TCBR strain may be due to either the one resistant allele (P45S) being homozygous or two or more co-inherited point mutations in the DLD gene that contribute to the strong phenotype for phosphine resistance.
Direct use of genetic markers can help in understanding the occurrence and distribution of resistance genes for managing phosphine-resistant pest populations (Chen et al. 2015). CAPS markers for detecting phosphine resistance in grain beetles are fast and inexpensive (Chen et al. 2015; Kaur et al. 2013). In addition, the technique can be readily applied to analyze dead or living beetles of any life stage (Daglish and Collins 1999; Emery et al. 2011). Application of both of these two CAPS markers could determine the rph2 R-allele frequency of the pest populations of T. castaneum that are known to carry them. Thus, our findings of an additional molecular marker for phosphine resistance provide important information for decision-making in managing phosphine resistance in storage insect pests. A low frequency of either or both markers suggests that a more effective use of phosphine could manage resistance, while a high frequency of resistance markers suggests that phosphine fumigation should be at least temporarily discontinued and suitable resistance management strategies should be adopted, such as use of an alternative fumigant or reduction in pest populations through other means.
Author contributions
ZH conceived the work with key inputs from TP, DS and YP. ZH did most experiments with key sequencing done by HJ. RG and BO provided critical insect strains for testing and contributed to other planning. GO and TP provided facilities and equipment for the work. ZH wrote the manuscript. All authors read and approved the manuscript.
References
Afful E, Elliott B, Nayak MK, Phillips TW (2018) Phosphine resistance in North American field populations of the lesser grain borer Rhyzopertha dominica (Coleoptera: Bostrichidae). J Econ Entomol 111(1):463–469. https://doi.org/10.1093/jee/tox284
Ahmad A, Ahmed M, Nourullah Ali GM, Abbas M, Arif S (2013) Monitoring of resistance against phosphine in stored grain insect pests in Sindh. Middle-East J Sci Res 16:1501–1507
Benhalima H, Chaudhry MQ, Mills KA, Price NR (2004) Phosphine resistance in stored-product insects collected from various grain storage facilities in Morocco. J Stored Prod Res 40:241–249
Cao Y, Son Y, Sun GY (2003) A survey of psocid species infesting stored grain in China and. Res. resistance to phosphine in field populations of Liposcelis entomophila (Enderlein) (Psocoptera: Liposcelididae), pp 662–667. In: Credland PF, Armitage DM, Bell CH, Cogan PM, Highley E (eds) Proceedings of the 8th international working conference on stored product protection, 22–26 July 2002, York, UK. CAB International, Wallingford, United Kingdom (ISBN 0851996914)
Cato A, Elliott B, Nayak MK, Phillips TW (2017) Geographic variation in phosphine resistance among North american populations of the red flour beetle (Coleoptera: Tenebrionidae). J Econ Entomol 110(3):1359–1365
Champ BR, Dyte CE (1976) Report of the FAO global survey of pesticide susceptibility of stored grain pests. FAO Plant Production and Series, 5, FAO, Rome
Chen Z, Schliaplius DI, Opit GP, Subramanyam B, Phillips TW (2015) Diagnostic molecular markers for phosphine resistance. PLoS ONE 10(3):0121343
Collins PJ, Daglish GJ, Nayak MK, Ebert PR, Schlipalius DI, Chen W, Pavic H, Lambkin TM, Kopittke R, Bridgeman BW (2001) Combating resistance to phosphine in Australia, pp 593–607. In: Donahaye EJ, Navarro S, Leesch JG (eds) International conference controlled atmosphere and fumigation in stored products, 29 October–3 November 2000, Fresno, CA. Executive Printing Services, Clovis, CA
Daglish GJ, Collins PJ (1999) Improving the relevance of assays for phosphine resistance. In: Zuxun J, Quan L, Yongsheng L, Xianchang T, Langhua G (eds) Proceedings of the 7th international working conference of stored product protection. Sichuan Publishing House of Science and Technology, Chengdu, Sichuan, People’s Republic of China, pp 584–593
Emery RN, Nayak MK, Holloway JC (2011) Lessons learned from phosphine resistance monitoring in Australia. Stewart Postharvest Rev 7:1–8
Food and Agriculture Organization (1975) Recommended methods for the detection and measurement of resistance of agricultural pests to pesticides. Tentative method for adults of some major pest species of stored cereals with methyl bromide and phosphine. FAO method no. 16. FAO Plant Prot Bull 23:12–25
Hagstrum DW, Phillips TW, Cuperus GW (2012) Stored product protection. Kansas State University. KSRE Publ. S-156. Manhattan, KS. 352 p
Jagadeesan R, Fotheringham A, Ebert PR, Schlipalius DI (2013) Rapid genome wide mapping of phosphine resistance loci by a simple regional averaging analysis in the red flour beetle, Tribolium castaneum. BMC Genom 14:650. https://doi.org/10.1186/1471-2164-14-650
Jagadeesan R, Collins PJ, Nayak MK, Schlipalius DI, Ebert PR (2016) Genetic characterization of field-evolved resistance to phosphine in the rusty grain beetle, Cryptolestes ferrugineus (Laemophloeidae: Coleoptera). Pestic Biochem Physiol 127:67–75. https://doi.org/10.1016/j.pestbp.2015.09.008Epub 2015 Sep 25
Jittanun C, Chongrattanameteekul W (2014) Phosphine resistance in Thai local strains of Tribolium castaneum (Herbst) and their response to synthetic pheromone. Kasetsart J Nat Sci 48:9–16
Katoh KK, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Kaur R, Daniels EV, Nayak MJ, Ebert PR, Schlipalius DI (2013) Determining changes in the distribution and abundance of a Rhyzopertha dominica phosphine resistance allele in farm grain storage using a DNA marker. Pest Man Sci 69:685–688
Kaur R, Subbarayalu M, Jagadeesan R, Daglish GJ, Nayak MK, Naik HR et al (2015) Phosphine resistance in India is characterized by a dihydrolipoamide dehydrogenase variant that is otherwise unobserved in eukaryotes. Heredity 115:188–194
Koçak E, Schlipalius DI, Kaur R, Tuck A, Ebert PR, Collins PJ, Yilmaz A (2015) Determining phosphine resistance in rust red flour beetle, Tribolium castaneum (Herbst.) (Coleoptera: Tenebrionidae) populations from Turkey. Türk Entomol Derg 39:129–136
Konemann CE, Hubhachen Z, Opit GP, Gautam S, Bajracharya NS (2017) Phosphine resistance in Cryptolestes ferrugineus (Coleoptera: Laemophloeidae) collected from grain storage facilities in Oklahoma, USA. J Econ Entomol 110(3):1377–1383
LeOra Software. (2005). PoloPlus user’s manual, version 2.0. LeOra Software, Petaluma, CA
Lorini I, Collins PG, Daglish GJ, Nayak MK, Pavic H (2007) Detection and characterization of strong resistance to phosphine in Brazilian Rhyzopertha dominica (F.) (Coleoptera: Bostrychidae). Pest Manag Sci 63:358–364
Lu Y, Zhang C, Wang Z, Yan X, Emery RN (2018) Rapid detection of phosphine resistance in the lesser grain borer, Rhyzopertha dominica (Coleoptera: Bostrychidae) from China using ARMS-PCR, pp 1043–1045. In: Adler C, Opit G, et al. (Eds) Proceedings of the 12th International working conference on stored product protection. Berlin, Germany October 7–11. Julius Kühn-Institut, Berlin
Nayak MK, Holloway JC, Emery RN, Pavic H, Bartlet J, Collins PJ (2013) Strong resistance to phosphine in the rusty grain beetle, Cryptolestes ferrugineus (Stephens) (Coleoptera: Laemophloeidae): its characterization, a rapid assay for diagnosis and its distribution in Australia. Pest Manag Sci 69:48–53
Nguyen TT, Collins PJ, Duong TM, Schlipalius DI, Ebert PR (2016) Genetic conservation of phosphine resistance in the rice weevil Sitophilus oryzae. J Hered 107(3):228–237. https://doi.org/10.1093/jhered/esw001
Opit GP, Phillips TW, Aikins MJ, Hasan MM (2012) Phosphine resistance in Tribolium castaneum and Rhyzopertha dominica from Stored Wheat in Oklahoma. J Econ Entomol 105:1107–1114
Oppert B, Guedes RNC, Aikins MJ, Perkin L, Chen Z, Phillips TW, Zhu KY, Opit GP, Hoon K, Sun Y, Meredith G, Bramlett K, Supunpong Hernandez N, Sanderson B, Taylor M, Dhingra D, Blakey B, Lorenzen M, Adedipe F, Arthur F (2015) Genes related to mitochondrial functions are differentially expressed in phosphine-resistant and -susceptible Tribolium castaneum. BMC Genom 16:968
Pimentel MA, Faroni LR, Tótola MR, Guedes RN (2007) Phosphine resistance, respiration rate and fitness consequences in stored-product insects. Pest Manag Sci 63(9):876–881
Pimentel MA, Faroni LR, Silva FH, Batista MD, Guedes RN (2010) Spread of phosphine resistance among Brazilian populations of three species of stored product insects. Neotrop Entomol 39(1):101–107
Rajendran S (1999) Phosphine resistance in stored grain insect pests in India, pp 635–641. In: Jin Z, Liang Q, Liang Y, Tan X, Guan L (Eds) Proceedings of the 7th international working conference on stored-product protection, 14–19 October 1998, Beijing, China. Sichuan Publishing House of Science and Technology, Chengdu, China
Ren YL, O’Brien LG, Whittle GP (1994) Studies on the effect of carbon dioxide in insect treatment with phosphine. In: Highley E, Wright EJ, Banks H, Champ BR (eds) Stored products protection. Proceedings of the 6th international conference on stored product protection, CAB Press, Wallingford, Oxon, UK, pp 173–177
Sağlam O, Edde PA, Phillips TW (2015) Resistance of Lasioderma serricorne (Coleoptera: Anobiidae) to fumigation with phosphine. J Econ Entomol 108:2489–2495
Schlipalius DI, Valmas N, Tuck AG, Jagadeesan R, Ma L, Kaur R, Goldinger A, Anderson C, Kuang K, Zuryn K, Mau YS, Cheng Q, Collins PJ, Nayak MK, Schirra HJ, Hilliard MA, Ebert PR (2012) A core metabolic enzyme mediates resistance to phosphine gas. Science 338:807–810
Schlipalius DI, Tuck AG, Jagadeesan R, Nguyen T, Kaur R, Subramanian S, Barrero R, Nayak MK, Ebert PR (2018) Variant linkage analysis using de Novo transcriptome sequencing identifies a conserved phosphine resistance gene in insects. Genetics 209:281–290
Tyler PS, Taylor RW, Rees DP (1983) Insect resistance to phosphine fumigation in food warehouses in Bangladesh. Internat Pest Cont 25:10–13
Yeam I, Kang BC, Lindeman W, Frantz JD, Faber N, Jahn N (2005) Allele-specific CAPS markers based on point mutations in resistance alleles at the pvr1 locus encoding eIF4E in Capsicum. Theor Appl Gen 112:178–186
Zettler JL, Cuperus GW (1990) Pesticide resistance in Tribolium castaneum (Coleoptera: Tenebrionidae) and Rhyzopertha dominica (Coleoptera: Bostrichidae) in wheat. J Econ Entomol 83:1677–1681
Acknowledgements
The authors appreciate suggestions and technical advice on genetics and molecular biology from Drs. Jeremy Marshall and Kun Yan Zhu. The authors also acknowledge the financial support of the Plant Biosecurity Cooperative Research Centre (Project No: PBCRC3035) established and supported under the Australian Government’s Cooperative Research Centre Program (http://www.crcplantbiosecurity.com.au). This paper is contribution number 19-176-J of the Kansas Agricultural Experiment Station.
Funding
This study was funded by the Australian Plant Biosecurity Center for Cooperative Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest with this agency, among themselves, or with other funding sources or institutions in regard to this article.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Since no human was involved in this study, there was no need to obtain informed consent from any individual participants included in the study.
Additional information
Communicated by C. G. Athanassiou.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hubhachen, Z., Jiang, H., Schlipalius, D. et al. A CAPS marker for determination of strong phosphine resistance in Tribolium castaneum from Brazil. J Pest Sci 93, 127–134 (2020). https://doi.org/10.1007/s10340-019-01134-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10340-019-01134-4